Abstract
We have taken a synthetic biology approach to the generation and screening of transcription factor binding sites for activity in human cells. All possible 10-mer DNA sequences were printed on microarrays as 100-mers containing 10 repeats of the same sequence in tandem, yielding an oligonucleotide library of 52,429 unique sequences. This library of potential enhancers was introduced into a retroviral vector and screened in multiple cell lines for the ability to activate GFP transcription from a minimal CMV promoter. With this method, we isolated 100 bp synthetic enhancer elements that were as potent at activating transcription as the WT CMV immediate early enhancer. The activity of the recovered elements was strongly dependent on the cell line in which they were recovered. None of the elements were capable of achieving the same levels of transcriptional enhancement across all tested cell lines as the CMV enhancer. A second screen, for enhancers capable of synergizing with the elements from the original screen, yielded compound enhancers that were capable of twofold greater enhancement activity than the CMV enhancer, with higher levels of activity than the original synthetic enhancer across multiple cell lines. These findings suggest that the 10-mer synthetic enhancer space is sufficiently rich to allow the creation of synthetic promoters of all strengths in most, if not all, cell types.
Keywords: Transcription , GFP reporter, RNA interference, synthetic biology
A limiting factor in the development of artificial genetic systems is the availability of promoter elements with potent activity in many cell types. DNA-based shRNA systems, for example, are known to be capable of greater levels of gene knockdown when shRNA transcripts are driven by stronger promoters. Many of the strong promoters currently in use have highly variable levels of transcription in different cellular contexts and show little or no expression in many cell lineages. This limits the utility of both loss-of-function (i.e., shRNA) and gain-of-function (i.e., cDNA expression) genetic screens in many cell types (1), as well as recombinant protein production. Synthetic promoters could conceivably drive transcription in cellular contexts in which current promoter options are not ideal. A well characterized synthetic promoter system has the potential to provide stronger expression, or expression levels that are precisely tailored to match a researcher’s specifications.
Eukaryotic promoters are generally described as having a core promoter near the site of transcription initiation and one or more enhancer elements that may be located more distantly (2). Although rationally designed strong synthetic core promoter elements have been described (3), currently the strongest eukaryotic enhancers are identical to sequences found in nature, and generally have only minimal modification (4, 5). Screening viral and mammalian genome sequences for preexisting enhancers has recovered the most potent transcriptional elements currently known. The enhancers that were discovered, however, were evolved rather than designed. A promoter that evolved in the context of an organism will possess levels of transcriptional activation tuned to maximize the fitness of their complete host organism rather than seeking the strongest possible transcription level. Overly strong transcription levels could easily be detrimental to the overall fitness of a virus. Through experimentation with chimeric transcriptional activators, the eukaryotic transcriptional machinery has been shown to be capable of far greater levels of transcription than are found from naturally occurring promoters (6).
If all contributions to transcriptional enhancers were known, promoters could be designed that would possess any desired characteristics of expression levels or tissue specificity. As a result of the small size, spatial effects, and complexity resulting from the combinatorial mechanism of action of transcription factor binding sites, discovery of the exact elements responsible for transcriptional activation has proven difficult. Many different approaches have been attempted to systematically derive the elements contributing to transcriptional activation (7, 8). However, because of the large sequence space of possible enhancer elements, these efforts have almost universally focused on combinations of known transcription factor binding sites.
Enhancer elements such as the CMV immediate early enhancer are typically larger than 400 bp in length. Random synthesis of such an element yields 4400 or 6.67 × 10240 possible combinations, far more than can be synthesized or screened. Enhancer elements are composed of many small transcription factor binding sites, with binding motifs typically less than 10 bp, making them more amenable to a complete and unbiased screening. Although the transcriptional enhancement resulting from a single transcription factor binding site is may be too low to reliably distinguish from background noise, previous studies have shown that robust transcription can be driven by repeated arrays of multiple binding sites (9, 10). The development of DNA synthesis on oligonucleotide microarrays (11, 12) enables the parallel generation of very large numbers of designed DNA sequences, which allows the systematic investigation of a large portion of transcription factor binding site sequence space. DNA repeat motifs printed on microarrays have been previously used to biochemically quantify the affinity of single transcription factors for many possible binding sites in high throughput (13). Although the high-throughput characterization of individual binding sites is an important resource for understanding transcription specificity, we sought to address the in vivo transcriptional activity that results from transcription factor binding. By designing sequences, synthesizing them in a massively parallel fashion on microarrays, cleaving these oligonucleotides from microarrays, and cloning them upstream of a reporter gene, it is possible to test the in vivo activity of a broad range of enhancer elements and select among them for specific properties.
Results
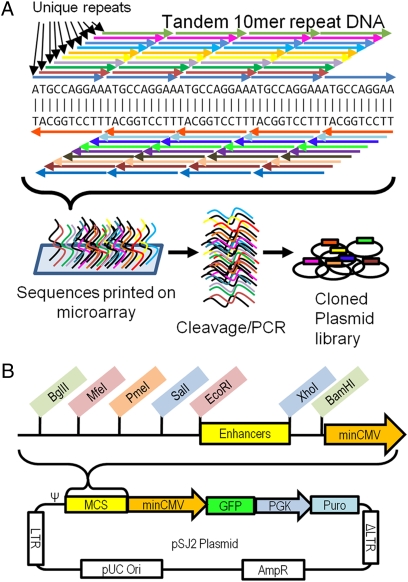
A synthetic enhancer library containing 10-mer repeats was generated by printing oligonucleotides on Nimblegen microarrays. Each sequence printed on the array consisted of ten 10-mer repeats to generate a 100-mer oligo that was flanked on either end by restriction and primer binding sites. Each 100-mer actually contains 10 different 10-mer repeats in the forward direction and 10 in the reverse. As a result of this feature, all possible combinations of 10-mer repeats could be printed in only 50,000 unique sequences, although some had only nine full repeats (Fig. 1A). The approximately 5 Mb of oligonucleotides were cleaved from the microarray using a brief treatment with a basic solution and PCR-amplified before restriction cloning into the vector pSJ2. The pSJ2 vector is a self-inactivating murine stem cell virus based vector containing a multiple cloning site (MCS) upstream of a minimal CMV promoter driving EGFP and a puromycin resistance marker. The use of a retroviral vector allows individual constructs to be integrated into cells at single copy and, when used at a low multiplicity of infection (MOI), mitigates the occurrence of multiple integrations that would confound promoter analysis. EGFP provides a marker for overall levels of transcription from the minimal CMV promoter that can be easily assayed by FACS. The use of a self-inactivating vector prevents promoter interference from the viral LTRs, which contain a deletion in the 3′ LTR resulting in complete inactivation of the retroviral promoter upon integration into the genome. The MCS was designed such that it contained multiple restriction sites with compatible ends for the subsequent insertion of multiple enhancers simultaneously (Fig. 1B), which becomes important later in combinatorial experiments. For example, the EcoRI-XhoI enhancer fragments can be recloned into MfeI-SalI or EcoRI SalI to make double inserts. Enhancers could also be excised as BamHI-EcoRI fragments and inserted back into BglII-MfeI cleaved fragments.
Fig. 1.
Design and synthesis of repeat library. (A) A single 10-mer DNA sequence printed in 10 head-to-tail repeats also contains 19 other potentially unique 10-mer repeats, for a total of 20 potentially unique sequence repeats in a 100-bp DNA fragment. DNA repeats were synthesized on microarrays, cleaved from the array surface, and PCR-amplified. The repeat oligonucleotides were cloned into a plasmid vector to generate a library of synthetic enhancers. (B) Map of plasmid vector pSJ2 for cloning/screening synthetic enhancers. Multiple restriction sites with compatible overhangs allow the cloning of multiple synthetic enhancers within the MCS. The MCS is upstream of a minimal CMV promoter driving GFP, and retroviral sequences are present for packaging as a self-inactivating retrovirus.
To test the feasibility of using the 10-mer repeat sequence library to screen for enhancer elements, the plasmid library of repeats was packaged as retrovirus, infected onto HeLa cells at an MOI of 0.1, and puromycin-selected (Fig. 2A). If strong enhancers are being sought, single copy infection is particularly important, as double integration events will give greater GFP signal than single integrations. This contaminates the resulting library population with many enhancers that are far weaker than desired. Although the minimal CMV promoter gave transcription levels that were not significantly greater than those of uninfected cells, FACS on the enhancer-containing HeLa cells revealed that greater than 15% of the 10-mer repeat library inserts were capable of giving transcriptional enhancement greater than the background level of a minimal CMV promoter (Fig. 2 B and C). Of the total population, fewer than 1% of cells were capable of driving GFP expression to levels that were within the range of WT CMV enhancer–driven constructs.
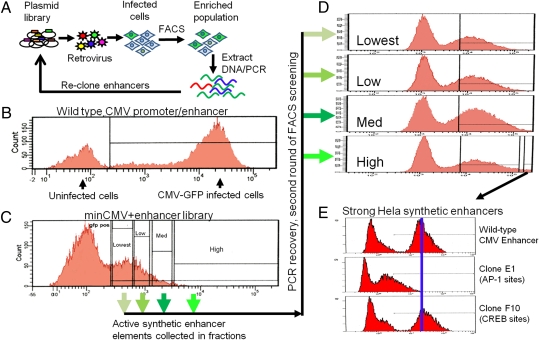
Fig. 2.
Screening synthetic enhancers for transcriptional activation. (A) The synthetic enhancer plasmid library is packaged as retroviruses and infected into tissue culture cells at an MOI of 0.1, puromycin-selected, and screened by FACS for cells expressing the desired level of GFP. After sorting and genomic DNA preparation, the synthetic enhancer inserts are PCR-amplified and recloned for subsequent rounds of screening. (B). Histogram of GFP fluorescence of HeLa cells expressing GFP under the control of the WT CMV promoter and enhancer. (C) Histogram of GFP fluorescence of HeLa cells expressing GFP driven by a minimal CMV promoter with the synthetic enhancer library cloned upstream. Populations indicated on the FACS histogram were separately sorted, and synthetic enhancers present were recloned and reinfected into HeLa cells. (D) Histograms of GFP fluorescence for cells infected in the second round of screening, with arrows indicating their parental population of cells from the first round of screening. (E) After two rounds of screening, clones were tested individually for transcriptional activity. A clone representing an enhancer with relatively weak activity (clone E1) and one with very potent activity (clone F10) are compared with the WT CMV promoter/enhancer. The predicted transcription factor binding sites for each synthetic enhancer are indicated.
GFP-expressing cells were sorted based on their level of fluorescence into four populations. The cells were expanded, genomic DNA extracted, and their 10-mer repeat inserts were amplified by PCR and recloned into pSJ2 as four separate libraries for a second round of screening. The second-round libraries were then packaged and infected onto HeLa cells again, and their GFP fluorescence assessed by flow cytometry (Fig. 2D). In the second round, the percentage of cells from the highest GFP expressing sublibrary showed an increased numbers of cells capable of achieving levels of GFP fluorescence on par with the WT CMV enhancer. As a result of different FACS settings, the histograms in Fig. 2D are not on the same scale as those in Fig. 2 B and C. The small fraction collected from the “high” population represents cells with fluorescence greater than the WT CMV enhancer median. To further enrich for strong enhancers, the highest expressing GFP population was sorted again and the inserts were recloned. The resulting library was run through a third round of screening, and again, as in the previous two rounds, clones that exceeded the mean GFP fluorescence of the WT CMV enhancer construct were selected.
After the third round of screening was complete, the remaining inserts were PCR-amplified and cloned as a plasmid library. Two 96-well plates of bacterial clones were then picked and plasmid DNA was sequenced. Sequencing revealed that many of the recovered clones contained highly similar sequences representing a few major classes of transcription factor binding sites. Individual representatives of the major classes of clones recovered were packaged as retroviruses and infected into cells. A variety of enhancer strengths were recovered in these individual clones. Strengths ranged from very low enhancement to enhancer activity on par with the WT CMV enhancer (Fig. 2E). Multiple related but distinct sequences were recovered in the final pool, supporting the reproducibility of the screen (Fig. S1).
The class of clones that contained consensus cAMP response element (CRE; bound by CREB protein) binding sites were recovered most frequently (Fig. 3A) and were found to exhibit the highest transcriptional enhancer activity (Fig. 2E and Fig. 3 A and B). Enhancers predicted to contain AP-1–binding sites were also frequently recovered, but were found to exhibit less activity than CRE site–containing elements. The recovered transcription factor–binding sites occurred in both orientations with respect to the promoter. The most potent of the recovered CRE enhancers contained approximately seven to 10 predicted CREB half-binding sites (14, 15), although some of the repeats were imperfect. To compare the activity of one of these that has at least seven CRE sites, which we call the F10 enhancer, against the activity of perfect CRE repeats, constructs were generated that contained two, four, six, eight, or 10 CRE consensus binding sites. These were tested against the F10 enhancer in HeLa cells. The F10 enhancer exhibited slightly less transcriptional activity than the construct with eight CRE repeats (Fig. S2B). Constructs with 10 CRE repeats were 60% stronger than CMV and were marginally stronger than clones with eight CRE repeats, indicating that transcriptional activation for CRE elements was approaching saturation in this particular context or that binding sites more distant than 100 bp were less effective. The recovered enhancers were tested for their transcriptional activity in other cell lines, as one of the most desirable characteristics of the CMV enhancer is its broad spectrum of activity in many cell lines. The CRE-based synthetic enhancers were found to have the strongest transcriptional enhancement in the HeLa cells where they were originally screened, with relatively little activity in other cell lines (Fig. S2B).
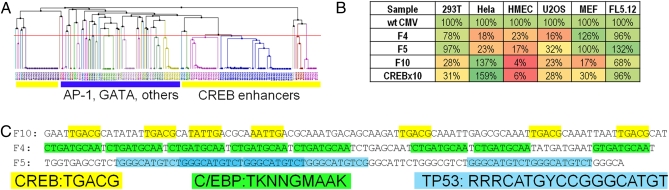
Fig. 3.
Analysis of sequence motifs present in most commonly recovered synthetic enhancers. (A) Unsupervised clustering of sequence homology between enhancer elements recovered from screening in HeLa cells. The yellow bars indicate clones with CREB binding elements, whereas the blue bar represents all other classes of enhancer elements. For a larger version, see Fig. S3. (B) Relative expression levels of selected synthetic enhancer elements across a panel of cell lines. Percentages are expressed as percent of WT CMV enhancer/promoter. (C) Predicted transcription factor binding motifs within selected enhancer elements. Yellow boxes indicate half-CREB binding sites, green boxes indicate C/EBP binding sites, and blue boxes indicate TP53 binding sites. Clone F10 was recovered from HeLa cells and clones F4 and F5 were recovered from 293T cells.
To recover transcriptional enhancer elements with stronger activity in other cell lineages and test their activity in combination with HeLa enhancers, five additional cell lines (293T, U2OS, mouse embryonic fibroblast, FL5.12, and human mammary epithelial cells) were screened with the enhancer library to isolate additional strong enhancers. Two rounds of FACS screening were performed in each cell line, with the 2% most fluorescent cells isolated in each round. After the second round of screening, a subset of enhancer inserts were cloned and individually screened for GFP fluorescence in all cell lines in 96-well plates. Clones exhibiting strong enhancer activity were sequenced and recloned into the pSJ2 MCS, downstream of the previously characterized HeLa-F10 enhancer that contains CREB protein binding sites. Profiling the recovered enhancers revealed at least two classes of enhancers: (i) CRE-based enhancers that were strong in HeLa cells but not in other lines and (ii) enhancers with strong activity across several of the cell types tested. FL5.12 cells exhibited very low transcription from the CMV minimal promoter, regardless of with which enhancer it was paired. The class of sequences that exhibited transcriptional activation in 293T and MEFs but not HeLa cells were represented by multiple different sequence motifs. Enhancers exhibiting this transcriptional pattern were most frequently predicted to contain C/EBP binding sites, but many other sites were also recovered, such as P53 sites, and sequences with no known transcription factor binding motifs (Fig. 3 B and C). The sequences of the recovered enhancers are shown in Table S1.
To determine whether the transcriptional level achieved by the strongest enhancer elements represented a fundamental limit of minimal CMV promoter, a second screen was performed to search for synthetic enhancers capable of acting synergistically or additively with already recovered clones. The original 50,000-sequence 10-mer repeat library was excised using EcoRI-XhoI and cloned into the MCS of EcoRI-SalI cut pSJ2 upstream of three different enhancers, representing each of the major sequence classes recovered in HeLa cells. The three resulting libraries were then screened for the strongest enhancers present with use of the same method as before. For already strong enhancers, such as clone F10, the addition of the library in the first round of screening gave little to no detectable transcriptional enhancement beyond their original levels. Enrichment was observed, however, for clones possessing greater transcriptional activity than the WT CMV enhancer after two additional rounds of screening and recloning. Multiple clones were found that possessed greater than twofold more transcriptional activity than the WT CMV enhancer upon picking individual clones to validate (Fig. 4A). Although the strongest double synthetic enhancer gave approximately twofold greater GFP transcription in HeLa cells than the WT CMV enhancer, it failed to achieve the same levels in other lines (Fig. 4B), although it did improve upon the levels of CRE-based enhancer alone. The sequences of the recovered double enhancers contained many of the same predicted binding sites as the individual HeLa enhancers, indicating that transcription was being improved by the same overall mechanisms, rather than specific synergy with the paired enhancer.
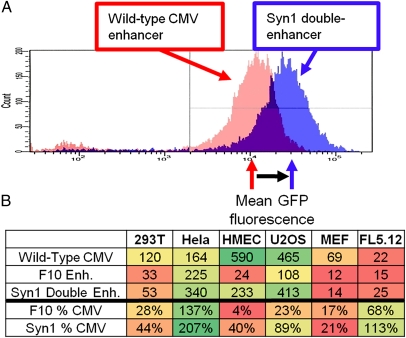
Fig. 4.
Screening for secondary enhancers of initial screen hits. (A) Double enhancer clone Syn1 recovered from the screen for secondary enhancers. The WT CMV enhancer is represented in red, the Syn1 double enhancer is represented in blue. (B) Median fluorescence values for the WT CMV, F10, and Syn1 enhancers. F10 and Syn1 are represented below as percentage of WT CMV values. Values colored in red represent low GFP fluorescence and values colored in green represent the highest values of fluorescence.
We sought to determine whether an enhancer with activity in HeLa cells could be combined with enhancers that were strong in other lines to yield enhancers that are functional in multiple lines. When the F10 enhancer was combined with enhancers recovered from 293T, U2OS, or HME cells, the strong transcriptional activity of the F10 enhancer in HeLa cells was masked by the addition of a downstream enhancer. The activity across the cell line panel of the resulting double enhancer invariably resembled the downstream enhancer most closely (Fig. S2).
Discussion
In this study we describe a method for the synthetic optimization of promoters. Using an unbiased screen of a synthetic 10-bp repeat sequence library we identified many synthetic enhancer elements capable of increasing transcription from a minimal CMV promoter. In several cases, the recovered synthetic enhancers had transcriptional activation activity on par with the WT CMV enhancer with only two rounds of enrichment. Thus a 100-bp synthetic enhancer can be engineered to match or exceed the promoter strength of the strongest known mammalian promoters.
We found that transcription levels from synthetic enhancers can be further modulated by altering the number of repeats present after an element with activity is isolated. The maximum of 10 repeats of transcription factor binding sites present in the synthetic enhancers were chosen for manageable microarray printing size. As evidenced by the improvement of recovered CREB enhancers by adding additional CREB elements, this was not always sufficient to achieve saturating levels of transcriptional activation. These experiments also show that very specific levels of transcription can easily be achieved by finding an active enhancer element through unbiased screening and increasing or decreasing the number of repeats. An “allelic series” of promoters of differing strengths could easily be generated for any cell line in this manner, allowing more careful fine-tuning of gene expression in genetic studies.
The elements recovered had a strong bias for the cell line in which they were initially screened, frequently exhibiting lower activity in other lines. Each cell line showed a distinct pattern of sequences that showed maximal activation reflecting the underlining complexity of the transcriptional milieu from cell type to cell type. From these data, it is clear that strong promoters will need to be designed for individual cell types depending on the particular application. This could be achieved more rapidly by the combination of FACS enrichment with massively parallel sequencing to determine how sequence space is distributed over bins of different expression levels, at the same time generating a fingerprint of the transcription factor activity pattern for that cell type. Characterization of the proteins binding specific enhancers, through MS or other means, could be instructive in determining the mechanism underlying the differential activity of enhancers across cell lines.
Although enhancers with significant levels of activity were recovered from most cell lines, transcriptional levels from all enhancers in the mouse pre–B-cell line FL5.12 were very low. Further, the WT CMV enhancer caused little to no increase in transcription from the minimal CMV promoter. Based on these data, it is possible that the minimal CMV promoter used for the screening is largely inactive in FL5.12 cells. However, we cannot rule out the possibility that other differences exist, such as effects on GFP folding or microRNA expression patterns that artificially mask the apparent transcription of this construct in FL5.12 cells. The WT CMV promoter is known to be poorly expressed in some stem cell lineages, although the factors responsible are unknown (16). This phenomenon may be specific for the CMV minimal promoter, as the CMV enhancer is functional in hematopoietic stem cells in concert with other promoters (17). These data suggest that a specific repressive factor may act upon the minimal CMV promoter. It may therefore be necessary to screen cell types with poor CMV activity with different minimal promoters to find their most active enhancer elements.
It is currently unclear what the rate-limiting factors are for gene expression, but they are likely to include RNA polymerase recruitment, chromatin modification, nucleosome remodeling, promoter clearance and pausing, abortive transcription, and rates of RNA polymerase transcription after initiation. Each of these could potentially be optimized to promote strong transcription, and a subset of these might be affected by the synthetic enhancers. In principle, if a particular pair of factors operate to promote transcription through different mechanisms, one would expect that they would behave synergistically. Our attempts to identify such synergy among different elements failed to discover strong synergy, although in all cases we could continue to marginally improve upon the promoter strength. For example, we observed that most of the recovered enhancers failed to show additive effects with the likely CREB-based F10 enhancer. This may be because we are nearing a theoretical limit to promoter strength or that we simply have not varied enough other promoter elements. It is also possible that the sequence space larger than 10 bp would contain additional elements capable of synergy. Adding the F10 enhancer, which was strong in HeLa cells, to an enhancer with activity in 293T cells yielded an enhancer that was strong in 293T but not HeLa. This indicates that the CREB-based F10 enhancer may be strongly dependent on close proximity to the minimal CMV promoter to activate transcription, or that the enhancer elements used for combinations interfered with each other. Although strict proximity requirements would make the design of promoters with combination enhancers more difficult, combinations could be designed that alternate the transcription factor sites with different spacing or in different orders to allow for more optimal promoter geometry. Screening for more precise spatial requirements or interference between enhancer elements could be performed by generating a library of many possible combinations and spatial arrangements of elements already known to possess activity from the first round of screening. It is possible that strong enhancer elements could be compressed substantially in size, or combined with minimal promoters that are smaller than CMV to generate very short complete promoter elements for specialized applications in which size may be an issue. With very small elements, it may be possible to knock-in promoters or enhancer elements at a specific locus using homologous recombination (18) with only synthetic oligonucleotides, without the difficulty of generating longer targeting constructs.
Although screening 10-mer sequence space is feasible, many transcription factors have binding sites that are larger than 10 bp. Although these sequences are too complex to screen in completely unbiased fashion, it is possible to use gapped or inverted repeats to take advantage of the symmetrical nature of transcription factor binding sites (19). With more complete, unbiased screening of potential binding sites, stronger enhancer elements or sites bound by heteromultimeric complexes could be discovered. Sequence space could also be rationally constrained by improving predictions of binding site specificity for known DNA binding factors in the human genome. Although predictions have improved markedly for zinc finger transcription factors (20), many other classes of DNA binding domains have unknown specificities. Predicting exact specificities, however, would be unnecessary for the purpose of designing new libraries. General predictions of which spatial arrangements of nucleotides are being “read” by proteins could be sufficient to reduce the complexity of libraries to screenable sizes.
The synthetic biology approaches described here could be applied to any biologic process mediated by short DNA/RNA binding sites. For example, the current library could be used to search for transcription factors regulated in response to a particular stimulus by FACS sorting with and without stimulus looking for a shift in the expression levels, much like we have done for protein stability (21). One could also take different enhancers and combine them with a synthetic minimal promoters either designed de novo or taken from the 20,000 known genes. This entire complement could then be used to look for optimization or for pairing between particular enhancers and minimal promoters. Synthetic repeat libraries could be placed in 5′ UTRs to look for sequences capable of enhancing splicing, translation, or even acting as internal ribosome entry site elements. Such libraries placed in 3′ UTRs could also be designed to act as microRNA targets to provide or to look for novel elements that confer stability, instability, or splicing regulation on a transcript through recruitment of RNA binding proteins.
Materials and Methods
Synthesis and Cloning of Synthetic Enhancer Library.
Fifty thousand 10-mer repeat sequences were synthesized on a NimbleGen microarray (Roche) flanked by EcoRI-XhoI sites and primer sequences. The sequences were cleaved from the array by a brief basic wash and PCR-amplified. The PCR amplification used the following primers: SJ2 forward, TCTAGGCGCCGGAATTAGAT; and SJ2 reverse, CGCCTACCTCGACATACGTT. PCR was performed at 95 °C for 5 min, at 55 °C for 30 s, and at 72 °C for 30 s with HS Taq according to the manufacturer’s recommendations (TaKaRa). Amplified product was digested overnight with EcoRI and XhoI and gel purified with a Qiaquick spin kit (Qiagen). The cut and gel-purified PCR material was then cloned into XhoI EcoRI cut λSM2C phage DNA and packaged using a λ-phage packaging extract (Epicentre). The resulting library of more than 107 recombinants was then amplified as phage and converted to plasmid form using a Cre recombinase expressing bacteria. The resulting plasmid library was amplified and prepped using a Maxiprep kit (Sigma). The XhoI-EcoRI fragment was then cut from this library, gel-purified, and cloned into XhoI-EcoRI cut pSJ2. The resulting library was in excess of 2 × 106 recombinants.
PCR Recovery and Recloning of Synthetic Enhancers.
Cells were trypsinized and resuspended at 10 million cells/mL in 10 mM Tris/10 mM EDTA, pH 8.0, and Proteinase K and SDS were added at final concentrations of 200 μg/mL and 0.5%, respectively. Lysis mixture was incubated overnight at 55 °C. Lysates were extracted twice with phenol/chloroform/isoamyl alcohol and once with chloroform in PhaseLock tubes (5 Prime). Residual chloroform was evaporated for 1 h at 55 °C and RNase A was added at 37°C for 1 h to remove any RNA remaining. Lysate was then extracted twice more with phenol/chloroform and precipitated with 1.5 volumes of ethanol. After centrifugation and washing with 70% ethanol, the resulting pellet was resuspended in 10 mM Tris, pH 8.0, 1 mM EDTA.
Purified genomic DNA then underwent PCR in 400-μL reactions containing 20 μg of DNA. One hundred nmol of each primer was added, and TaKaRa HS Taq was used at the manufacturer’s recommended concentrations of buffer and dNTPs. Primers were SJ2FWD and SJ2REV, and the PCR program was 95 °C for 5 min, 55 °C for 30 s, and 72 °C 30 s for 35 cycles. The resulting PCR product was precipitated and gel-purified for the correct size using a TAE agarose gel and Qiaex II gel extraction kit (Qiagen). The gel-purified PCR product was digested with EcoRI and XhoI overnight, and gel-purified again before ligation into EcoRI-XhoI cut pSJ2. The ligation mixture was electrotransformed into MegaX DH10B ultracompetent Escherichia coli (Invitrogen) and plated on four LB-carbenicillin 150-mm plates. Colonies were scraped from the plates and grown in liquid culture for 8 h before analysis with a Maxiprep kit (Sigma). For cloning of second-site library into clones F10, A11, and E1, the complete synthetic enhancer library was digested with SalI and EcoRI and cloned into XhoI-EcoRI digested pSJ2 already containing the listed inserts.
Informatics.
Transcription factor motif searches were performed using the TFSearch (http://www.cbrc.jp/htbin/nph-tfsearch) and PMatch (http://www.gene-regulation.com/cgi-bin/pub/programs/pmatch/bin/p-match.cgi) programs. The dendrogram of sequence similarities and multiple alignment of CRE sequences were generated using ClustalW/Jalview (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914803107/DCSupplemental.
References
- 1.Baup D, et al. Variegation and silencing in a lentiviral-based murine transgenic model. Transgenic Res. 2009 doi: 10.1007/s11248-009-9318-4. Aug 23. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 3.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 4.Foecking MK, Hofstetter H. Powerful and versatile enhancer-promoter unit for mammalian expression vectors. Gene. 1986;45:101–105. doi: 10.1016/0378-1119(86)90137-x. [DOI] [PubMed] [Google Scholar]
- 5.Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981;290:304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- 6.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 7.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 11.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 12.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fink JS, et al. The CGTCA sequence motif is essential for biological activity of the vasoactive intestinal peptide gene cAMP-regulated enhancer. Proc Natl Acad Sci USA. 1988;85:6662–6666. doi: 10.1073/pnas.85.18.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig JC, et al. Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J Biol Chem. 2001;276:11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 16.Chung S, et al. Analysis of different promoter systems for efficient transgene expression in mouse embryonic stem cell lines. Stem Cells. 2002;20:139–145. doi: 10.1634/stemcells.20-2-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramezani A, Hawley TS, Hawley RG. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 18.Kandavelou K, et al. Targeted manipulation of mammalian genomes using designed zinc finger nucleases. Biochem Biophys Res Commun. 2009;388:56–61. doi: 10.1016/j.bbrc.2009.07.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandell JG, Barbas CF., III Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.