Abstract
The identification and characterization of tumor-associated antigens (TAAs) and their use in antigen mini-arrays for cancer immunodiagnosis has been of interest recently as an approach to cancer detection. In this study, autoantibodies in sera from a patient with HCC were used as probes to immunoscreen a HepG2 cDNA expression library for the identification of TAAs involved in malignant liver transformation. Recombinant proteins from two genes identified in this manner, Sui1 and RalA were expressed, purified and used as antigens in immunoassays to detect the presence of antibodies in sera from 77 patients with HCC, 30 with chronic hepatitis (CH), 30 with liver cirrhosis (LC) and 82 normal human sera (NHS). The prevalence of antibody to Sui1 and RalA in HCC were 11.7% (9/77) and 19.5% (15/77), respectively, which were significantly higher than prevalence in liver cirrhosis (3.3% and 3.3%), chronic hepatitis (0% and 0%) and normal human sera (0% and 0%). When Sui1 and RalA were added to a panel of eight other TAAs used in a previous study, the final cumulative prevalence of anti-TAA antibodies in HCC to the 10 TAA array was raised to 66.2% (51/77). The specificity for HCC compared with LC, CH and NHS, was 66.7%, 80.0%, and 87.8%, respectively. When anti-TAA was added to abnormal serum AFP as combined diagnostic markers, it raised the diagnostic sensitivity from 66.2% to 88.7%. AFP and anti-TAA were independent markers and the simultaneous use of these two markers significantly resulted in the increased sensitivity of HCC detection.
Keywords: Autoantibodies, Tumor-associated antigens (TAAs), Alpha-fetoprotein (AFP), Immunodiagnostic markers, Hepatocellular carcinoma
1. Introduction
Autoimmune responses have been frequently observed in patients with malignancies and have been postulated to be driven by tumor-associated antigens (TAAs) which might be involved in cellular functions related to tumorigenesis [1]. The identification of appropriate panels of tumor antigens which elicit humoral immune responses may have utility in cancer screening, diagnosis, determining recurrence as well as monitoring prognosis. Such antigens may also have utility in preparation of tumor antigen vaccines in immunotherapy against cancers as well as helping to define factors involved in the multiple stages of tumorigenesis and in drug design strategy. In recent years, the molecular cloning of tumor antigens recognized by autoantibodies has opened a new era in tumor immunology and the list of defined immunogenic human tumor antigens is growing rapidly. Interpreting the specificity of an observed humoral or cellular immune response to tumor antigens has become a critical issue in tumor immunology [1,2].
Hepatocellular carcinoma (HCC) is a malignancy with very poor prognosis. The majority of people with HCC will die within one year of its detection. The high case-fatality rate can in part be attributed to the lack of sensitive and specific diagnostic methods for early detection. A feature of HCC is that antecedent liver cirrhosis and chronic hepatitis are common precursor conditions and during transition to malignancy some patients develop autoantibodies which were not present during the preceding chronic liver disease phase [3–7]. The immune system appears to have the ability to recognize malignancy and respond to cellular factors related to the transformation process. The basis of this notion is that autoantibody changes during progression from chronic liver disease to HCC could be related to aberrant cellular mechanisms stimulating immune responses, and therefore autoantibodies can be used as probes to identify cell proteins or other agents which are involved in the transformation process. The identification and characterization of HCC-associated TAAs and their autoantibodies provide a way to find potential markers for early detection of HCC and targets for immunotherapy of HCC.
A major issue in the field of cancer immunodiagnosis is the definition of what constitutes a TAA. It is erroneous to include all cellular antigens identified by autoantibodies in cancer sera as TAAs since some autoantibodies may exist in conditions that pre-date malignancy. This was particularly evident in several studies of subjects with HCC where serial serum samples were available several years before malignancy when these subjects had conditions such as chronic hepatitis and liver cirrhosis [3–7]. Autoantibodies to cellular components were readily detected by Western Blotting during the pre-malignant conditions of chronic hepatitis and liver cirrhosis but the interesting phenomenon was that coincident with or closely preceding the clinical detection of HCC, novel autoantibodies were detected in some patients by Western blotting and immunofluorescence imaging [3–7]. In cases where the novel antigen–antibody systems were characterized, many antigens turned out to be cellular components which have been described to be aberrantly expressed in cancer [7–10]. Failing to recognize the likelihood of pre-malignancy circulating antibodies would result in the inclusion of many antigens erroneously as TAAs, especially if serum at a single time point from a cancer subject was used to characterize the antigens since this might include both cancer-related and unrelated antigens. Some studies have reported more than 2000 tumor-related antigens which were identified with sera from cancer patients [11,12]. It would appear that further studies might be needed to establish the true tumor-related antigens and to exclude those that might have been present prior to malignancy. An approach which is used in this study is the confirmation of tumor-relatedness of the antigens by testing against non-cancer sera which in the case of HCC include chronic hepatitis and liver cirrhosis.
In the present study, autoantibodies in HCC serum were used as probes to immunoscreen a HepG2 cDNA expression library and a group of antigens was expressed from the cDNA clones and analyzed to select those which might be tumor-related. Two antigens were further evaluated by inclusion into a TAA mini-array employed in a previous study [13]. The results indicated that both the sensitivity and specificity of TAA panels were enhanced. Combined use of anti-TAAs and elevated AFP further increased the diagnostic sensitivity for HCC.
2. Materials and methods
2.1. Sera and antibodies
Sera from 77 patients with HCC, 30 patients with chronic hepatitis (CH), 30 patients with liver cirrhosis (LC), and 82 normal human sera (NHS) were obtained from the serum bank of the Tumor Cell Engineering Laboratory of Xia'men University (Fujian Province, PR China). All HCC patients were diagnosed according to the criteria described [14]. Of 77 HCC patients, 73 (94.8%) were histologically confirmed. Fifty-seven (74.0%) were males, and 20 (26.0%) were females. Mean age was 58.0 ± 12.8 years (range, 24–78 years). Fifty-eight (75.3%) patients were HBV positive, 8 (10.4%) patients were HCV positive, and 4 (5.2%) patients were positive for both HBV and HCV. Fifty-two (67.5%) patients had a previous history of chronic hepatitis, 13 (16.9%) patients with liver cirrhosis, and 12 (15.6%) patients had no previous history of either chronic hepatitis or liver cirrhosis. In the present study, 62 sera were available for AFP testing. The AFP test kit was provided by Gen-Way Biotech (San Diego, CA). The results showed that 38 of 62 (61.3%) had abnormal levels (>100 ng/ml) whereas 24 (38.7%) had normal levels (<100 ng/ml). Based on guidelines for liver cancer, out of 71 HCC patients with available data on clinical stages, 25 (35.2%) patients were in clinical stage I, 16 (22.5%) in stage II, 22 (31.0%) in stage III, 8 (11.3%) in stage IV. All HCC sera were collected at the time of initial cancer diagnosis, when the patients had not yet received chemotherapy or radiation therapy. Patients with CH and LC were followed for at least 18 months after the date of blood collection to exclude individuals with primary biliary cirrhosis and asymptomatic or clinically undetectable HCC. Normal human sera were collected during annual health examinations of people from the same locality who had no obvious evidence of malignancy. Due to regulations concerning studies of human subjects, the subjects' name and identification number were blinded to investigators, and some clinical information for sera used in this study was not currently available. The HCC serum which was used to immunoscreen the cDNA expression library was obtained from William Beaumont Medical Center in El Paso, Texas. This study was approved by the Institutional Review Board of the University of Texas at El Paso (IRB-UTEP) and collaborating institutions. Monoclonal mouse anti-RalA was commercially available (BD Biosciences, San Diego, CA). Anti-Sui1 was raised by immunization of rabbit with full length recombinant Sui1 protein (ProSci incorporated, Poway, CA).
2.2. Immunoscreening HepG2 cDNA expression library
The HepG2 cDNA expression library constructed in the Uni-ZAP XR vector system was purchased from Stratagene Inc., La Jolla, CA. The HCC serum was diluted 1:200 and used as a probe for initial immunoscreening of the HepG2 cDNA library as previously described [9,10]. Before screening, the HCC serum was extensively adsorbed against wild-type Uni-ZAP XR phage infected Escherichia coli cell lysates to reduce nonspecific antibody binding. The absorbed serum was used to immunoscreen 4.0 × 105 recombinant plaques. All screenings were performed on duplicate isopropyl β-d-thiogalactoside (IPTG) pre-impregnated nitrocellulose filters. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Caltag, Burlingame, CA) at 1:3000 dilution was used as secondary antibody and immunoreactive clones were detected by autoradiography using chemiluminescence (Pierce Biotechnology, Rockford, IL). Double positive phage clones in the first screening were subsequently purified to 100% purity by further screening. cDNAs from positive clones were isolated and converted to pBluescript phagemid by in vivo excision using the ExAssist helper phage (Stratagene Inc., La Jolla, CA) with E. Coli SOLR strain as recommended in the manufacture's instructions. The phagemids were purified, amplified and used for sequence analysis. All cDNA inserts were analyzed by restriction mapping and sequencing.
2.3. cDNA sequencing and sequence analysis
The cDNA inserts of the pBluescript phagemid were sequenced with T3 and T7 primers by the dideoxy chain termination method using SequiTherm EXCEL™ II DNA Sequencing Kit (Epicentre Technologies, Madison, WI, USA). DNA sequences were read by e-Seq™ DNA sequencing software (Li-COR Biosciences, Lincoin, Nebraska USA). All cDNA sequences were analyzed by BLAST search with known sequence databases.
2.4. PCR and subcloning of full-length cDNAs from EP (El Paso)-HCC-1 (Sui1) and EP-HCC-13 (RalA)
As shown in Table 1, cDNAs from two identified genes EP-HCC-1 (Sui1) and EP-HCC-13 (RalA), which were obtained from human expressed sequence tag (EST) database (Genbank Accession numbers: CX163967, BM560822), were used as template in PCR. One pair of forward and reverse primers was designed for each gene of EP-HCC-1 (Sui1) and EP-HCC-13 (RalA). For EP-HCC-1 (Sui1): Sui1-F, 5′-TTGGATCCATGTCCGCTATCCAGAAC-3′ and Sui1-R, 5′-TTCTCGAGCACTTAAGCTTCAGTGAGC-3′. For EP-HCC-13 (RalA): RalA-F, 5′-TTGGATCCATGGCTGCAAATAAGCCCAAG-3′ and RalA-R, 5′-TTCTCGAGAAGAAAGGAGTTTGGGCTTTG-3′. The primers were incorporated with BamHI and XhoI sites. Each PCR reaction was performed in a total volume of 50 μl including 2 μl plasmid cDNA (0.2–0.3 μg/μl), 80 pmol primers (4 μl each), 5U Taq polymerase (Qiagen, Valencia, CA). 1 μl of 10 μm dNTPs (Qiagen, Valencia, CA), 5 μl 10× PCR buffer (500 mM KCL; 100 Mm Tris–HCl, Ph 8.3; and 0.1% gelatin), and 4 μl of 25 mM Mgcl2 (Promega Biotec, Madison, WI, USA). PCR was programmed using a thermocycler (Eppendorf, Westbury, NY). The reaction was performed at 95 °C for 3 min, followed by 35 cycles at 95 °C for 1 min, 59 °C for 1 min, 72 °C for 1 min, and a final incubation at 72 °C for 10 min. PCR products were analyzed by 1% agarose gel electrophoresis.
Table 1.
Nine cancer-related genes identified from HepG2 cDNA library screening.
| Designation | Insert size (kb) | Identity (access no.) | cDNA size (kb) | Functional significance and cancer association |
|---|---|---|---|---|
| EP-HCC-1 | 0.65 | Translation initiation factor, hu-sui1 (BC008710) | 1.32 | Associated with apoptosis; related to HCC [18,19] |
| EP-HCC-5 | 0.88 | Homo sapiens interferon gamma receptor 2 (BC003264) | 2.77 | Associated with the inhibition of tumor cell growth [20] |
| EP-HCC-6 | 0.96 | Homo sapiens ferritin (BC016354) | 0.90 | Regulator of cellular differentiation. Associated with increased risk of liver and breast cancers [21,22] |
| EP-HCC-7 | 0.68 | Serine proteinase inhibitor; pigment epithelium derived factor, PEDF (BC000522) | 1.45 | Associated with apoptosis; plays a role in the antiangiogenic properties of HCC [23,24] |
| EP-HCC-8 | 0.89 | Antitrypsin-alpha 1-AT (NM_001002235) | 1.59 | Serine proteinase inhibitor, Associated with increased risk of cirrhosis and primary liver cancer [25] |
| EP-HCC-9 | 0.76 | TAP2, a transporter associated with antigen processing (AB073779) | 2.11 | Plays a key role in the adaptive immune response; TAP2 functions as a tumor suppressor gene; related to HCC [26] |
| EP-HCC-10 | 0.56 | Homo sapiens upstream transcription factor 2-USF2 (BC042655) | 2.80 | Antiproliferative activities and regulation of several genes involved in tumorigenesis [27] |
| EP-HCC-12 | 0.89 | Homo sapiens fibronectin 1-FN1 (BC016875) | 1.51 | Associated with cell proliferation and apoptosis, have a role in the pathogenesis of papillary thyroid carcinoma [28,29] |
| EP-HCC-13 | 0.79 | v-Ral simian leukemia viral oncogene homolog A (Ras related)-RalA (BC039858) | 2.77 | Oncogene; it may play a role in cell motility and tumor metastasis [30–33] |
2.5. Expression and purification of EP-HCC-1 (Sui1) and EP-HCC-13 (RalA) recombinant proteins
For the expression and purification of recombinant protein, the full-length cDNAs of Sui1 and RalA obtained from PCR were subcloned into the pET28 expression vector which is designed to produce a fusion protein with N-terminal 6× histidine and T7 epitope tags. Recombinant proteins were further expressed in E. coli BL21 (DE3) and purified using nickel column chromatography. The protocol used for high-level expression and purification of 6× His-tagged proteins were performed as described (QIAGEN Inc., Valencia, CA). Elution buffer (8 M urea, 0.1 M NaH2-PO4, 0.01 M Tris, pH4.5) was used to elute the recombinant protein. The purified recombinant proteins were further analyzed by electrophoresis on SDS–PAGE and confirmed by Western Blot using anti-Sui1 and anti-RalA antibodies as well as the HCC serum antibody which was used for immunoscreening.
2.6. Enzyme-linked immunosorbent assay (ELISA)
Purified recombinant proteins were diluted in phosphate-buffered saline (PBS) to a final concentration of 0.5 μg/ml and coated onto a 96 well microtiter plate (Dynatech Laboratories, Alexandria, VA). Human sera diluted 1:200 were incubated in the antigen-coated wells. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Caltag, Burlingame, CA) and the substrate 2, 2′-azinobis (3-ethylbenzthiazoline-6-sulfonic acid) (Boehringer Manheim GmbH, Mannheim, Germany) were used as detecting reagents. Each serum sample was tested at least two times, and the average OD value at 405 nm was used for data analysis. The cutoff value designating positive reaction was the mean OD of 82 normal human sera (NHS) + 3SD. Each run of ELISA included 8 NHS selected to represent a range of absorbance above and below the mean of 82 normal human sera, and whose OD average is close to the mean OD of 82 normal human sera. The inclusion of these normal controls is to normalize all absorbance values in other runs of ELISA. All positive sera were further confirmed by Western blotting. The detailed protocol of ELISA was used as described by Rubin [15].
2.7. Western blotting and slot blot
Western Blotting was performed essentially as described by Chan and Pollard [16]. Purified recombinant EP-HCC-1 (Sui1) and EP-HCC-13 (RalA) proteins were electrophoresed on SDS–PAGE and transferred to nitrocellulose paper. After pre-blocking with PBS containing 5% nonfat dry milk and 0.05% Tween-20 (PBST) for 30 min at room temperature, the nitrocellulose strips were incubated for 90 min at room temperature with a 1:200 dilution of serum. HRP-conjugated goat anti-human IgG (Caltag, Burlingame, CA) was applied as secondary antibody at a 1:3000 dilution. Immunoreactive bands were detected using the ECL kit (Amersham, Arlington Heights, IL) according to the manufacturer's instructions. Slot Blot was performed in exactly the same way as Western Blotting except that the purified recombinant proteins were applied directly onto the nitrocellulose membrane at the concentration of 100 ng per well using a vacuum source. Ten different proteins were loaded into 10 different wells so that the immunoreactivity of each HCC serum against 10 antigens can be tested in one blot simultaneously.
2.8. Absorption of autoantibodies with recombinant proteins
Sera (2.5 μl) were diluted in 500 μl PBS, and incubated for 2 h at room temperature with 15 μg recombinant proteins. The mixture was centrifuged at 10,000g for 10 min, and the supernatant was used for ELISA analysis with recombinant protein as coating antigen. The experiment was repeated four times, and average OD values were used for data analysis.
2.9. Statistical analysis
To determine whether the frequencies of autoantibody to TAAs in each cohort of patients' sera were significantly higher than that in sera from normal individuals and other controls, the frequencies of antibody were compared using the Chi-squared (χ2) test with Yate's correction, and two significant levels (0.05 and 0.01) were used. Methods for calculating sensitivity/specificity, positive predictive value/negative predictive value were used as described [17].
3. Results
3.1. Identification of potential HCC-related TAAs by immunoscreening cDNA expression library
In this study, we used a HCC serum with high titer autoantibodies against HepG2 cells by immunofluorescence and Western Blot analysis. The patient was a 64 year old white male with HCC but with no past history of HBV or HCV infection and was not alcoholic by history. By immunoscreening a HepG2 cDNA expression library, we identified 30 reactive cDNA clones. All these 30 reactive clones were amplified, purified and sequenced. After the analysis of all cDNA sequences by BLAST search against known sequence databases, 24 of these clones were found to have been functionally characterized and the remaining six clones were unknowns. Among the 24 known genes, two genes were identical to each other and encoded the protein of fibronectin1. Three mitochondrial genes showed sequence identity to each other. Thus, 21 independent genes were identified. Eleven of the 21 genes are involved in cell apoptosis, transcription, differentiation and proliferation as well as antigen processing. As shown in Table 1, of interest was that nine of them (EP-HCC-1, 5–9, 10, 12, 13) have been reported to be related to cancer [18–23]. In particular, 3 (EP-HCC-1, EP-HCC-7, EP-HCC-9) of these nine cancer-related proteins were reported to be associated with HCC.
3.2. Prevalence of antibodies to Sui1 and RalA in HCC
The purified recombinant Sui1 and RalA proteins were used as coating antigens in ELISA to detect antibodies against these two putative TAAs. The ELISA results were also confirmed by Western blotting analysis. Sera from 77 patients with HCC, 30 patients with liver cirrhosis (LC), 30 patients with chronic hepatitis (CH) and 82 normal human sera were analyzed. As demonstrated in Table 2, the prevalence of antibody to Sui1 was 11.7% (9/77), and to RalA was 19.5% (15/77) in HCC, which were significantly higher than in sera with LC, CH, and normal human sera (p < 0.01). Fig. 1 shows the range of OD values. Fig. 2A and B shows that representative HCC sera with positive reaction to Sui1 or RalA in ELISA also showed strongly reactive bands in Western Blot compared to two normal sera. The specificity of recombinant Sui1 or RalA in ELISA was further confirmed by antibody absorption study. Three HCC sera which showed positive reaction with Sui1 and three HCC sera which showed positive reaction with RalA in ELISA and Western Blot were pre-incubated with recombinant Sui1 or RalA and subsequently subjected to ELISA. Two normal human sera were also tested as controls. As shown in Fig. 2 (C, D), reactivity of HCC sera decreased substantially by pre-incubation with Sui1 or RalA. In contrast, the reactivity did not change much in normal control sera. This preliminary result suggested that these two newly identified HCC-related antigens might be used as potential serological markers in HCC.
Table 2.
Frequencies of antibodies to 10 TAAs.
| Type of serum | No. sera | No. and percentage of autoantibodies toa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imp1 | p62 | Koc | p53 | CyclinB1 | C-myc | p16 | survivin | Any of 8 TAAs | Sui1 | RalA | Any of 10 TAAs | ||
| HCCb | 77 | 14c (18.2) |
13c (16.9) |
16c (20.8) |
14c (18.2) |
11c (14.3) |
9c (11.7) |
15c (19.5) |
9d (11.7) |
46c (59.7) |
9d (11.7) |
15c (19.5) |
51c (66.2) |
| LC | 30 | 0 (0) | 1 (3.3) | 4d (13.3) | 2 (6.7) | 0 (0) | 1 (3.3) | 4d (13.3) | 0 (0) | 9 (30) | 1 (3.3) | 1 (3.3) | 10 (33.3) |
| CH | 30 | 3 (10) | 0 (0) | 2 (6.7) | 1 (3.3) | 0 (0) | 0 (0) | 2 (6.7) | 0 (0) | 6 (20) | 0 (0) | 0 (0) | 6 (20) |
| NHS | 82 | 2 (2.4) | 1 (1.2) | 1 (1.2) | 2 (2.4) | 2 (2.4) | 0 (0) | 1 (1.2) | 2 (2.4) | 10 (12.2) | 0 (0) | 0 (0) | 10 (12.2) |
Cutoff value: mean + 3SD of NHS.
Abbreviations: HCC, hepatocellular carcinoma; LC, liver cirrhosis; CH, chronic hepatitis; NHS, normal human sera.
p values relative to NHS: p < 0.01.
p < 0.05.
Fig. 1.
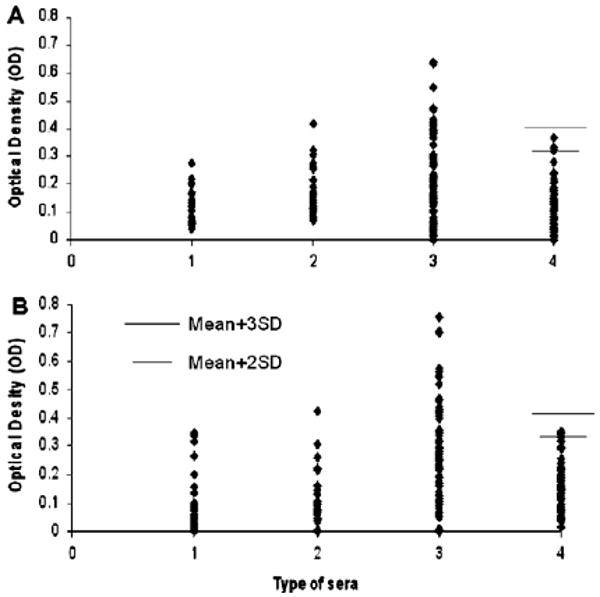
Titer of autoantibodies to EP-HCC-1 (Sui1) and EP-HCC-13 (RalA) in HCC, liver cirrhosis (LC), chronic hepatitis (CH) and NHS. The range of antibody titers to Sui1 and RalA is expressed as optical density (OD) obtained from ELISA. The mean + 2SD and + 3SD of 82 normal human sera are shown in relationship to all serum samples. Panel A: Antibody titer to EP-HCC-1 (Sui1). Panel B: Antibody titer to EP-HCC-13 (RalA). Lane 1, CH; lane 2: LC; lane 3: HCC; lane 4: NHS.
Fig. 2.
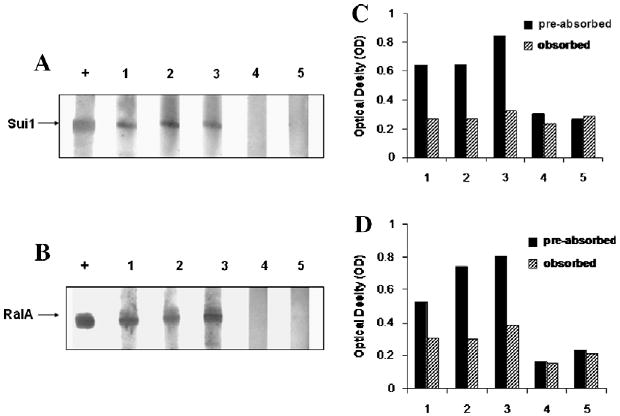
Representative Western Blot showing reactivity of ELISA positive HCC sera with recombinant proteins Sui1 (A) or RalA (B). To confirm the specificity of antibodies in positive sera for ELISA, all the positive sera were subjected to antibody absorption with recombinant protein Sui1 or RalA, followed by ELISA to evaluate the reduction of OD values (C, D). Panels A and C: The lane with “+” indicates anti-Sui1 polyclonal antibody used as positive control; lanes 1–3, three HCC positive sera; lanes 4 and 5, two normal human sera used as negative control. Panels B and D: The lane with “+” indicates anti-RalA monoclonal antibody used as positive control; lanes 1–3: three HCC positive sera; lane 4 and 5, two normal human sera used as negative control. The residual OD value after absorption with purified antigens is comparable to the “background” OD value shown by normal sera.
3.3. Evaluation of the addition of Sui1 and RalA to an array of multiple TAAs in enhancing antibody detection for diagnosis of HCC
As shown in Table 2, antibodies to eight TAAs (IMP1, p62, Koc, p53, c-myc, cyclin B1, survivin and p16), which had been previously used in antigen mini-arrays for the detection of antibodies in HCC [13] and other types of cancer [34,35], were used to test the group of 77 HCC serum, 30 LC sera and 30 CH serum samples in the current study. The prevalence of antibody to individual TAAs ranged from 11.7% to 20.8%. When the two newly identified TAAs (Sui1 and RalA) were added to the panel of 8 TAAs, the cumulative prevalence was 66.2% (51/77), which was significantly higher than the cumulative positive antibody reaction in LC (33.3%), CH (20%) and normal individuals (12.2%). Compared to the cumulative prevalence (59.7%, 46/77) of antibody to eight TAAs used in the previous panel of TAA array in HCC study, the increase in prevalence was 6.5%. Thus the two newly identified antigens incrementally raised the sensitivity of the 10 TAA array as a diagnostic modality for HCC. The association between anti-TAA antibodies and clinical stages in HCC sera was also analyzed. The results showed that 15 (60.0%), 11 (68.8%), 17 (77.3%) and 6 (75.0%) sera were positive with any of the 10 anti-TAA antibodies in clinical stage I, II, III and IV, respectively. There were no statistical differences for anti-TAA frequencies among clinical stages, but this could be due to the small size of the samples.
In order to address the question of how good the mini-array of multiple TAAs was in separating people with and without HCC, parameters such as sensitivity/specificity and positive predictive value/negative predictive value were calculated and summarized in Table 3. As described above, the sensitivity of the 10 TAA arrays for detecting HCC was 66.2%. Relative to LC, the specificity, positive predictive value and negative predictive value were 66.7%, 83.6% and 43.5%, respectively. Relative to CH, they were 80.0%, 89.5% and 48.0%, and relative to NHS, they were 87.8%, 83.6% and 73.5% (see Table 3 for calculations).
Table 3.
Evaluation of diagnostic values of anti-TAA antibodies to a panel of 10 TAAsa.
| Any anti-TAA positive | All anti-TAAs negative | Total | |
|---|---|---|---|
| HCC | 51 (A) | 26 (B) | 77 |
| LC | 10 (C1) | 20 (D1) | 30 |
| CH | 6 (C2) | 24 (D2) | 30 |
| NHS | 10 (C3) | 72 (D3) | 82 |
Relative to LC: Sensitivity (%) = A/(A + B) = 51//77 = 66.2%. Specificity (%) = D1/(C1 + D1) = 20/30 = 66.7%. Positive predictive value (PPV, %) = A/(A + C1) = 51/61 = 83.6%. Negative predictive value (NPV, %) = D1/(B + D1) = 20/46 = 43.5%.
Relative to CH: Sensitivity (%) = A/(A + B) = 51//77 = 66.2%. Specificity (%) = D2/(C2 + D2) = 24/30 = 80.0%. Positive predictive value (PPV, %) = A/(A + C2) = 51/57 = 89.5%. Negative predictive value (NPV, %) = D2/(B + D2) = 24/50 = 48.0%.
Relative to NHS: Sensitivity (%) = A/(A + B) = 51//77 = 66.2%. Specificity (%) = D3/(C3 + D3) = 72/82 = 87.8%.Positive predictive value (PPV, %) = A/(A + C3) = 51/61 = 83.6%. Negative predictive value (NPV, %) = D3/(B + D3) = 72/98 = 73.5%.
Diagnostic values relative to each cohort were calculated as follows.
3.4. Simultaneous use of both AFP and anti-TAAs as markers in HCC detection
Serum alpha fetoprotein (AFP) is a fetal glycoprotein produced by yolk sac and fetal liver and has been commonly used as a serum marker for diagnosis of HCC. The sensitivity of AFP has been reported to be about 60% in HCC [36]. The elevation of AFP is also associated with benign hepatic diseases such as acute and chronic viral hepatitis as well as toxic liver injury. Serum AFP levels remain within normal limits in most patients who have small HCC nodules (<20 mm) or well-differentiated HCC [37,38]. Moreover, elevated serum AFP levels reflect hepatic regeneration after destruction of hepatocytes in patients with chronic hepatitis and liver cirrhosis [39]. In the present study, 62 of 77 HCC sera were available for further AFP analysis. The sensitivity of simultaneous use of both AFP and anti-TAAs as markers in HCC detection is demonstrated in Table 4. Thirty-eight of 62 (61.3%) HCC sera had abnormal serum AFP level (>100 ng/ml). The sensitivity of AFP alone as marker in HCC detection was consistent with a previous report [36]. Of interest was that 17 of 24 (70.8%) HCC sera with normal range of serum AFP levels (<100 ng/ml) were anti-TAA positive. If both anti-TAA and AFP were simultaneously used as diagnostic markers, 55 (15 + 23 + 17) of 62 (88.7%) HCC patients could be correctly identified. Elevated AFP and anti-TAA appear to be independent but supplementary serological markers for the diagnosis of HCC.
Table 4.
Sensitivity of combined use of both AFP and anti-TAAs as markers in HCC detectiona.
| Anti-TAAs | |||
|---|---|---|---|
| Positive | Negative | Total | |
| Serum AFP levels: | |||
| >100 ng/ml | 23 (A) | 15 (B) | 38 |
| <100 ng/ml | 17 (C) | 7 (D) | 24 |
| Total | 40 | 22 | 62 |
Sensitivity (%) = (A + B + C)/(A + B + C + D) = 55/62 = 88.7%.
4. Discussion
Profiling the immune responses in cancer patients and identifying the targeted autoantigens can provide valuable information regarding the intracellular molecules participating in the malignant transformation process. The possible mechanisms underlying the production of these autoantibodies remain unknown, but is likely to be related to the dysregulation of the function of these molecules or alteration of their molecular structure or location [40,41]. In the present study, two (EP-HCC-1, EP-HCC-13) of 27 cDNA clones (21 known, 6 unknown) isolated by cDNA library screening with HCC serum antibodies were further investigated. EP-HCC-1 is identical to translation initiation factor Hu-Sui1 or eIF1 (eukaryotic translation initiation factor 1) which is a translation initiation factor that helps to maintain the integrity of translation initiation by combining with elongation initiation factor 2 (eIF-2) to enable initiator tRNA (met) to establish ribosomal recognition of an AUG codon [42]. By inhibiting frame shifting, it ensures the fidelity of multiple steps in translation [43]. Down regulation of Sui1 promotes the translation of growth stimulatory molecules and/or prevents the translation of proteins that mediate growth suppression [44]. Sui1 was also a novel gene associated with apoptosis, and was shown to be a tumor suppressor in hepatocarcinogenesis [18,19]. Lian and coworkers have reported that expression of Sui1 was suppressed by hepatitis B virus × antigen in X-antigen transgenic HepG2 cells. In the same study, Sui1 was also found to be highly expressed in non-tumor liver but not in liver tumor tissues. Introduction of Sui1 into HepG2 cells can inhibit cell growth and partially suppress tumor formation in nude mice [18]. The data in our study also demonstrated that the frequency of autoantibody to Sui1 in HCC sera was significantly higher than in other conditions such as liver cirrhosis, chronic hepatitis, and normal human sera. One of the possibilities is that the high prevalence of autoantibody response in HCC might be associated with the mutation or aberrant expression of Sui1. It has been suggested that Sui1 is a negative growth regulator, and the aberrant Sui1 allows for unregulated cell growth and contributes to the development of HCC [18].
EP-HCC-13 was identical to a Ras related oncogene RalA. Ral proteins are small GTPase molecules that work down-stream of the Ras protein signaling pathway [45]. The monomeric RAL (RAS-like) GTPases have been indirectly implicated in mitogenic regulation and Ras-mediated oncogenic transformation [46]. It was shown that RalA and RalB collaborate to maintain tumorigenicity through the regulation of both cell proliferation and cell survival signaling pathways [47]. RalA was more commonly activated compared to other major Ras effectors pathways in distinct cancer cell lines, and knockdown of RalA expression impeded, if not abolished the ability of human cancer cells to form tumors. Thus activation of RalA plays a critical role in Ras-induced tumorigenesis [30]. RalA was also reported to play an important role in EGF-mediated cell motility and has potential contribution to tumor metastasis in human cancer [31–33]. Together with the evidence provided by our study that the frequency of autoantibody to RalA was significantly higher in HCC sera (19.5%) compared to other conditions such as liver cirrhosis, chronic hepatitis and normal human sera, it suggests RalA may play an important role in HCC tumorigenesis and this role is stimulating a response by the immune system.
As demonstrated in many other studies, cancer has long been recognized as a multi-step and multi-factor process which involves not only genetic changes conferring growth advantage but also factors which disrupt regulation of growth and differentiation. This biological and molecular nature of cancer was well demonstrated by the frequent observation that most cancer patients produce autoantibodies to several TAAs [4], and different types of cancer showed distinct profiles of autoantibody in which specific TAAs were preferentially targeted [13,34,35]. It indicates that a well defined autoantibody profile for a specific cancer might help to reveal the unique factors or molecules that might be involved in the tumorigenesis and provides a powerful tool to enhance cancer diagnosis.
Although anti-TAA antibodies have been extensively investigated and evaluated as serological markers for cancer diagnosis, the enthusiasm for this approach has been tempered by low sensitivity when individual antigen–antibody systems were used. The evidence from our study also showed that the prevalence of antibody to any individual antigen from the selected TAA array was variable and ranged from 10% to 20% in HCC, and does not reach the level of sensitivity to be routinely useful in diagnosis of HCC. With the consideration of the drawback in single TAA-antibody system and the feature of unique anti-TAA antibody profiles for each specific cancer, we have demonstrated in this study that sensitivity of antibody screening can be optimized. With a panel of 10 TAAs selected on the basis of association with tumorigenic pathways, there was a statistically significant number of positive antibody reactions in HCC patients but not in patients with chronic hepatitis and normal individuals. Wang et al. have developed a phage-display library derived from prostate cancer tissue and a phage protein microarray, to analyze autoantibodies in sera from prostate cancer patients and controls [48]. In this study, a 22-phage-peptide detector was constructed for prostate cancer serum screening, with 81.6% sensitivity and 88.2% specificity. All these studies including our previous [13,34] and current studies, strongly support the hypothesis that “customized” TAA arrays enhance autoantibody detection in cancer and constitute promising and powerful tools for the immunoserological diagnosis of certain types of cancer, such as liver and prostate cancer.
In conclusion, the data in our study suggests that a uniquely constituted antigen panel or array of selected TAAs provides an additional and supplementary approach to the diagnosis of HCC, especially for AFP-negative cases. This type of study also highlights a promising direction for the development of biomarkers of disease, especially cancers [49]. The sensitivity and specificity of this approach is contingent on the careful selection of the most favorable combinations of TAAs. Further studies on other cDNA clones identified in this study may help us to define more proteins as candidate antigens for the formulation of other cancer TAA mini-arrays to supplement other approaches to detection of tumors.
Acknowledgments
We thank Drs. Thomas M. Wertin and Daniel Kim at William Beaumont Army Medical Center, El Paso, Texas, for providing serum samples which were used for cDNA library screening in this study as well as Ms. Roxanne Megliorino at University of Texas at El Paso for assistance with some of the molecular techniques in this study. This work was supported by National Institutes of Health (NIH) Grants #2S06GM008012, 5G12RR08124, and CA56956, and China 863 Program #2006AA02A40, and China NSF Grant #30872962.
Footnotes
Conflicts of interest: None declared.
References
- 1.Tan EM, Zhang J. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev. 2008;222:328–340. doi: 10.1111/j.1600-065X.2008.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disis ML, Bernhard H, Jaffee EM. Use of tumor-responsive T cells as cancer treatment. Lancet. 2009;373:673–683. doi: 10.1016/S0140-6736(09)60404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai H, Nakano Y, Kiyosawa K, Tan EM. Increasing titers and changing specificities of antinuclear antibodies in patients with chronic liver disease who develop hepatocellular carcinoma. Cancer. 1993;71:26–35. doi: 10.1002/1097-0142(19930101)71:1<26::aid-cncr2820710106>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 4.Zhang JY, Zhu W, Imai H, Kiyosawa K, Chan EKL, Tan EM. De-novo humoral immune responses to cancer-associated autoantigens during transition from chronic liver disease to hepatocellular carcinoma. Clin Exp Immunol. 2001;125:3–9. doi: 10.1046/j.1365-2249.2001.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covini G, Muhlen CAV, Pacchetti S, Colombo M, Chan EKL, Tan EM. Diversity of antinuclear antibody responses in hepatocellular carcinoma. J Hepatol. 1997;26:1255–1265. doi: 10.1016/s0168-8278(97)80460-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JY, Wang X, Peng XX, Chan EKL. Autoimmune responses in Chinese hepatocellular carcinoma (HCC) J Clin Immunol. 2002;22:98–105. doi: 10.1023/a:1014483803483. [DOI] [PubMed] [Google Scholar]
- 7.Imai H, Chan EKL, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Invest. 1993;92:2419–2426. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muro Y, Chan EK, Landberg G, Tan EM. A cell-cycle nuclear autoantigen containing WD-40 motifs expressed mainly in S and G2 phase cells. Biochem Biophys Res Commun. 1995;207:1029–1037. doi: 10.1006/bbrc.1995.1288. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JY, Chan EKL, Peng XX, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med. 1999;189:1101–1110. doi: 10.1084/jem.189.7.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soo Hoo L, Zhang JY, Chan EKL. Cloning and characterization of a novel 90 kDa ‘companion’ auto-antigen of p62 overexpressed in cancer. Oncogene. 2002;21:5006–5015. doi: 10.1038/sj.onc.1205625. [DOI] [PubMed] [Google Scholar]
- 11.Old LJ, Chen YT. New paths in human cancer serology. J Exp Med. 1998;187:163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jongeneel V. Towards a cancer immunome database. Cancer Immunol. 2001;1:3. [PubMed] [Google Scholar]
- 13.Zhang JY, Megliorino R, Peng XX, Tan EM, Chen Y, Chan EKL. Antibody detection using tumor-associated antigen mini-array in immunodiagnosing human hepatocellular carcinoma. J Hepatol. 2007;46:107–114. doi: 10.1016/j.jhep.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson PJ, Leung N, Cheng P, Welby C, Leung WT, Lau WY, Yu S, Ho S. ‘Hepatoma-specific’ alphafetoprotein may permit preclinical diagnosis of malignant change in patients with chronic liver disease. Brit J Cancer. 1997;75:236–240. doi: 10.1038/bjc.1997.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin RL. Enzyme-linked immunosorbent assays for antibodies to native DNA histones and (H2A-H2B)-DNA. In: Rose NR, Conway de Macario E, Folds JD, Lane HC, Nakamura RM, editors. Manual of Clinical Laboratory Immunology. fifth. American Society for Microbiology; Washington, DC: 1997. pp. 935–941. [Google Scholar]
- 16.Chan EKL, Pollard KM. Detection of autoantibodies to ribonucleoprotein particles by immunoblotting. In: Rose NR, Conway de Macario E, Folds JD, Lane HC, Nakamura RM, editors. Manual of Clinical Laboratory Immunology. fifth. American Society for Microbiology; Washington, DC: 1997. pp. 928–934. [Google Scholar]
- 17.Gordis L. Assessing the validity and reliability of diagnostic and screening tests. In: Gordis L, editor. Epidemiology. second. W.B. Saunders Company; Philadelphia, Pennsylvania: 2000. pp. 63–81. [Google Scholar]
- 18.Lian Z, Pan J, Liu J, Zhang SM, Zhu MH, Arbuthnot P, Kew M, Feitelson MA. The translation initiation factor, hu-Sui1 may be a target of hepatitis B × antigen in hepatocarcinogenesis. Oncogene. 1999;18:1677–1687. doi: 10.1038/sj.onc.1202470. [DOI] [PubMed] [Google Scholar]
- 19.Chen CP, Liu J, Wu KC, Fan DM. Effect of C2 gene on human gastric cancer cell cycle. Clin J Surg. 2001;10:224–227. [Google Scholar]
- 20.Kortylewski M, Komyod W, Kauffmann ME, Bosserhoff A, Heinrich PC, Behrmann I. Interferon-mediated growth regulation of melanoma cells: involvement of STAT1-dependent and STAT1-independent signals. J Invest Dermatol. 2004;122:414–422. doi: 10.1046/j.0022-202X.2004.22237.x. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs A, Jones B, Ricketts C, Bulbrook RD, Wang DY. Serum ferritin concentration in early breast cancer. Brit J Cancer. 1976;34:286–290. doi: 10.1038/bjc.1976.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kew MC, Torrance JD, Derman D, Simon M, Macnab GM, Charlton RW, Bothwell T. Serum tumor ferritins in primary liver cancer. Gut. 1978;19:294–299. doi: 10.1136/gut.19.4.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto K, Ishikawa H, Nishimura D, Hamasaki K, Nakao K, Eguchi K. Antiangiogenic property of pigment epithelium-derived factor in hepatocellular carcinoma. Hepatology. 2004;40:252–259. doi: 10.1002/hep.20259. [DOI] [PubMed] [Google Scholar]
- 24.Takenaka K, Yamagishi S, Jinnouchi Y, Nakamura K, Matsui T, Imaizumi T. Pigment epithelium-derived factor (PEDF)-induced apoptosis and inhibition of vascular endothelial growth factor (VEGF) expression in MG63 human osteosarcoma cells. Life Sci. 2005;77:3231–3241. doi: 10.1016/j.lfs.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson S, Carlson J, Velez R. Risk of cirrhosis and primary liver cancer in alpha 1-antitrypsin deficiency. New Engl J Med. 1986;314:736–739. doi: 10.1056/NEJM198603203141202. [DOI] [PubMed] [Google Scholar]
- 26.Kurokohchi K, Carrington M, Mann DL, Simonis TB, Alexander-Miller MA, Feinstone SM, Akatsuka T, Berzofsky JA. Expression of HLA class I molecules and the transporter associated with antigen processing in hepatocellular carcinoma. Hepatology. 1996;23:1181–1188. doi: 10.1002/hep.510230537. [DOI] [PubMed] [Google Scholar]
- 27.Szentirmay MN, Yang HX, Pawar SA, Vinson C, Sawadogo M. The IGF2 receptor is a USF2-specific target in nontumorigenic mammary epithelial cells but not in breast cancer cells. J Biol Chem. 2003;278:37231–37240. doi: 10.1074/jbc.M305791200. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Chen X, Guo D, Hong Q, Fu B, Ding R, Yu L, Hou K, Feng Z, Zhang X, Wang J. Knockdown of fibronectin induces mitochondria-dependent apoptosis in rat mesangial cells. J Am Soc Nephrol. 2005;16:646–657. doi: 10.1681/ASN.2004060445. [DOI] [PubMed] [Google Scholar]
- 29.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, Pellegata NS, de la Chapelle A. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci USA. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Ward Y, Wang W, Woodhouse E, Linnoila I, Liotta L, Kelly K. Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol Cell Biol. 2001;21:5958–5969. doi: 10.1128/MCB.21.17.5958-5969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gildea JJ, Harding MA, Seraj MJ, Gulding KM, Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;4:982–985. [PubMed] [Google Scholar]
- 33.Tchevkina E, Agapova L, Dyakova N, Martinjuk A, Komelkov A, Tatosyan A. The small G-protein RalA stimulates metastasis of transformed cells. Oncogene. 2005;24:329–335. doi: 10.1038/sj.onc.1208094. [DOI] [PubMed] [Google Scholar]
- 34.Zhang JY, Casiano AC, Peng XX, Koziol JA, Chan EKL, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomarkers Prev. 2001;12:136–143. [PubMed] [Google Scholar]
- 35.Koziol JA, Zhang JY, Casiano CA, Peng XX, Chan EKL, Tan EM. Recursive partitioning for tumor classification using a panel of recombinant tumor-associated antigens. Clin Cancer Res. 2003;9:5120–5151. [PubMed] [Google Scholar]
- 36.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S108–112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 37.Ryder SD, Rizzi PM, Volkmann M, Metivier E, Pereira LM, Galle PR, Naoumov NV, Zentgraf H, Williams R. Use of specific ELISA for the detection of antibodies directed against p53 protein in patients with hepatocellular carcinoma. J Clin Pathol. 1996;49:295–299. doi: 10.1136/jcp.49.4.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo F, Wada K, Nagato Y, Nakajima T, Kondo Y, Hirooka N, Ebara M, Ohto M, Okuda K. Biopsy diagnosis of well-differentiated hepatocellular carcinoma based on new morphologic criteria. Hepatology. 1989;9:751–755. doi: 10.1002/hep.1840090516. [DOI] [PubMed] [Google Scholar]
- 39.Gebo KA, Chander G, Jenckes MW, Ghanem KG, Herlong HF, Torbenson MS, El-Kamary SS, Bass EB. Screening tests for hepatocellular carcinoma in patients with chronic hepatitis C: a systematic review. Hepatology. 2002;36(5 Suppl 1):S84–92. doi: 10.1053/jhep.2002.36817. [DOI] [PubMed] [Google Scholar]
- 40.Tan EM. Autoantibodies in pathology and cell biology. Cell. 1991;67:841–842. doi: 10.1016/0092-8674(91)90356-4. [DOI] [PubMed] [Google Scholar]
- 41.Tan EM, Chan EK, Sullivan KF, Rubin RL. Antinuclear antibodies (ANNs): diagnostically specific immune markers and clues toward the understanding of systemic autoimmunity. Clin Immunol Immunopathol. 1998;47:121–141. doi: 10.1016/0090-1229(88)90066-9. [DOI] [PubMed] [Google Scholar]
- 42.Yoon HJ, Donahue TF. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui Y, Gonzalez CI, Kinzy TG, Dinman JD, Peltz SW. Mutation in the MOF2/Sui1 gene affects both translation and nonsense-mediated mRNA decay. RNA. 1999;5:794–804. doi: 10.1017/s1355838299982055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chin LS, Singh SK, Wang Q, Murray SF. Identification of okadaic-acid-induced genes by mRNA differential display in glioma cells. J Biomed Sci. 2000;7:152–159. doi: 10.1007/BF02256622. [DOI] [PubMed] [Google Scholar]
- 45.Wolthuis RM, Zwartkruis F, Moen TC, Bos JL. Ras-dependent activation of the small GTPase Ral. Curr Biol. 1998;8:471–474. doi: 10.1016/s0960-9822(98)70183-6. [DOI] [PubMed] [Google Scholar]
- 46.Reuther GW, Der CJ. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr Opin Cell Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 47.Yuchen C, Michael AW. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, et al. New Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 49.Paradis V, Bedossa P. In the new area of noninvasive markers of hepatocellular carcinoma. J Hepatol. 2007;46:9–11. doi: 10.1016/j.jhep.2006.10.006. [DOI] [PubMed] [Google Scholar]


