Seeing is believing. This old saying sums up one of the main goals in biology and biophysics: to watch and ultimately understand the molecular processes that sustain life. Thanks to advances in both experimentation and simulation, we are rapidly gaining a molecular-level understanding of key biomolecular processes. Nevertheless, experiments remain largely limited to the identification and characterization of intermediates along a reaction path, requiring significant populations and sufficiently long lifetimes to be detectable. Ultimately, a full understanding of the molecular mechanisms will also require the characterization of the transitions between the intermediates. One might even argue that the molecular mechanism is contained in the transition state separating the intermediates. But despite much recent progress [e.g., with an upper bound determined for the transition time in protein folding from fluorescence spectroscopy (1)], our mechanistic understanding of biomolecular transitions is derived primarily from indirect measurements, such as phi-value analysis (2), leaving the transitions themselves largely in the dark. In this issue of the PNAS, Vreede et al. (3) show how modern molecular simulation techniques can be used to shed light on molecular transitions, even those occurring on a millisecond timescale far beyond that of standard molecular dynamics calculations.
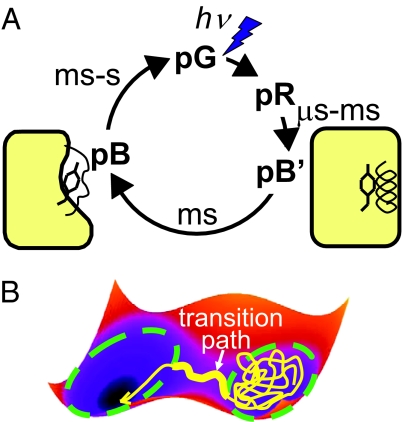
Vreede et al. (3) explore a key step in the sequence of light-induced conformational changes of photoactive yellow protein (PYP). PYP is a light receptor thought to be used by the bacterium Halorhodospira halophila to detect and ultimately avoid blue light. Packed into the hydrophobic core of PYP is a covalently attached chromophore that undergoes a trans-to-cis isomerization upon absorption of light (Fig. 1A). As the first major intermediate, the pR state forms on a nanosecond timescale. In pR, the tight packing of the chromophore with its aromatic ring limits the extent of the initial conformational changes in response to photoabsorption (7). On a timescale of ≈500 μs (4), a proton is internally transferred from Glu-46 to the chromophore to form the pB′ intermediate. After a few milliseconds (4), the mechanical stress built up by the trans-cis isomerization is relieved, producing dramatic changes of the protein structure in the resulting pB intermediate, including partial unfolding (8). This transition from pB′ to pB is the focus of the simulations by Vreede et al. (3).
Fig. 1.
(A) Simplified photocycle of PYP, adapted from ref. 4. (B) Transition path sampling (5, 6). The trajectory (yellow) on the simplified energy surface spends most of the time “waiting” in the reactant state. The transition path is the short trajectory segment (thick yellow line) connecting the reactant and product states, whose boundaries are indicated by dashed lines.
How can Vreede et al. (3) use molecular simulations, each covering only a few nanoseconds, to study a process that occurs on a timescale of milliseconds? The trick is the realization that during this millisecond time, each individual PYP protein is primarily just waiting around for a series of random thermal kicks that convert the pB′ state to pB. But the actual transition of an individual PYP molecule from pB′ to pB can be many orders of magnitude faster than the rate of interconversion (1) (Fig. 1B). The goal then becomes to sample only the relatively fast transition paths that connect the reactant and product states, avoiding the long waiting times in the reactant state pB′.
An ensemble of such reactive trajectories can be created efficiently by transition-path sampling (5, 6). In molecular dynamics simulations, the interactions of the molecular system, including the solvent, are represented by an energy function that includes terms for bonding, electrostatics, and van der Waals interactions. The system moves on this energy surface according to classical Newtonian dynamics. In transition-path sampling (5, 6), reactive trajectories are created with exactly the same distribution as in very long equilibrium simulations. Configuration space is divided into reactant and product states, separated by a transition region (Fig. 1B). Transition paths are the trajectory segments that exit from one state, pass through the transition region, and directly reach the other state. Importance sampling techniques are used to harvest these rare reactive trajectories. In a particularly efficient Monte Carlo move, trajectories are shot forward and backward (with reversed velocities) from a randomly selected branch point along an existing transition path. If one of the trajectory segments reaches the reactant state and the other the product state, the combined trajectory is accepted.
With this powerful transition-path sampling scheme, Vreede et al. (3) achieved efficiency gains of nearly six orders of magnitude, based on the experimental rates. Their ensemble of trajectories consists only of nanosecond transition paths and leaves out the millisecond waiting times in the metastable pB′ initial state. Along the transition from pB′ to pB, the simulations showed a series of additional intermediates. In a first step, the helix α3 unfolded partially, followed by its complete unfolding. This local loss of structure in the long-lived signaling state pB is consistent with a series of experiments (9), including NMR (8). However, the extent of the structural disorder seems to depend on the environment (9), being largely suppressed in protein crystals (10, 11). In a bifurcating path, the simulations showed that helix unfolding was followed by the exposure of either Glu-46 or the chromophore to solvent (3). Interestingly, only the first branch was productive in the sense that the system reached the pB signaling state with α3 partially unfolded and both Glu-46 and the chromophore exposed to solvent; along the other branch, the system remained stuck in a metastable state on the simulation timescale.
Sometimes, seeing is not enough. The transition paths provide a “movie” of the PYP conformational change. However, even with this movie, the transition can appear too complex to be easily understood. Recent theoretical developments (12 –14) provide a framework for organizing the information contained in the transition paths and for constructing quantitative descriptions of the reaction mechanism. To measure the progress of a reaction, one typically uses a reaction coordinate. However, finding a good reaction coordinate for a protein conformational change poses a formidable challenge. In essence, one would like to define a function r(x) of the high-dimensional configuration space of the protein [and possibly also the solvent (15)] that condenses all transition states onto a hypersurface r ‡ = r(x). With function space being so enormous (a well-known problem in quantum chemistry), the construction of reaction coordinates relies largely on physical intuition. Nevertheless, significant progress has been made by optimizing trial coordinates in a reduced function space (16 –18). For example, Vreede et al. (3) find that a linear combination of helix root–mean-square distance, a backbone hydrogen bond distance, and an amino acid pair distance capture the transition state for the initial unfolding of helix α3. At the bifurcation in the pathway, the deciding factor for following the productive branch was the breaking of a salt bridge between Asp-20 and Lys-55. These results illustrate how path sampling can be used to extract biomolecular reaction mechanisms in unprecedented detail.
But should we trust the simulations? Molecular dynamics simulations have already provided important insights into the early events of PYP photoactivation (19, 20). Quite generally, the quality of simulation force fields has much improved. Nevertheless, many challenges remain, and the simulations of Vreede et al. (3) will have to be validated against experiments. Whereas some detailed components of the reaction coordinates (e.g., a specific hydrogen bond) may not be easily probed, the importance of the salt bridge between Asp-20 and Lys-55 in steering the protein onto different pathways during the pB′-to-pB transition could be tested in mutation experiments. This interplay between simulation, theory, and experimentation promises to elucidate not only the mechanistic aspects of PYP function, but also biomolecular processes in general.
Acknowledgments
G.H. is supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The author declares no conflict of interest.
See companion article on page 2397.
References
- 1.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc Natl Acad Sci USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. 1. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 3.Vreede J, Juraszek J, Bolhuis PG. Predicting the reaction coordinates of millisecond light-induced conformational changes in photoactive yellow protein. Proc Natl Acad Sci USA. 2010;107:2397–2402. doi: 10.1073/pnas.0908754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan DH, Philip A, Hoff WD, Mathies RA. Time-resolved resonance Raman structural studies of the pB intermediate in the photocycle of photoactive yellow protein. Biophys J. 2004;86:2374–2382. doi: 10.1016/S0006-3495(04)74294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellago C, Bolhuis PG, Csajka FS, Chandler D. Transition path sampling and the calculation of rate constants. J Chem Phys. 1998;108:1964–1977. [Google Scholar]
- 6.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: Throwing ropes over rough mountain passes in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 7.Genick UK, Soltis SM, Kuhn P, Canestrelli IL, Getzoff ED. Structure at 0.85 Å resolution of an early protein photocycle intermediate. Nature. 1998;392:206–209. doi: 10.1038/32462. [DOI] [PubMed] [Google Scholar]
- 8.Rubinstenn G, et al. Structural and dynamic changes of photoactive yellow protein during its photocycle in solution. Nat Struct Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 9.Hellingwerf KJ, Hendriks J, Gensch T. Photoactive yellow protein, a new type of photoreceptor protein: Will this yellow lab bring us where we want to go? J Phys Chem A. 2003;107:1082–1094. [Google Scholar]
- 10.Genick UK, et al. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 11.Ihee H, et al. Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds. Proc Natl Acad Sci USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hummer G. From transition paths to transition states and rate coefficients. J Chem Phys. 2004;120:516–523. doi: 10.1063/1.1630572. [DOI] [PubMed] [Google Scholar]
- 13.Berezhkovskii A, Szabo A. One-dimensional reaction coordinates for diffusive activated rate processes in many dimensions. J Chem Phys. 2005;122:014503. doi: 10.1063/1.1818091. [DOI] [PubMed] [Google Scholar]
- 14.Weinan E, Vanden-Eijnden E. Towards a theory of transition paths. J Stat Phys. 2006;123:503–523. [Google Scholar]
- 15.Miller TF, Vanden-Eijnden E, Chandler D. Solvent coarse-graining and the string method applied to the hydrophobic collapse of a hydrated chain. Proc Natl Acad Sci USA. 2007;104:14559–14564. doi: 10.1073/pnas.0705830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Best RB, Hummer G. Reaction coordinates and rates from transition paths. Proc Natl Acad Sci USA. 2005;102:6732–6737. doi: 10.1073/pnas.0408098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma A, Dinner AR. Automatic method for identifying reaction coordinates in complex systems. J Phys Chem B. 2005;109:6769–6779. doi: 10.1021/jp045546c. [DOI] [PubMed] [Google Scholar]
- 18.Peters B, Beckham GT, Trout BL. Extensions to the likelihood maximization approach for finding reaction coordinates. J Chem Phys. 2007;127:034109. doi: 10.1063/1.2748396. [DOI] [PubMed] [Google Scholar]
- 19.Groenhof G, et al. Photoactivation of the photoactive yellow protein: Why photon absorption triggers a trans-to-cis isomerization of the chromophore in the protein. J Am Chem Soc. 2004;126:4228–4233. doi: 10.1021/ja039557f. [DOI] [PubMed] [Google Scholar]
- 20.Lee IR, Lee W, Zewail AH. Primary steps of the photoactive yellow protein: Isolated chromophore dynamics and protein directed function. Proc Natl Acad Sci USA. 2006;103:258–262. doi: 10.1073/pnas.0510015103. [DOI] [PMC free article] [PubMed] [Google Scholar]