Abstract
Dorsal organizer formation is one of the most critical steps in early embryonic development. Several genes and signaling pathways that positively regulate the dorsal organizer development have been identified; however, little is known about the factor(s) that negatively regulates the organizer formation. Here, we show that Setdb2, a SET domain-containing protein possessing potential histone H3K9 methyltransferase activity, restricts dorsal organizer development and regulates left–right asymmetry by suppressing fibroblast growth factor 8 (fgf8) expression. Knockdown of Setdb2 results in a massive expansion of dorsal organizer markers floating head (flh), goosecoid (gsc), and chordin (chd), as well as a significant increase of fgf8, but not fgf4 mRNAs. Consequently, disrupted midline patterning and resultant randomization of left–right asymmetry are observed in Setdb2-deficient embryos. These characteristic changes induced by Setdb2 deficiency can be nearly corrected by either overexpression of a dominant-negative fgf receptor or knockdown of fgf8, suggesting an essential role for Setdb2–Fgf8 signaling in restricting dorsal organizer territory and regulating left–right asymmetry. These results provide unique evidence that a SET domain-containing protein potentially involved in the epigenetic control negatively regulates dorsal organizer formation during early embryonic development.
Keywords: set domain, histone methylation, epigenetics, embryonic development, zebrafish
Under bilateral external symmetry, vertebrates conserve a left–right asymmetry placement of internal organs and nervous system. By using mouse, chicken, Xenopus, and zebrafish as animal models, the developmental and molecular mechanisms of how left–right asymmetry is established during embryogenesis have been well characterized in the past few years: two embryonic structures—the node [also known as Kupffer’s vesicle (KV) in zebrafish] and the midline, mainly consisting of notochord and floorplate—are considered essential for maintaining the left–right asymmetry (1, 2). Inside the KV, cilia are specifically organized to generate a counterclockwise fluid flow, by which the first laterality signal is propagated to the left lateral plate mesoderm (LPM), resulting in an asymmetric gene expression on the left side (3, 4). The southpaw (member of nodal-related subfamily of TGF-β superfamily) is the earliest known gene that is asymmetrically expressed in the left LPM at 10–12 somite stage to initiate the left–right signaling through inducing its downstream targets lefty2 and pitx2 in the left LPM, and lefty1 in the axial midline that acts as a molecular barrier to prevent the left-sided signals from leaking to the other side. This signaling cascade is highly conserved during vertebrate evolution (5).
It has been shown that the KV is originated from the dorsal forerunner cells (DFCs), a group of noninvoluting cells at the leading edge of the embryonic dorsal organizer or shield, which also produces the midline/notochord (6, 7). Eighty-five years ago Spemann and Mangold originally identified the dorsal organizer in amphibian through transplanting the dorsal lip of blastopore to the ventral region of a host embryo, which led to a secondary axis formation. Since then, the dorsal organizer has been well studied in many systems including Xenopus and zebrafish, and it has been found that many inducers released by the dorsal organizer are essential for patterning the embryonic midline (8, 9) and that developmental defects in the midline are usually accompanied by KV abnormality and disturbed left–right asymmetry (10, 11). Genes or signaling transduction pathways that positively regulate the formation of the dorsal organizer have been identified, including homeobox genes dharma/bozozok and iroquois3 and the T-box gene eomesodermin (12 –14), as well as Wnt/β-catenin signaling transduction pathway (15). Despite these progresses, little is known about the negative regulator of dorsal organizer development during embryogenesis (8, 9).
It has been shown that epigenetic mechanisms involving histone methylation play a critical role in establishing and maintaining heritable programs of gene expression during cellular differentiation and early embryonic development (16, 17). A family of histone methyltransferases (HMT) that catalyzes histone methylations at lysine residues contains a SET domain, which was originally identified in the members of the Su(var) family, polycomb group (PcG), and trithorax group (trxG) and was named after the genes Su(var)3–9, Enhancer of Zeste [E(z)], and trithorax (trx) (18). Although SET domain-containing proteins and histone methylations have been implicated in many embryonic developmental processes, the evidence of involvement of SET domain-containing genes in regulating dorsal organizer formation and left–right asymmetry is still lacking. In this study, we report that a SET domain-containing protein Setdb2 possesses potential transcriptional repression activity through catalyzing trimethylation at histone H3 lysine 9 (H3K9me3) and negatively regulates dorsal organizer formation by suppressing the expression of fibroblast growth factor 8 (fgf8), a gene that has been shown to be involved in organizer induction, cilia formation, and left–right asymmetry (19 –21).
Results
Setdb2 Mediates H3K9me3 Histone Methyltransferase Activity in Vivo.
The zebrafish setdb2 (SET domain bifurcated 2) gene was originally identified in our large-scale sequencing database and genomewide survey and developmental expression mapping of zebrafish SET domain-containing genes (22, 23). Zebrafish Setdb2 protein exhibits 32% amino acid identities to mouse and human counterparts and 34% identity to Xenopus (Fig. S1). Two remarkably conserved regions were observed: a methyl-CpG binding domain (MBD) and a bifurcated SET domain with an adjacent pre-SET domain (Fig. 1A and Fig. S1). The MBD domain usually coexisting with additional domains (bromodomain, SET domain, and PHD finger) has been reported to specifically bind to the methylated CpG dinucleotides, resulting in chromatin compaction and transcriptional repression (24). The SET domain has been shown playing an essential role in regulating gene transcription through catalyzing methylation of the lysine residues of histones, with the exception of only H3 lysine 79 (23). The structural features indicate that the Setdb2 protein may function as a transcriptional regulator through its MBD and SET domains.
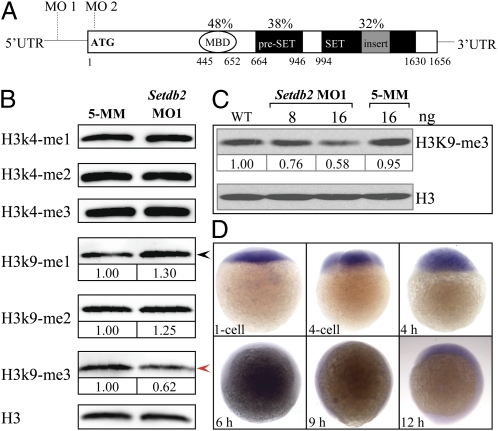
Fig. 1.
Setdb2 is a maternally expressed gene in zebrafish and possesses H3K9me3 activity in vivo. (A) Evolutionarily conserved domain architecture of zebrafish Setdb2 protein. The Setdb2 protein is composed of a methylated DNA binding domain (MBD), a pre-SET domain, and a bifurcated SET domain. The MO1 and MO2 indicate the positions of sequences targeted by the morpholino oligonucleotides. The percentages indicate the amino acid identities between human and zebrafish Setdb2. The Arabic numbers denote the position of each domain. (B) Western blot analyses of histone methylation levels in the control and the Setdb2-deficient embryos at 10 hpf. Red and black arrows denote a reduction and an increase of the level of H3K9me3 and H3K9me1, respectively. (C) Dose-dependent reduction of H3K9me3 level in the embryos injected with indicated amounts of setdb2 morpholino. (D) Spatial and temporary expression of setdb2 during early embryonic development. The Arabic numbers at the Bottom denote the extent of methylation modification compared with histone H3 level by densitometry.
If Setdb2 protein was involved in the histone methylation at lysine residues, one would expect that the histone methylation levels might be affected by Setdb2 deficiency. To test this hypothesis, we first designed two morpholino oligonucleotides (MOs), MO1 and MO2, to target the 5′-UTR and the sequences surrounding ATG of setdb2, respectively (Fig. 1A). To test the knockdown efficiency and specificity of these two morpholinos, we coinjected 8 ng of either MO1, 5-mismatch MO1 control, or MO2 with 400 pg of EGFP mRNA reporter containing MO-targeting sequences into one-cell-stage embryos. The results indicate that both MO1 and MO2 morpholinos were able to efficiently and specifically block the translation of the EGFP reporter (Fig. S2).
Next, we performed Western blot analyses to determine the levels of histone 3 methylations with a panel of antibodies against methylated H3 (mono-, di-, and trimethylations of H3K4, H3K9, and trimethylation of H3K27) in the protein lysates extracted from 10-hours postfertilization (hpf) embryos injected with 8 and 16 ng of either setdb2 MO1 or 16 ng of 5-mismatch (5-MM) control morpholino oligonucleotides. The results showed that the level of H3K9 trimethylation was significantly reduced in a dose-dependent fashion (Fig. 1B, red arrow, and Fig. 1C), accompanied by a concomitant 1.3-fold increase in the level of H3K9 monomethylation in the Setdb2-deficient embryos (Fig. 1B, black arrow). No changes in the levels of other H3 methylations were detected (Fig. 1B). These results suggest that the Setdb2 possesses H3K9me3 activity and may function as a negative transcriptional regulator of gene expression in vivo (25).
Setdb2 mRNA Is Maternally Provided and Ubiquitously Expressed Throughout Embryogenesis.
To examine the spatial and temporal expression of setbd2 during embryogenesis, whole-mount mRNA in situ hybridization (WISH) assays were performed using digoxigenin-labeled antisense RNA probe. The setdb2 expression was readily detected between one-cell stage and 18 hpf (Fig. 1D and Fig. S3A). By 24–120 hpf, setdb2 mRNA expression was faintly observed (Fig. S3A), which is consistent with the semiquantitative RT-PCR results (Fig. S3B). The results suggest that Setdb2 might play a role in early embryonic development.
Knockdown of Setdb2 Randomizes Visceral and Diencephalic Asymmetry.
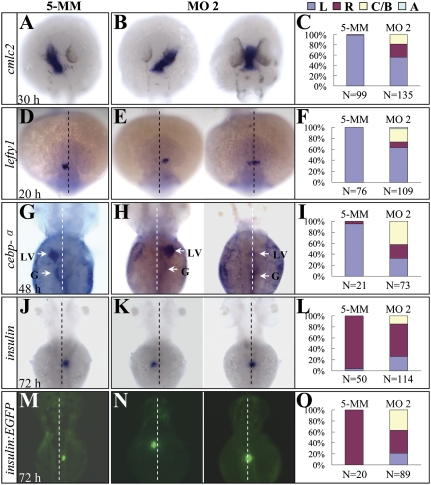
During the processes of analyzing the molecular and morphological phenotypes in Setdb2-deficient embryos, we unexpectedly found randomization of heart laterality as evidenced by WISH assay using cmlc2 as a probe at 30 hpf (Fig. 2 A and B). Approximately 26 and 29% of the MO1- and MO2-injected embryos showed a right-sided heart tube, and 19 and 13% of MO1- and MO2-injected embryos had the heart tube positioned to the ventral midline (Fig. 2 B and C and Fig. S4A). In contrast, only 1.1% of embryos injected with a 5-MM control morpholino developed a right-sided heart tube (Fig. 2 A and C). Furthermore, the expression of lefty1, a well-known gene expressed on the left side of diencephalons at 20 hpf (26), was also randomized in the Setdb2-deficient embryos (Fig. 2 D–F and Fig. S4B). Consistent with these observations, the expression domain of cebpa (denotes left-sided liver and gut at 48 and 72 hpf) and insulin (denotes right-sided pancreas at 72 hpf) were also randomized (Fig. 2 G–L and Fig. S4 C and D). The abnormal left-sided location of fluorescent pancreas was also detected in the Tg(insulin:EGFP) transgenic embryos injected with setdb2 MO2 (Fig. 2 M–O and Fig. S4E). These results indicated that Setdb2 had a global effect on laterality of visceral organs and nervous system.
Fig. 2.
Organ laterality is randomized in setdb2-knockdown embryos. (A–L) Whole-mount mRNA in situ hybridization and statistical analyses of visceral and diencephalic laterality in the 5-MM control and setdb2 MO2-injected embryos at the indicated developmental stages with probes cmlc2 (A–C), lefty1 (D–F), cebpα (G–I, arrows), and insulin (J–L). (M–O) Determination of pancreatic laterality in 72-hpf Tg(insulin:EGFP) transgenic embryos injected with 5-MM control and setdb2 MO2. Dashed lines denote the embryonic midline. LV, liver; G, gut; L, Left side; R, Right side; C/B, central or bilateral; A, absent. All embryos are dorsal views with head to the top.
Effects of Setdb2 Knockdown on Asymmetric Gene Expression.
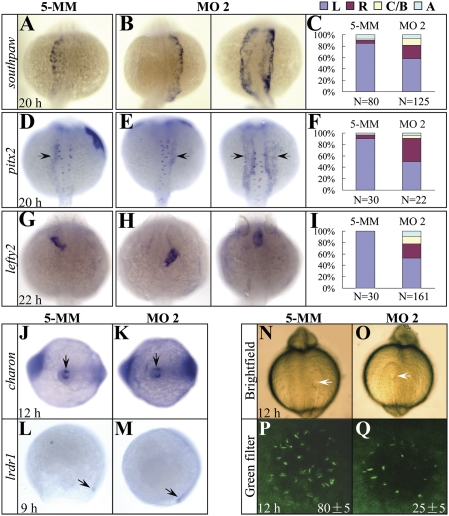
The earliest known molecular event to initiate the cascade of left–right asymmetry is the asymmetric expression of the nodal gene southpaw and its downstream targets pitx2, lefty1, and lefty2 (5, 27). We examined the expression of southpaw, pitx2, and lefty2 by WISH in the Setdb2-knockdown embryos at 20–22 hpf and found that the asymmetric expressions of all three genes were randomized (Fig. 3 A–I). The result suggests that setdb2 acts upstream of southpaw signaling to regulates the left–right asymmetry at a very early developmental stage.
Fig. 3.
The expression of asymmetric genes is randomized and cilia development in the Kupffer’s vesicle is disrupted in setdb2-knockdown embryos. (A–I) Whole-mount mRNA in situ hybridization and statistical analyses of the expression of asymmetric genes southpaw (A–C), pitx2 (D–F, arrows), and lefty2 (G–I) in the 5-MM control and setdb2 MO2-injected embryos at the indicated developmental stages. L, Left side; R, Right side; C/B, central or bilateral; A, absent. All embryos are dorsal views with head to the top. (J–M) WISH analyses of charon (J and K, arrows) and lrdr1 expression (L and M, arrows) in the 5-MM control and setdb2 MO2-injected embryos. (N–Q) Morphology of Kupffer’s vesicle (N and O, arrows) and confocol analyses of ciliary development (P and Q, green) as revealed by immunostaining with anti-acetylated tubulin antibody. The numbers in P and Q denote the mean ± SD. Embryos are ventral views in J, K, N, and O and lateral views in L and M.
Effects of Setdb2 Knockdown on KV-Derived Cilia and Midline Development.
KV is the zebrafish homologous organ of mouse and chicken node, which is a transient embryonic structure located at the end of the notochord and adjacent to the yolk cells between late gastrulation stage and somite stage (4). Previous studies have shown that abnormalities in KV development (4) or defects in the cilia formation and function within the KV disrupt left–right asymmetry (20). In setdb2 morphants, the morphology of KV was indistinguishable from the control embryos as evidenced by appropriate expression of either charon, a gene expressing around KV and essential for left–right asymmetry establishment (28), or lrdr1 that is required for ciliary motility (4) (Fig. 3 J–O, arrows). However, the number of cilium within the KV was significantly reduced in the setdb2 morphants (25 ± 5 vs. 80 ± 5, n = 5/5; Fig. 3 P and Q), suggesting a reduction of the number and functional compromise of ciliated cells.
Because the precursors of ciliated cells are a group of ∼20–30 cells known as dorsal forerunner cells (DFCs), which locate at the leading edge of the developing notochord (7), and because the notochord has been known to be an important barrier for preventing the laterality signals from leaking to the other side (1, 2, 29), we performed WISH analyses with several midline markers oep (30), shh (31), and ntl (10) to examine the notochord and midline structures in the setdb2-knockdown embryos. The results showed that the midline was shorter and wider in the Setdb2-deficent embryos at 10 hpf, compared to the control embryos (Fig. S5 A, B, E, F, I, and J, n = 47/60). Interestingly, in ∼10% of Setdb2-deficent embryos, an incomplete secondary axis was observed (Fig. S5 F and J, Right). Furthermore, undulations of the midline were frequently observed between 18 and 28 hpf ((Fig. S5 C, D, G, H, K, and L, n = 85/110). Because two midline genes ntl and shh were colocalized within the defective midline of Setdb2-deficient embryos (Fig. S6A; n = 18/25), and the convergence and extension (CE) movements were also inhibited in the setdb2 morphants as revealed by lateral expansion of myoD-positive somites (Fig. S6B; n = 20/28), it suggests that the abnormal expression patterns of midline genes are a consequence of defective midline development due to inappropriate CE movement.
Previous studies have shown that several signaling transduction pathways and genes are essential for midline development, CE movements, or subsequent left–right specification (11, 32). We examined the components of noncanonical WNT pathway (wnt4/5/8/11, wnt11r, duboraya, dvl2, dvl3, fzd2, kny, rock2, and rhoA) (32, 33), TGF-β superfamily (bmp4, cyclops, squint, and smad2) (30, 34), T-box gene family (tbx16 and tbxc) (35, 36) and other reported genes involved in the regulation of left–right asymmetry such as alk4, ipk1, pkd2, and pdip5 (37 –40) with WISH to determine the expression levels of these genes. No detectable changes were observed between the setdb2-knockdown and control embryos (Fig. S6 C–L).
Enlargement of Dorsal Organizer in the setdb2-Knockdown Embryos.
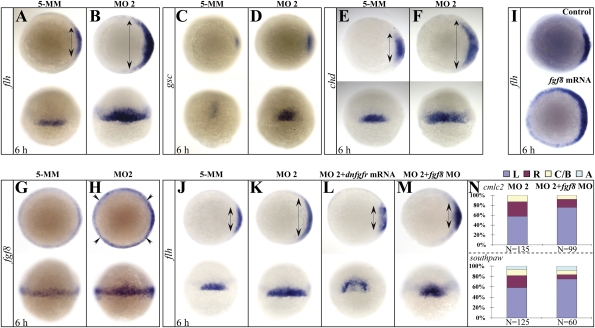
What developmental and molecular mechanisms might account for the abnormal midline development, decreased cilium number, and randomization of left–right asymmetry induced by Setdb2 deficiency? It has been shown that physiological midline development depends on the normal function and formation of the Spemann organizer (8, 9). We first examined the expression patterns of three well-known organizer genes floating head (flh), goosecoid (gsc), and chordin (chd) (12) at 6 hpf, and found that the expression territories of all three genes were significantly enlarged in the setdb2-knockdown embryos compared to the control embryos (Fig. 4 A–F, n = 45/60). To examine whether the earlier step of gastrulation is affected by setdb2 deficiency, the expression pattern of two additional genes wnt8a and fgf8 were analyzed by WISH at 5.3 hpf, and no detectable changes were observed (Fig. S7 A–D), suggesting that the expanded dorsal organizer is not a consequence of abnormalities in the earlier stage of gastrulation. The results suggest that Setdb2 acts as a negative regulator to restrict the size of the dorsal organizer in a relatively specific manner.
Fig. 4.
Setdb2 restricts dorsal organizer formation and regulates left–right asymmetry through suppressing Fgf8 activity. (A–F) WISH analyses of organizer markers flh (A and B), gsc (C and D), and chd (E and F) in the 5-MM control and setdb2 MO2-injected embryos at 6 hpf. Double-headed arrows denote territories of dorsal organizer. Embryos are animal views in the Upper panels, dorsal views in the Bottom panels. (G–I) WISH analyses of fgf8 expression (G and H) in the 5-MM control and setdb2 MO2-injected embryos at 6 hpf and effects of fgf8 overexpression on the dorsal organizer development as revealed by flh expression (I). Embryos are animal views in panels G and H (Upper) and I and dorsal views in panels G and H (Bottom). (J–M) WISH analyses of flh in the 6-hpf embryos injected with 5-MM control (J), setdb2 MO2 (K), setdb2 MO2 plus dominant-negative fibroblast growth factor receptor (dnfgfr) mRNAs (L), and setdb2 MO2 plus fgf8 morpholino oligonucleotides (M). (N) Statistic analyses of heart laterality marker cmlc2 and asymmetric gene southpaw in 6-hpf embryos injected with setdb2 MO2 and setdb2 MO2 plus fgf8 morpholino. Embryos are animal views in the Upper Panels and dorsal views in the Lower Panels.
To further understand the molecular mechanisms of how Setdb2 affects dorsal organizer formation, we examined the expression of several previously reported organizer-regulating genes such as eomesodermin, dharma, iroquois3 (12 –14), and members of the fgf family (19) at shield stage (6 hpf). Although no changes in the levels of eomesodermin, dharma, iroquois3, and fgf4 expressions were observed between control and setdb2-knockdown embryos (Fig. S7 E and F), the levels of fgf8 expression were significantly increased at 6 and 22 hpf upon Setdb2 deficiency (Fig. 4 G and H and Fig. S8; n = 30/40).
We next microinjected 300 pg of fgf8 mRNAs into one-cell-stage embryos and found that the dorsal organizer was also significantly expanded as revealed by an enlarged flh expression domain, suggesting that fgf8 may be a downstream target of setdb2 to mediate the restrictive effects of Setdb2 on dorsal organizer development (Fig. 4I, n = 15/20). To test this hypothesis, we microinjected setdb2 morpholino with dominant-negative fgf receptor (dnfgfr) mRNA (300 pg), which has been shown to efficiently abolish fgf8 function (41), and found that the expanded dorsal organizer induced by Setdb2 deficiency was nearly corrected to normal size (Fig. 4 J–L, n = 25/30). Consistent with this result, knockdown of fgf8 with a specific morpholino oligonucleotide (42) also restores the dorsal organizer to normal size in the Setdb2-deficient embryos (Fig. 4M, n = 30/35). Furthermore, the randomization of the expression of either cmlc2 or southpaw was also corrected in the setdb2–fgf8 double morphants (Fig. 4N). Taken together, these results suggest that the Fgf8 is a bona fide downstream target of Setdb2 to regulate the development of dorsal organizer and left–right asymmetry.
Discussion
In the past few years, epigenetic regulation has been shown to be essential for early embryonic development (16, 17). The development and formation of the dorsal organizer is one of the most critical developmental events during early embryonic development and is responsible for neural induction, mesoderm and endoderm patterning, midline and axis formation, and subsequent left–right asymmetry establishment (43, 44). Although many signaling pathways and genes have been found to be involved in the regulation of organizer development (12 –15), whether or not epigenetic regulators participate in this process is largely unknown. In this study, we provided unique evidence, to our best knowledge, that the setdb2, an epigenetic regulator responsible for repressive H3K9me3, plays an important role in negatively regulating dorsal organizer formation and contributes to the establishment of left–right axis through suppressing fgf8 signaling. Interestingly, setdb1, a paralog of setdb2, also possesses H3K9me3 activity and is required for peri-implantation development (23, 45). The possibility that other H3K9 methyltransferases may participate in the regulation of dorsal organizer and determination of L–R asymmetry cannot be excluded.
The establishment of left–right axis has been molecularly attributed to the asymmetric distribution of nodal signaling genes such as southpaw, which is required for initiating subsequent asymmetric location of downstream targets such as lefty1/2 and pitx2. However, elegant studies have shown that the function and number of cilium in the developing KV during gastrulation act as an earlier physiological gatekeeper to maintain the asymmetric distribution of these nodal signaling factors through a mechanistic counterclockwise flow (4). Furthermore, several groups have convincingly shown that the formation of the dorsal organizer, which is induced by a complex interaction between signaling pathways, plays a critical role in appropriate patterning of midline and the KV-derived cilia development (4, 7). Because loss-of-function of setdb2 not only causes the randomizations of visceral and asymmetric gene laterality, but also results in the decreased number of cilia in the KV, disrupted midline patterning, and enlarged dorsal organizer, these results, along with previous findings, prompt us to propose that the establishment of left–right axis is a consequence not only of a highly coordinated process associated with distribution of asymmetry-initiating factors, but also a stepwise development and physiological interactions of early embryonic structures(organizer→midline→KV→cilia).
Although it has been well established that the members of fgf signaling (fgf3, fgf4, and fgf8) are essential for the development of the dorsal organizer and the establishment of left–right asymmetry (19, 46), little is known about the upstream signaling or factor that regulates the fgf expression at the transcriptional level through the histone methylation-mediated epigenetic mechanism. A unique finding of this study is the identification of fgf8, but not fgf4, as a downstream target of Setdb2. The facts that the Setdb2 knockdown specifically upregulates fgf8 and that the suppression of fgf8 function by either overexpression of dominant-negative fgfr mRNAs or fgf8 knockdown nearly corrected both the expansion of the organizer and randomization of left–right asymmetry induced by Setdb2 deficiency, strongly suggest that the Setdb2 acts upstream to regulate the fgf8 expression. Future studies are needed to clarify whether the Setdb2 is recruited to the promoter of the fgf8 gene to directly exert its H3K9me3 and transcriptional repression activity and which transcriptional factor might mediate the proper positioning of Sedb2 to this promoter, especially given that histone methyltransferases have not been shown to directly bind promoters due to lack of sequence-specific DNA-binding domains.
Materials and Methods
Zebrafish Strains.
Zebrafish stocks were maintained at 28.5 °C under standard aquaculture conditions. Embryos were staged by hours postfertilization as described previously (47). The Tg(insulin:EGFP) transgenic zebrafish lines were established in our laboratory.
Plasmid Constructs and RNA Probes.
Full-length coding sequence for setdb2 and fragments of alk4, bmp4, dharma, dub, dvl2, dvl3, fzd2, gsc, ipk1, iroquois3, kny, pdip5, pkd2, rock2, smad2, and tbxc were cloned into pCS2+ or pGEM− Teasy vector. The plasmids southpaw, lefty1, lefty2, pitx2, lrdr1, charon, oep, shh, ntl, wnt4, wnt5a, wnt8, wnt11, wnt11-related, rhoA, cyclops, squint, tbx16, flh, chordin, eomesodermin, dnfgfr4, and fgf8 are generous gifts from other laboratories (Acknowledgments). The digoxigenin- (Dig-) or fluorescein-labeled probes were transcribed from linearized plasmids.
WISH.
WISH was performed as described previously (48). The stained embryos were mounted in 4% methylcellulose and photographed using the Nikon SMZ1500 stereomicroscope.
Morpholino and mRNA Injections.
Antisense MOs against either the 5′-UTR (MO1: 5′-CAGGGTTCAGGAGATTTTAATGACT-3′; 5-mismatch to MO1: 5′-CAcGGTTgAGGAcATTTaAATcACT-3′) or the start codon of the setdb2 gene (MO2: 5′-TTGCTGTGTCGGCTTCAGTCTCCAT-3′), as well as the MO against the start codon of the fgf8 gene (fgf8 MO: 5′-GAGTCTCATGTTTATAGCCTCAGTA-3′) were obtained from Gene Tools. Capped mRNAs were transcribed with mMESSAGE mMACHINE Kit (Ambion). All MOs and mRNAs were injected at the one-cell stage.
Immunofluorescence and Confocal Microscopy.
Whole-mount immunofluorescence with primary antibody against mouse acetylated tubulin (1:500 dilution, Sigma) and Alexa Fluor 488-labeled secondary antibody (donkey anti-mouse, 1:500 dilution, Invitrogen) was performed as described previously (3).
The stained embryos were mounted in 1% low-melting point agarose for visualization of KV and the images were collected using a Leica TCS-LSI confocal microscope equipped with 5× objective. The 488-nm laser line was used for excitation of the Alexa Fluor 488.
Western Blot.
Embryos at 10 hpf were deyolked as described previously (49). Embryos were homogenized in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM ethylenediaminetetra-acetic acid, 10% glycerol, and 0.1% Triton X-100) containing protease inhibitor mixture and phosphatase inhibitor (Roche Diagnostics) as described previously (48). Mouse anti-histone H3 antibody (1:10,000) and a panel of antibodies against methylated histone 3 (H3K4, H3K9, and H3K27) (1:1,000) and a horseradish peroxidase-conjugated secondary antibody (1:10,000) were used. Band densities were quantified with Image-Pro Plus software.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Yost at University of Utah (Salt Lake City, UT) for providing southpaw, cyclops, squint, lefty1, lefty2, pitx2, and lrdr1 plasmids; Dr. Michael Rebagliati at University of Iowa (Iowa City, IA) for shh, ntl, and tbx16 plasmids; Dr. Masahiko Hibi at the Center for Developmental Biology in RIKEN (RIKEN, Japan) for the charon plasmid; Dr. Wu Han Xiao at the Institute of Hydrobiology in Wuhan (HuBei Province, China) for wnt4, wnt5a, wnt11, and wnt11-related plasmids; Dr. Randall T. Moon at the Howard Hughes Medical Institute (Chevy Chase, MD) for the wnt8 plasmid; Dr. Boon Chuan Low at National University of Singapore Singapore, for the rhoA plasmid; Dr. Ashley Bruce at University of Toronto (Toronto, Canada) for the eomesodermin plasmid; Dr. Steve Wilson at University College London (London, UK) for flh, chordin, and fgf8 plasmids; and Dr. Igor B. Dawid at National Institutes of Health (Bethesda, MD) for the dnfgfr4 plasmid. We thank Xia Ming and Jin Hao for excellent technical assistance. We thank Dr. Xiao Yan Ding at Shanghai Institutes for Biological Sciences and all members of the laboratory for helpful discussions. The work was supported in part by the National Basic Research Program of China (2007CB947003), National Natural Science Foundation of China (30525019 and 30830047), and the Science and Technology Commission of Shanghai Municipality (09XD1404700), the National High Tech Program for Biotechnology (863, 2006AA02A405); the Chinese National Key Basic Research Project (973, 2010CB529200); the Key Discipline program of Shanghai Municipal Education Commission (Y0201); the Grant for Innovation Group of the National Natural Science Foundation of China (30821063); the Shanghai Municipal Commission for Science and Technology (06DZ2202, 07DZ05908, 08DZ2200100).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914396107/DCSupplemental.
References
- 1.Capdevila J, Vogan KJ, Tabin CJ, Izpisúa Belmonte JC. Mechanisms of left-right determination in vertebrates. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- 2.Raya A, Izpisúa Belmonte JC. Left-right asymmetry in the vertebrate embryo: From early information to higher-level integration. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 3.Kreiling JA, Williams G, Creton R, Prabhat Analysis of Kupffer’s vesicle in zebrafish embryos using a cave automated virtual environment. Dev Dyn. 2007;236:1963–1969. doi: 10.1002/dvdy.21191. [DOI] [PubMed] [Google Scholar]
- 4.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 5.Shen MM. Nodal signaling: Developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- 6.Cooper MS, D’Amico LA. A cluster of noninvoluting endocytic cells at the margin of the zebrafish blastoderm marks the site of embryonic shield formation. Dev Biol. 1996;180:184–198. doi: 10.1006/dbio.1996.0294. [DOI] [PubMed] [Google Scholar]
- 7.Lee JD, Anderson KV. Morphogenesis of the node and notochord: The cellular basis for the establishment and maintenance of left-right asymmetry in the mouse. Dev Dyn. 2008;237:3464–3476. doi: 10.1002/dvdy.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Robertis EM, Larraín J, Oelgeschläger M, Wessely O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat Rev Genet. 2000;1:171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niehrs C. Regionally specific induction by the Spemann-Mangold organizer. Nat Rev Genet. 2004;5:425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- 10.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 11.Oishi I, Kawakami Y, Raya A, Callol-Massot C, Izpisúa Belmonte JC. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat Genet. 2006;38:1316–1322. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- 12.Bruce AE, et al. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130:5503–5517. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- 13.Fekany K, et al. The zebrafish bozozok locus encodes Dharma, a homeodomain protein essential for induction of gastrula organizer and dorsoanterior embryonic structures. Development. 1999;126:1427–1438. doi: 10.1242/dev.126.7.1427. [DOI] [PubMed] [Google Scholar]
- 14.Kudoh T, Dawid IB. Role of the iroquois3 homeobox gene in organizer formation. Proc Natl Acad Sci USA. 2001;98:7852–7857. doi: 10.1073/pnas.141224098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xanthos JB, et al. The roles of three signaling pathways in the formation and function of the Spemann Organizer. Development. 2002;129:4027–4043. doi: 10.1242/dev.129.17.4027. [DOI] [PubMed] [Google Scholar]
- 16.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 17.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Tschiersch B, et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maegawa S, Varga M, Weinberg ES. FGF signaling is required for beta-catenin-mediated induction of the zebrafish organizer. Development. 2006;133:3265–3276. doi: 10.1242/dev.02483. [DOI] [PubMed] [Google Scholar]
- 20.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regan JC, Concha ML, Roussigne M, Russell C, Wilson SW. An Fgf8-dependent bistable cell migratory event establishes CNS asymmetry. Neuron. 2009;61:27–34. doi: 10.1016/j.neuron.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song HD, et al. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci USA. 2004;101:16240–16245. doi: 10.1073/pnas.0407241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun XJ, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3:e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4:1. doi: 10.1186/1471-2164-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 26.Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–240. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- 27.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto H, et al. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development. 2004;131:1741–1753. doi: 10.1242/dev.01070. [DOI] [PubMed] [Google Scholar]
- 29.Danos MC, Yost HJ. Role of notochord in specification of cardiac left-right orientation in zebrafish and Xenopus. Dev Biol. 1996;177:96–103. doi: 10.1006/dbio.1996.0148. [DOI] [PubMed] [Google Scholar]
- 30.Warga RM, Kane DA. One-eyed pinhead regulates cell motility independent of Squint/Cyclops signaling. Dev Biol. 2003;261:391–411. doi: 10.1016/s0012-1606(03)00328-2. [DOI] [PubMed] [Google Scholar]
- 31.Ertzer R, et al. Cooperation of sonic hedgehog enhancers in midline expression. Dev Biol. 2007;301:578–589. doi: 10.1016/j.ydbio.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Matsui T, et al. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev. 2005;19:164–175. doi: 10.1101/gad.1253605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu S, Liu L, Korzh V, Gong Z, Low BC. RhoA acts downstream of Wnt5 and Wnt11 to regulate convergence and extension movements by involving effectors Rho kinase and Diaphanous: Use of zebrafish as an in vivo model for GTPase signaling. Cell Signal. 2006;18:359–372. doi: 10.1016/j.cellsig.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Chen JN, et al. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- 35.Dheen T, et al. Zebrafish tbx-c functions during formation of midline structures. Development. 1999;126:2703–2713. doi: 10.1242/dev.126.12.2703. [DOI] [PubMed] [Google Scholar]
- 36.Griffin KJ, Amacher SL, Kimmel CB, Kimelman D. Molecular identification of spadetail: Regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development. 1998;125:3379–3388. doi: 10.1242/dev.125.17.3379. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, et al. ALK4 functions as a receptor for multiple TGF beta-related ligands to regulate left-right axis determination and mesoderm induction in Xenopus. Dev Biol. 2004;268:280–294. doi: 10.1016/j.ydbio.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Schottenfeld J, Sullivan-Brown J, Burdine RD. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development. 2007;134:1605–1615. doi: 10.1242/dev.02827. [DOI] [PubMed] [Google Scholar]
- 40.Hoshijima K, Metherall JE, Grunwald DJ. A protein disulfide isomerase expressed in the embryonic midline is required for left/right asymmetries. Genes Dev. 2002;16:2518–2529. doi: 10.1101/gad.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong SK, Dawid IB. FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc Natl Acad Sci USA. 2009;106:2230–2235. doi: 10.1073/pnas.0812880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fürthauer M, Van Celst J, Thisse C, Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131:2853–2864. doi: 10.1242/dev.01156. [DOI] [PubMed] [Google Scholar]
- 43.Nieto MA. Reorganizing the organizer 75 years on. Cell. 1999;98:417–425. doi: 10.1016/s0092-8674(00)81971-6. [DOI] [PubMed] [Google Scholar]
- 44.Nascone N, Mercola M. Organizer induction determines left-right asymmetry in Xenopus. Dev Biol. 1997;189:68–78. doi: 10.1006/dbio.1997.8635. [DOI] [PubMed] [Google Scholar]
- 45.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamauchi H, Miyakawa N, Miyake A, Itoh N. Fgf4 is required for left-right patterning of visceral organs in zebrafish. Dev Biol. 2009;332:177–185. doi: 10.1016/j.ydbio.2009.05.568. [DOI] [PubMed] [Google Scholar]
- 47.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 48.Fu YF, et al. Mir-144 selectively regulates embryonic alpha-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113:1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- 49.Link V, Shevchenko A, Heisenberg CP. Proteomics of early zebrafish embryos. BMC Dev Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.