Abstract
4EBP1 is phosphorylated by the mTORC1 kinase. When mTORC1 activity is inhibited, hypophosphorylated 4EBP1 binds and sequesters eIF4E, a component of the mRNA cap-binding complex, and blocks translation. As a consequence, mTORC1 activity is needed to maintain active translation. The human cytomegalovirus pUL38 protein preserves mTORC1 activity, keeping most of the E4BP1 in the infected cell in a hyperphosphorylated, inactive state. Here we report that a second viral protein, pUL69, also antagonizes the activity of 4EBP1, but by a separate mechanism. pUL69 interacts directly with eIF4A1, an element of the cap-binding complex, and the poly(A)-binding protein, which binds to the complex. When pUL69 accumulates during infection with wild-type virus, 4EBP1 is excluded from the complex. However, 4EBP1 is present in the cap-binding complex after infection with a pUL69-deficient virus, coincident with reduced accumulation of several late virus-coded proteins. We propose that pUL69 supports translation in human cytomegalovirus-infected cells by excluding hypophosphorylated 4EBP1 from the cap-binding complex.
Keywords: translational control, two-hybrid screen, eukaryotic initiation factor 4A, poly(A)-binding protein
Human cytomegalovirus (HCMV) is a ubiquitous β-herpesvirus. Although infection is generally asymptomatic in healthy children and adults, HCMV is a leading cause of birth defects and a major cause of morbidity and mortality in immunocompromised individuals (1). The viral genome is estimated to contain nearly 200 genes, many of which interact with cell-coded proteins to modulate cell functions. Some HCMV-coded genes have homologs in all known herpesviruses, others are unique to the β-herpesvirus subfamily, and several are exclusive to cytomegaloviruses.
The HCMV UL69-coded protein, pUL69, is a member of a protein family for which representatives are found in all herpesviruses. Its homologs include ICP27 in HSV type 1 and EB2 in Epstein-Barr virus. Members of this family share a domain with conserved amino acid sequence. For pUL69, this domain is located in the central portion of the protein, and several protein-protein interactions have been mapped to this region, including the ability of pUL69 to multimerize (2).
pUL69 is a phosphoprotein that is packaged in HCMV virions (3). The earliest functional analysis of pUL69 showed that it enhances expression of reporter constructs controlled by a variety of promoters within transfected cells (4). It is not known whether pUL69 influences transcription directly, but such a role would be consistent with its ability to interact with hSPT6 (5), a cell protein involved in transcriptional elongation and chromatin remodeling, and cyclin T1 (6), a component of another transcriptional elongation complex. pUL69 also modulates cell cycle progression. Expression of the protein outside the context of an infection induces a cell cycle block late in G1 (7), and a mutant virus unable to express pUL69 fails to efficiently induce the G1 block seen after infection of fibroblasts with wild-type virus (8). The mechanism by which pUL69 blocks cell cycle progression is not known. It may result from interaction with cellular cyclin-dependent kinases, such as CDK1, which phosphorylates pUL69 (6), or with the HCMV-coded pUL97 kinase (9). pUL97, which has a structural resemblance to cellular cyclin-dependent kinases, hyperphosphorylates and inactivates the cellular retinoblastoma protein, which favors cell cycle progression (10, 11). Although the interaction of pUL97 and cellular cyclin-dependent kinases with pUL69 might simply reflect phosphorylation of pUL69 by the kinases, it is possible that the associations also modulate kinase behavior, influencing cell cycle progression.
The most intensively studied activity of pUL69 is its ability to facilitate the export of unspliced mRNA from the nucleus to cytoplasm (12), a function that it shares with its counterparts in other herpesviruses (13 –15). pUL69 shuttles between nucleus and cytoplasm (16), binds RNA (17), and interacts with the DExD/H-box RNA helicase UAP56 or the related URH49 protein (12), cellular proteins involved in RNA transport. The pUL69 transport function is stimulated by both cellular cyclin-dependent kinases (6) and pUL97 (9).
Here we report an additional activity of pUL69: it excludes 4EBP1 from the mRNA cap-binding complex. 4EBP1 is a target of the mTOR kinase (18); when mTOR activity is inhibited, hypophosphorylated 4EBP1 binds and sequesters eIF4E, inhibiting translation (19). Consistent with a role of pUL69 in translational regulation, changes in the location of specific RNAs within polysome profiles and in the levels of their protein products were evident in wild-type as compared to pUL69-deficient virus-infected cells.
Results
HCMV-Coded pUL69 Interacts with eIF4A1 and Poly(A)-Binding Protein.
To generate hypotheses for mechanisms underlying pUL69 function, we performed a yeast two-hybrid screen to identify potential interacting proteins. The UL69 ORF was used as bait and a cDNA library containing equal parts of cDNAs isolated from total cell RNA at 6, 24, and 48 hpi with HCMV was used as prey. Yeast cells were cotransfected with bait and prey plasmids, and selected for reconstitution of Gal4 activity by requiring simultaneous activation of four Gal4-responsive genes. Positive cDNA clones were sequenced; and cDNAs expressing ≥ 3-fold more α-galactosidase, which is controlled by Gal4, in the presence than in the absence of the bait plasmid were considered to potentially encode proteins interacting with pUL69 (Table 1).
Table 1.
Candidate pUL69 interaction partners
| NCBI GeneID | Gene symbol | α-gal Units pGADT7cDNA + pMG1 | α-gal Units pGADT7cDNA |
| 1973 | EIF4A1 | 0.35 | 0.029 |
| 26986 | PABPC1 | 0.199 | <0.001 |
| 7936 | RDBP | 0.333 | 0.025 |
| 7936 | RDBP | 0.75 | 0.035 |
| 64784 | CRTC3 | 0.169 | <0.001 |
| 26747 | NUFIP1 | 1.651 | <0.001 |
| 5411 | PNN | 0.262 | 0.008 |
| 2237 | FEN1 | 3.805 | 0.111 |
| 8650 | NUMB | 0.041 | <0.001 |
| 4600 | MX2 | 0.417 | 0.076 |
| 4600 | MX2 | 0.161 | <0.001 |
| 51079 | NDUFA13 | 0.133 | <0.001 |
| 1508 | CTSB | 0.609 | <0.050 |
| 1508 | CTSB | 0.916 | 0.008 |
| 1513 | CTSK | 0.225 | <0.001 |
| 55558 | PLXNA3 | 0.127 | 0.042 |
| 55558 | PLXNA3 | 0.715 | 0.009 |
| 51389 | RWDD1 | 0.758 | <0.001 |
| 2885 | GRB2 | 0.782 | <0.001 |
| 2885 | GRB2 | 0.565 | <0.001 |
| 10188 | TNK2 | 0.735 | <0.001 |
| 2597 | GAPDH | 0.537 | 0.073 |
| 10330 | CNPY2 | 0.682 | <0.001 |
| 1278 | COL1A2 | 0.479 | 0.017 |
| 3077530 | HCMV.UL82 | 0.017 | <0.001 |
Each human gene encoding a possible pUL69 interacting protein is identified by its National Center for Biotechnology Information (NCBI) gene ID and gene symbol. α-galactosidase (α-gal) units are reported for extracts of yeast receiving the cDNA prey plasmid plus pUL69 bait plasmid (pGADT7cDNA + pMG1) or the cDNA prey plasmid alone (pGADT7cDNA).
Although we readily detected the interaction of pUL69 with a known binding partner, SPT6 (5), in a reconstruction experiment (data not shown), we did not detect SPT6 or several other reported partners (6, 12) in our screen. We assayed ∼3.2 × 105 primary yeast transformants, and consequently we would not expect to identify all interactions mediated by proteins that are encoded by low abundance mRNAs. Nevertheless, our assay identified 20 interacting protein segments that robustly induced Gal4 activity, and 5 of them were identified twice (Table 1). The putative interaction partners included both nuclear and cytoplasmic proteins, consistent with the ability of pUL69 to shuttle between the two compartments (16).
Our attention was drawn to the predicted interaction of pUL69 with poly(A)-binding protein C1 (PABPC1) and eukaryotic initiation factor 4A1 (eIF4A1), because the HSV pUL69 homolog, ICP27, has been shown to interact with initiation factors, including PABP (20), and facilitate the translation of a subset of virus-coded mRNAs (21 –23). PABPC1 and eIF4A1 are both constituents of the mRNA cap-binding complex, raising the possibility that pUL69 interacts with them to modulate translation.
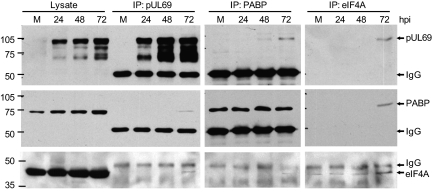
To confirm the prediction that pUL69 binds to PABPC1 and eIF4A1, extracts were prepared at various times after infection, immunoprecipitated with antibody to each of the three proteins, and the precipitates were analyzed by Western blot using the same set of antibodies (Fig. 1). Analysis of whole-cell lysates by Western blot identified multiple pUL69-specific species, as reported previously (7), and also demonstrated that the level of PABPC1, but not eIF4A1, increases several-fold as the infection progresses, which also has been reported (24). Extracts prepared at 72 hpi demonstrated the anticipated interactions. PABPC1 and eIF4A1 were coprecipitated with pUL69 antibody, pUL69 was coprecipitated with antibody to PABPC1 or eIF4A1, and antibody to each cellular factor coprecipitated the other factor.
Fig. 1.
pUL69 interacts with PABPC1 and eIF4A1. MRC5 cells were mock infected (M) or infected with TNwt and harvested at the times indicated. Whole-cell lysates or immunoprecipitated (IP) proteins were analyzed by Western blot using antibodies to the indicated proteins. The position of protein markers (identified by their mass in kilodaltons) is shown to the left of the panels, and IgG designates the Ig heavy chain.
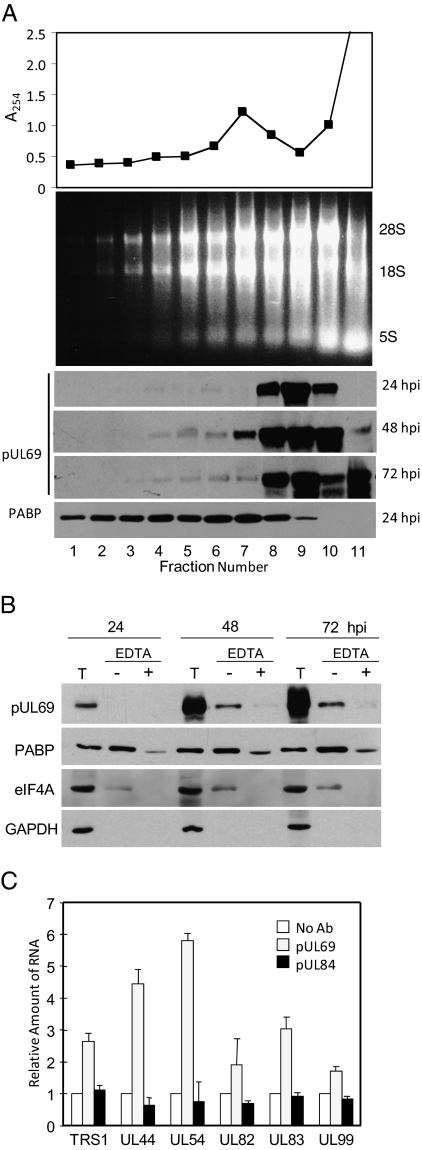
Given the interaction of pUL69 with translation factors, we tested the possibility that pUL69 is associated with polysomes. Cytoplasmic extracts were prepared at 72 hpi, subjected to centrifugation in a sucrose gradient, and fractionated. Protein was monitored by UV absorbance to identify the monosome peak in fractions 5 to 8 (Fig. 2A, Top). Similarly, ribosomal RNAs were monitored by electrophoretic analysis, and they extended through the gradient to fraction 2 near the bottom, indicating that fractions 1 to 4 are heavily enriched for polysomes (Fig. 2A, Middle). Western blot assays demonstrated that PABPC1 and a minor portion of pUL69 were present in the polysome-containing region of the gradient (Fig. 2A, Bottom).
Fig. 2.
pUL69 associates with polysomes. MRC5 cells were harvested at 72 h after infection with TNwt. (A) Polysome preparation and analysis. Cytoplasmic extracts of infected MRC5 cells were prepared and fractionated by centrifugation on a 10 to 50% sucrose gradient. The bottom of gradient is to the left, and fractions 1 to 4 correspond to polysomes. (Top) UV absorbance profile of gradient fractions; (Middle) Analysis of total RNA present in gradient fractions by electrophoresis in an agarose gel; (Bottom) Western blot analysis of proteins in gradient fractions using antibodies for pUL69 and PABPC1. (B) Disruption of polysomes displaces pUL69 from rapidly sedimenting fractions. Cytoplasmic extracts were treated with EDTA (+) or left untreated (−) and subjected to centrifugation; proteins in fractions 1 to 4 containing polysomes were precipitated and analyzed by Western blot by using antibodies for pUL69, PABPC1 and eIF4A1. T, total cytoplasmic extract. (C) pUL69 interacts with viral mRNAs. Cytoplasmic extracts were subjected to immunoprecipitation by using an antibody to pUL69 pUL84 or they were mixed with protein A/G Sepharose without first receiving a primary antibody (No Ab). mRNA was isolated from the immunoprecipitates and quantified by RT-PCR using sequence-specific primers.
To confirm that pUL69 is associated with polysomes and not some other rapidly sedimenting structure, we divided cytoplasmic extracts into two portions and treated one with 50-mM EDTA to disrupt polysomes (25) before centrifugation (Fig. 2B). pUL69 was present in the polysome fractions at 48 and 72 hpi in the absence of EDTA, but treatment with the chelator almost completely removed pUL69 from this region of the gradient, arguing that pUL69 at this position traveled with polysomes. Similar results were obtained with EDTA treatment for eFF4A1, and PABPC1 was partially shifted out of the polysome region.
We also asked if virus mRNAs could be coimmunoprecipitated with pUL69 (Fig. 2C). Precipitations were performed using cytoplasmic extracts of cells prepared at 72 hpi, and RNAs were assayed by qRT-PCR. Six HCMV RNAs, including representatives of all three kinetic classes of HCMV gene expression, were tested, and all were coprecipitated with pUL69. They were not precipitated if pUL69-specific antibody was left out of the reaction or if a nonspecific antibody was used.
These experiments demonstrate that pUL69 binds to elements of the mRNA cap protein complex. Consistent with this interaction, the viral protein is present on polysomes and associates with a variety of HCMV mRNAs.
pUL69 Excludes 4EBP1 from the Cap-Binding Complex, Facilitating Accumulation of HCMV Proteins.
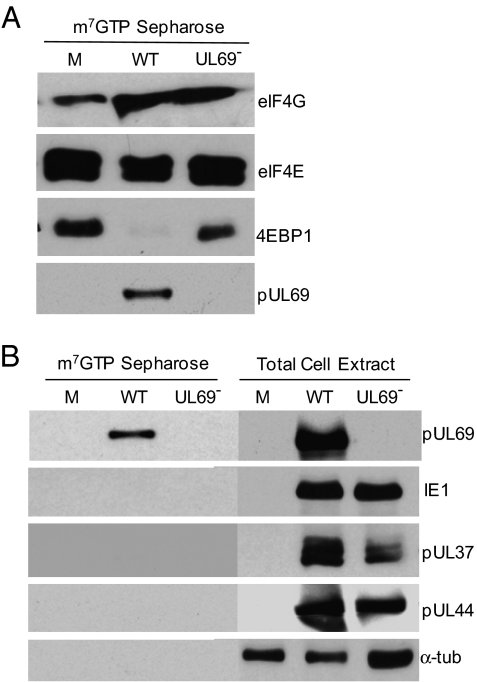
To investigate the consequences of the interaction of pUL69 with constituents of the cap-binding complex, we employed TNsubUL69, a pUL69-deficient HCMV mutant (8). We prepared extracts from cells infected with the mutant or its parent, TNwt, and used m7GTP Sepharose to capture elements of the cap-binding complex to confirm that pUL69 interacted with the complex and to test whether the viral protein altered its composition (Fig. 3A). Two components of the complex were assayed, eIF4E and EIF4G, and both were captured from extracts of mutant and wild-type virus-infected cells. pUL69 was captured from wild-type but not mutant samples and, strikingly, 4EBP1 was captured with components of the complex from extracts of cells infected with the mutant but not wild-type virus. As a control for specificity, three additional virus-coded proteins were tested, and none were captured (Fig. 3B, Left). These proteins were assayed by Western blot from cell extracts, and each was produced by both viruses, confirming that the mutant virus produced all proteins assayed except for pUL69 (Fig. 3B, Right).
Fig. 3.
UL69 excludes 4EBP1 from the cap-binding complex. Cells were mock infected (M) or infected with TNwt (WT) or TNsubUL69 (UL69−). (A) m7GTP Sepharose chromatography. At 72 hpi, extracts were prepared and proteins bound to m7GTP Sepharose were analyzed by Western blot using antibodies to the indicated proteins. (B) Specificity of m7GTP Sepharose interactions. (Left) Experiment was as described in A. (Right) Total cell extracts were analyzed by Western blot. α-tub, α-tubulin.
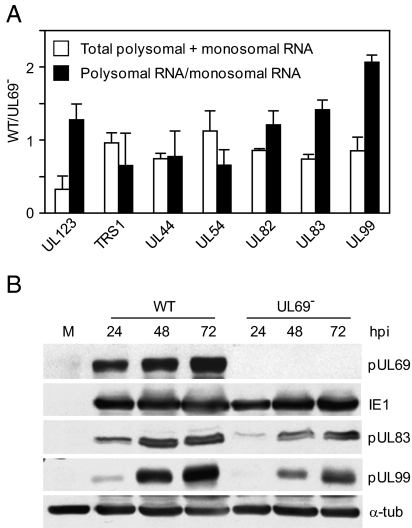
Because 4EBP1 inhibits translation when it interacts with the eIF4E constituent of the cap-binding complex (19), we anticipated that its presence among the components of the complex captured from cells infected with the pUL69-deficient virus might reflect less active translation in mutant, as compared to wild-type, virus-infected cells. To investigate this possibility, we asked whether pUL69 induces a larger portion of viral mRNA molecules to become associated with polysomes within infected cells. Extracts were prepared at 72 hpi, their components were separated by centrifugation, and RNA was isolated from fractions 1 to 4, the polysome region, and fractions 5 to 8, the monosome region (Fig. 2A). Then, qRT-PCR assays were performed to determine the total amount of several different virus-coded RNAs in the two regions of the gradients and the relative portion of each RNA in the two fractions (Fig. 4A). An increase in the ratio of RNAs in the polysome versus monosome fractions reflects an increase in the efficiency of translation. When the amount of RNA present in the two regions was totaled, most RNAs were found to be present at similar levels after infection with the mutant or wild-type. The major exception to this generalization was UL123 RNA, which encodes the IE1 protein: 3-fold less UL123 RNA was present in wild-type than in mutant virus-infected samples. Mutant samples contained higher overall levels of IE1 RNA; however, it was less efficiently translated in the absence of pUL69, as demonstrated by its decreased association with polysomes. Several late RNAs, most notably those encoded by UL83 and UL99, also accumulated to somewhat higher levels in mutant virus-infected cells. Despite their increased levels, these virus-coded late RNAs were not as efficiently incorporated into polysomes during infection with the mutant virus, suggesting that their translation is decreased in the absence of pUL69. This prediction proved to be correct (Fig. 4B). Despite the fact that there was more total UL83 and UL99 RNA in pUL69-deficient virus-infected cells, pUL83 and pUL99 accumulated to higher levels in wild-type virus-infected cells. The enhanced accumulation of the late proteins after infection with the wild-type virus is consistent with the elevated portion of RNAs in polysome fractions of wild-type as compared to mutant virus-infected cells. We did not observe enhanced accumulation of IE1 protein after infection with wild-type as compared to mutant virus, but in this case the elevated levels of the RNA in mutant virus-infected cells likely obscured diminished translation, resulting from decreased polysome association.
Fig. 4.
UL69 effects supports translation of viral mRNAs. Cells were mock infected (M) or infected with TNwt (WT) or TNsubUL69 (UL69-). (A) At 72 hpi, extracts were fractionated as described in Fig. 2. Fractions containing monosome (fractions 5–8) or polysome-associated (fractions 1–4) RNAs were pooled and RNA extracted. Each sample was analyzed by qRT-PCR for UL123, TRS1, UL44, UL54, UL83, and UL99 RNA. The total (monosome + polysome fractions) amount of each indicated virus-coded RNA was determined relative to GAPDH RNA in WT versus UL69--infected cells (open bars), and the ratio of polysome-associated to monosome-associated RNA was calculated for each RNA in TNwt versus TNsubUL69-infected cells (solid bars). (B) At the indicated times, mock or infected cell extracts were assayed by Western blot with antibodies for the indicated proteins. α-tub, α-tubulin.
We conclude that pUL69 excludes 4EBP1 from the cap-binding complex, and this facilitates translation of at least some HCMV-coded mRNAs.
Discussion
Our study demonstrates that pUL69 interacts with elements of the mRNA cap-binding complex. A two-hybrid screen identified eIF4A1 and PABPC1 as putative partners that are directly contacted by the viral protein (Table 1). eIF4A1 is a constituent of the eIF4F cap-binding complex and PABPC1 binds directly to the complex (26). These predicted interactions were confirmed by coimmunoprecipitation analyses (Fig. 1). EIF4A1 and PABPC1 are both members of multigene families, and it is possible that pUL69 interacts with additional family members as well, but this was not tested. As anticipated for a protein that interacts with these factors, pUL69 sedimented with polysomes and antibody to pUL69 coprecipitated viral mRNAs (Fig. 2). The RNA association could result from the interaction of pUL69 with cap elements, its innate RNA-binding ability (17), or a combination of both. Importantly, E4BP1, which inhibits translation (19), was not captured from extracts of wild-type virus-infected cells with m7GTP Sepharose, but it was captured from extracts of cells infected with a pUL69-deficient virus (Fig. 3). Again, there are multiple 4EBP family members, and we monitored only 4EBP1. Consistent with the ability of pUL69 to block the interaction of 4EBP1 with other components of the cap-binding complex, a larger percentage of UL83 and UL99 RNA was associated with polysomes (Fig. 4A) and the two proteins accumulated to a higher level (Fig. 4B) after infection with wild-type virus, as compared to infection with a pUL69-deficient mutant. As yet, we do not know whether pUL69 affects translation globally or if it enhances translation of a subset of genes, as reported for pUL69 homologs encoded by HSV (21 –23) and Epstein-Barr virus (27).
Although we have not measured the affinity with which pUL69 interacts with eIF4A1 and PABPC1, the association might be fairly weak, because a small portion of the host factors were coprecipitated with the relatively abundant pUL69 (Fig. 1) and a small portion of the viral protein cosedimented with polysomes (Fig. 2A). However, in spite of this apparently weak interaction, pUL69 very effectively excluded 4EBP1from m7GTP Sepharose (Fig. 3A). A low affinity association could serve a timing function, delaying maximal exclusion of 4EBP1 from the cap-binding complex until the late stage of infection, when pUL69 and late viral mRNAs have accumulated to high levels, which would allow for more efficient translation of mRNAs made later in the infection cycle, many of which encode abundant virion proteins.
4EBP1 interacts with eIF4E and inhibits translation by interfering with the ability of eIF4E to bind to eIF4G within the m7G-cap-binding complex. The activity of 4EBP1 is regulated by phosphorylation. Hypophosphorylated 4EBP1 binds to eIF4E and blocks assembly of the cap-binding complex; mTORC1 phosphorylates 4EBP1, and the hyperphosphorylated factor doesn’t bind to eIF4E or inhibit translation (18). We have previously reported that HCMV-coded pUL38 maintains activity of mTORC1 within infected cells, so that the majority of 4EBP1 is hyperphosphorylated (28). However, a minor portion of 4EBP1 remains hypophosphorylated in infected cells (28), and this is presumably the 4EBP1 that is antagonized by pUL69. It is well established that 4EBP1 is not degraded after HCMV infection (24, 29), so it is likely that pUL69 blocks access of 4EBP1 to the m7G-cap complex through its contacts with eIF4A1 and PABPC1. Alternatively, the presence of pUL69 in the complex could make 4EBP1 more susceptible to phosphorylation, which would then cause its exclusion. Although primarily cytoplasmic proteins, a portion of 4EBP1 (30), eIF4E (31), PABPC1 (32, 33), and pUL69 (16) are present in the nucleus, so it is possible that pUL69 prevents association of 4EBP1 with cap-associated proteins in the nucleus, even before the mRNA is transported to the cytoplasm. An interaction of pUL69 with nuclear RNAs through PABPC1 could also contribute to its role in mRNA nucleo-cytoplasmic transport (12).
As noted above, the HSV pUL69 homolog, ICP27, has been shown to interact with PABPC1 and eIF4G (20); and, like pUL69 (12), ICP27 (13), and the Epstein-Barr pUL69 homolog, EB2 (see ref. 27 and references therein) regulate the export of mRNA from the nucleus and stimulate translation. These similarities raise the possibility that ICP27, EB2, and other members of this pan-herpesvirus protein family also prevent 4EBP1 from associating with the m7G-cap-binding complex.
Does pUL69 block cell cycle progression in late G1 (7, 8) as a consequence of its ability to regulate translation? Translational effects could alter the levels of cell cycle regulatory factors within the infected cell, but we have not yet tested this possibility. It is intriguing to note, however, that the two-hybrid screen generated a novel hypothesis for how pUL69 might inhibit cell cycle progression. The IFN-inducible, MX2-coded MxB protein was identified twice in the screen. There are two human Mx genes, MX1 and MX2, encoding the MxA and MxB proteins, respectively. Both proteins are IFN-inducible dynamin-like GTPases; and the MxA protein is known to inhibit numerous RNA viruses, whereas MxB has not been shown to have antiviral activity (34). We have reported previously that MxB RNA is present in uninfected fibroblasts and its levels increase after infection (35). Dominant-negative MxB mutants and RNAi knockdown of MxB cause a delay in G1 to S cell cycle progression (36). Conceivably, then, pUL69 inhibits cell cycle progression by interfering with the activity of MxB protein.
Materials and Methods
Human Cells, Viruses, and Antibodies.
Human MRC-5 fibroblasts were propagated in medium containing 10% FCS. A plaque-purified derivative of the HCMV Towne strain (37), TNwt, was propagated in MRC-5 fibroblasts, and a pUL69-deficient mutant, TNsubUL69, was grown by complementation in MRC-5 cells expressing pUL69 (8). The pfu/particle ratio for TNsubUL69 was measured as described (38), and found to be ∼10-fold higher than for its wild-type parent (data not shown). Consequently, cells were infected at a multiplicity of 3 pfu per cell with wild-type and an equivalent number of mutant virus particles in all experiments. Under these infection conditions, the two viruses produced equal numbers of cells expressing the virus-coded IE1 immediate-early protein and pUL83 late protein at 24 hpi (data not shown). Antibodies to the following proteins were used: 4EBP1 (9452) and eIF4E (9742) from Cell Signaling Technologies; eIF4G (610536) from BD Biosciences; eIF4A1 (sc-14211) and PABPC1 (sc-32318) from Santa Cruz Biotechnology; GAPDH (G8795) and α-tubulin (T9026) from Sigma-Aldrich; IE1 (39); pUL37 × 1 (40) pUL69 (7); pUL44 (CA006, Virusys); pUL83 (41); and pUL99 (38).
Yeast Two-Hybrid Assay.
The two-hybrid assay (42, 43) was performed using GAL4 fusion proteins and Saccharomyces cerevisiae strain AH109 (Matchmaker Gal4 Yeast Two-Hybrid System, Clontech). The UL69 ORF was cloned in frame with the Gal4 DNA-binding domain in the bait plasmid pGBKT7 to produce pMG1, and its sequence confirmed. Correct expression of the fusion protein within yeast was verified by Western blot assay; and the bait plasmid alone failed to activate the production of α-galactosidase, whose expression is controlled by a Gal4-responsive promoter. A cDNA library (ZAP cDNA Library Construction Kit, Stratagene) was produced from equal parts of polyadenylated RNA isolated from human fibroblasts at 6, 24, and 72 h after infection with HCMV at a multiplicity of 3 pfu per cell; it was cloned into the prey plasmid pGADT7, containing the Gal4 activation domain, to generate a library termed pGADT7cDNA.
To identify putative interactors, cells were cotransformed with pMG1 and the cDNA library, and the cultures were selected for Gal4 activity by requiring simultaneous activation of four Gal4-responsive genes. Fifty-three clones were identified in the screen, and sequence analysis demonstrated that 13 either lacked an insert or contained an unknown DNA segment. The remaining 40 clones were assayed for α-galactosidase expression levels in cells containing or lacking the pMG1 bait plasmid. For classification as a putative interaction, the required ratio of expression in the presence versus absence of bait was arbitrarily set at ≥3.
Polysome Isolation.
At various times after infection with TNwt, MRC-5 cells were maintained in medium containing or lacking 0.1 mM cycloheximide for 10 min at 37 °C. After washing, cells were pelleted by centrifugation, resuspended in ice-cold lysis buffer [1.5 mM MgCl2; 15 mM Tris, pH 7.5; 1.5 mM MgCl2, 150 mM NaCl; 10 mM DTT; 1% Triton X; protease inhibitor mixture (Roche Applied Science); 50 U/mL RNasin (Promega); 0.1 mM cycloheximide or 50 mM EDTA], incubated on ice for 10 min, lysed by using a homogenizer, and then nuclei and insoluble material were pelleted by centrifugation. Supernatants were loaded onto a 10 to 50% sucrose gradient containing 1.5 mM MgCl2, 15 mM Tris (pH 7.5), 1.5 mM MgCl2, 150 mM NaCl and subjected to centrifugation in an SW41 Ti ultracentrifuge rotor (part number 331362, Beckman Coulter, Brea, CA) for 2 h at 250,000 × g. Gradients were fractionated and mRNA was isolated with TRIzol reagent (Invitrogen). For protein analysis, 20% TCA was added to each fraction and incubated on ice for 15 min, precipitated proteins were pelleted by centrifugation and rinsed with ice-cold acetone twice, and then proteins were dissolved in alkaline sample buffer (50 mM Tris, pH 8.0; 2% SDS; 100 mM DTT; 10% glycerol).
m7GTP Sepharose Chromatography.
Chromatography was as described previously (44): 5 × 106 infected fibroblasts were dissolved in 1-mL lysis buffer [50 mM hepes, pH 7.4; 150 mM NaCl; 2mM EDTA; 2 mM Na3VO4; 25 mM glycerophosphate; mini protease inhibitor mixture (Roche Applied Science); 0.5% Nonidet P-40], the lysate was precleared for 20 min at 4 °C with 30 μL (settled bed volume) Sepharose 4B, and then incubated for 1 h at 4°C with 50 μL (settled bed volume) m7GTP Sepharose 4B (GE Healthcare). Beads were washed four times with lysis buffer, resuspended in SDS sample buffer, boiled for 5 min, and insoluble debris was removed by centrifugation before Western blot analysis.
Analysis of RNA and Proteins.
To quantify viral RNA levels by real-time RT-PCR, qRT-PCR, cDNAs were synthesized from RNAs treated with TURBO DNase (Ambion) by using TaqMan reverse-transcription reagents and random hexamers (Applied Biosystems). The amplification reaction was performed with SYBR green PCR master mix (Applied Biosystems) and primers specific to UL123 (5′-GCCTTCCCTAAGACCACCAAT-3′ and 5′-ATTTTCTGGGCATAAGCCATAATC-3′); UL44 (5′-TACAACAGCGTGTCGTGCTCCG-3′ and 5′-GGCGTGAAAAACATGCGTATCAAC-3′), UL54 (5′-CCCTCGGCTTCTCACAACAAT-3′ and 5′-CGAGTTAGTCTTGGCCATGCAT-3′), UL82 (5′-TGGACCTCGGTACCAACATA-3′ and ACCGAGACCGTGTTGTTTTC-3′), UL83 (5′-GGGACACAACACCGTAAAGCCG-3′ and 5′-CGTGGAAGAGGACCTGACGATGAC-3′), UL99 (5′-GTGTCCCATTCCCGACTCG-3′ and 5′-TTCACAACGTCCACCCACC-3′), TRS1 (5′-ACAGAACCACCGTTGACTCC-3′ and 5′-AACACCGTTTTCTTCCAACG-3′) or glyce-raldehyde-3-phosphate dehydrogenase (GAPDH) (5′-CTGTTGCTGTAGCCAAATTC-GT-3′ and 5′-ACCCACTCCTCCACCTTTGAC-3′).
Immunofluorescent analysis (40) and immunoprecipitations (45) were performed as described previously. For Western blot analysis, proteins were transferred from polyacrylamide gel to polyvinylidene difluoride membranes, and membranes were blocked with 5% nonfat milk in Tris-buffered saline containing 0.05% Tween 20. Proteins were labeled with primary antibodies and then incubated with a secondary goat anti-mouse antibody, which was visualized by using the ECL detection reagent (GE Healthcare).
Acknowledgments
This study was supported by National Institutes of Health Grant CA85786.
Footnotes
The authors declare no conflict of interest.
References
- 1.Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Lischka P, Thomas M, Toth Z, Mueller R, Stamminger T. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J Gen Virol. 2007;88:405–410. doi: 10.1099/vir.0.82480-0. [DOI] [PubMed] [Google Scholar]
- 3.Winkler M, Stamminger T. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J Virol. 1996;70:8984–8987. doi: 10.1128/jvi.70.12.8984-8987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winkler M, Rice SA, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler M, aus Dem Siepen T, Stamminger T. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J Virol. 2000;74:8053–8064. doi: 10.1128/jvi.74.17.8053-8064.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rechter S, et al. Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 and modulate its nuclear localization and activity. J Biol Chem. 2009;284:8605–8613. doi: 10.1074/jbc.M805693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi ML, Blankenship C, Shenk T. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc Natl Acad Sci USA. 2000;97:2692–2696. doi: 10.1073/pnas.050587597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas M, et al. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J Gen Virol. 2009;90:567–578. doi: 10.1099/vir.0.005827-0. [DOI] [PubMed] [Google Scholar]
- 10.Hume AJ, et al. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science. 2008;320:797–799. doi: 10.1126/science.1152095. [DOI] [PubMed] [Google Scholar]
- 11.Prichard MN, et al. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J Virol. 2008;82:5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol. 2006;26:1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandri-Goldin RM. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci. 2008;13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- 14.Batisse J, Manet E, Middeldorp J, Sergeant A, Gruffat H. Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J Virol. 2005;79:14102–14111. doi: 10.1128/JVI.79.22.14102-14111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J Biol Chem. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 16.Lischka P, Rosorius O, Trommer E, Stamminger T. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 2001;20:7271–7283. doi: 10.1093/emboj/20.24.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toth Z, Lischka P, Stamminger T. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 2006;34:1237–1249. doi: 10.1093/nar/gkl007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 19.Jackson RJ, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr Opin Genet Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 20.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology. 2004;330:487–492. doi: 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Ellison KS, Maranchuk RA, Mottet KL, Smiley JR. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J Virol. 2005;79:4120–4131. doi: 10.1128/JVI.79.7.4120-4131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontaine-Rodriguez EC, Knipe DM. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol. 2008;82:3538–3545. doi: 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larralde O, et al. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J Virol. 2006;80:1588–1591. doi: 10.1128/JVI.80.3.1588-1591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh D, Perez C, Notary J, Mohr I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol. 2005;79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burny A, Huez G, Marbaix G, Chantrenne H. On a messenger ribonucleoprotein complex from rabbit reticulocytes. Biochim Biophys Acta. 1969;190:228–231. doi: 10.1016/0005-2787(69)90176-2. [DOI] [PubMed] [Google Scholar]
- 26.Rhoads RE. eIF4E: new family members, new binding partners, new roles. J Biol Chem. 2009;284:16711–16715. doi: 10.1074/jbc.R900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricci EP, et al. Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res. 2009;37:4932–4943. doi: 10.1093/nar/gkp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moorman NJ, et al. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78:11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong L, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA. 2008;14:1318–1327. doi: 10.1261/rna.950608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–247. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 33.Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haller O, Staeheli P, Kochs G. Interferon-induced Mx proteins in antiviral host defense. Biochimie. 2007;89:812–818. doi: 10.1016/j.biochi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King MC, Raposo G, Lemmon MA. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc Natl Acad Sci USA. 2004;101:8957–8962. doi: 10.1073/pnas.0403167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plotkin SA, Furukawa T, Zygraich N, Huygelen C. Candidate cytomegalovirus strain for human vaccination. Infect Immun. 1975;12:521–527. doi: 10.1128/iai.12.3.521-527.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharon-Friling R, Goodhouse J, Colberg-Poley AM, Shenk T. Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci USA. 2006;103:19117–19122. doi: 10.1073/pnas.0609353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowak B, et al. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology. 1984;132:325–338. doi: 10.1016/0042-6822(84)90039-4. [DOI] [PubMed] [Google Scholar]
- 42.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 43.Luban J, Goff SP. The yeast two-hybrid system for studying protein-protein interactions. Curr Opin Biotechnol. 1995;6:59–64. doi: 10.1016/0958-1669(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 44.Walsh D, Mohr I. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 2004;18:660–672. doi: 10.1101/gad.1185304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]