Abstract
ORF50 protein activates KSHV lytic cycle. The promoter of an early lytic cycle gene ORF47, encoding envelope protein gL, is activated by an interaction between ORF50 protein and RBP-Jκ. In ORF47p only one of two sequences fitting the consensus RBP-Jκ recognition site strongly binds RBP-Jκ and confers a response to ORF50 protein. Flanking sequences 5’ to the RBP-Jκ binding site are required to confer a maximal response to ORF50 protein. Not all mutant ORF50 response elements in the ORF47p that are bound by RBP-Jκ are sufficient to confer maximal ORF50 responsiveness. Four sequences containing an RBP-Jκ site and flanking sequences characteristic of the ORF50 response element in ORF47p were identified in human DNA. All bound RBP-Jκ, but only one responded robustly to ORF50 protein. We propose models for the possible function of ancillary sequences flanking the RBP-Jκ-binding element which are crucial for mediating ORF50 transactivation.
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV), human herpesvirus 8, is implicated in the pathogenesis of Kaposi’s sarcoma, primary effusion lymphoma and multicentric Castleman’s disease (Boshoff et al., 1995; Cesarman et al., 1995; Chang et al., 1994; Soulier et al., 1995). Based on similarities in nucleotide sequence, genome organization, and biologic properties, KSHV is classified as a member of the gamma subfamily of the Herpesviridae, which also includes the transforming viruses Epstein-Barr virus (EBV) and Herpesvirus saimiri (Chang et al., 1994; Russo et al., 1996). Like all herpesviruses, KSHV has two distinct phases in its life cycle, latency and lytic replication (Miller et al., 1997; Renne et al., 1996). Although the physiological signals that stimulate the switch between latency and lytic-cycle gene expression of KSHV are not fully understood, chemical agents such as 12-O-tetradecanoylphorbol-13-acetate or n-butyrate significantly disrupt viral latency and initiate the lytic cycle (Miller et al., 1997). Upon chemical induction, the lytic cycle is expressed in a temporally ordered program, which includes immediate-early, delayed-early and late gene expression (Miller et al., 1997; Sun et al., 1999). Among these viral genes, the immediate-early genes encode regulatory proteins that up- and down-regulate the expression of viral and cellular genes and therefore play a crucial role in the switch from latency to lytic replication (Zhu et al., 1999).
The protein encoded by open reading frame 50 (ORF50) of KSHV genome is an immediate-early product that is sufficient to trigger the viral lytic cascade to completion in latently infected cells (Lukac et al., 1999; Lukac et al., 1998; Sun et al., 1998). ORF50 protein is a potent transcriptional activator that contains an N-terminal basic DNA-binding domain and a C-terminal acidic activation domain (Chang and Miller, 2004; Chang et al., 2002; Lukac et al., 1999; Wang et al., 2001). Transient transfection experiments have demonstrated that ORF50 protein activates the promoters of a number of viral genes that are expressed during early stage of lytic cycle, including its own gene, polyadenylated nuclear (PAN) RNA, K12 (kaposin), ORF57, K8, K9 (vIRF), ORF21 (thymidine kinase), K5, K6 (vMIP-1), ORF6 (single-stranded DNA binding protein), K14 (vOX-2), ORF74 (vGPCR), K2 (vIL-6) and ORF59 (Chang et al., 2002; Chang et al., 2005; Chen et al., 2000; Deng et al., 2002; Deng et al., 2000; Haque et al., 2000; Jeong et al., 2001; Liu et al., 2008; Lukac et al., 2001; Lukac et al., 1999; Song et al., 2001; Wang and Yuan, 2007; Zhang et al., 1998). In addition to these delayed-early genes, ORF50 protein also activates the promoter of a late gene (gB) in transient reporter assays (Damania et al., 2004; Ziegelbauer et al., 2006).
Although the molecular mechanism by which ORF50 protein activates its target genes during the lytic cycle has been extensively investigated, several details still remain to be addressed (Chang et al., 2005). The most complicated observation is that many ORF50 response elements (ORF50 REs) identified in target promoters do not share conserved DNA sequences (Chang et al., 2005; Lukac et al., 2001; Song et al., 2003; Ueda et al., 2002). Currently, two subclasses of the ORF50 REs that contain homologous sequences have been grouped together and their modes of response to ORF50 protein have been proposed. Activation of one class of targets, such as the PAN and K12 genes, operates mainly by ORF50 protein directly binding to promoter DNA (Chang et al., 2002; Song et al., 2002). The related 26-nt response elements in the PAN and K12 promoters serves a direct binding target for ORF50 protein (Chang et al., 2002; Song et al., 2002). Activation of the second class of target promoters by ORF50 protein is mediated through a cellular DNA-binding protein RBP-Jκ (Chang et al., 2005; Liang et al., 2002). RBP-Jκ, also known as CSL or CBF1, is a sequence-specific DNA-binding protein, which is a downstream effector of the Notch signaling pathway (Lai, 2002). The RBPJκ-dependent mechanism has been demonstrated in several ORF50 target promoters, such as ORF57, ORF6, vMIP-1, K14/ORF74, ORF50, K8 and ORF59 (Chang et al., 2005; Liang et al., 2002; Liang and Ganem, 2003; Liang and Ganem, 2004; Liu et al., 2008; Wang and Yuan, 2007). All the ORF50 REs identified from these promoters contain a conserved or related RBP-Jκ recognition sequence, GTGGGAA (Tun et al., 1994). In the case of the ORF57 and vMIP-1 promoters, mutation of the RBPJκ-binding site dramatically abolished the response to ORF50 protein (Chang et al., 2005). Furthermore, Liang et al. (2002) first demonstrated that ORF50 protein failed to activate the ORF57 and ORF6 promoters in RBP-Jκ deficient cells, while such activation was restored by co-transfection with an RBP-Jκ expression vector. Since ORF50 protein can directly interact with RBP-Jκ protein (Carroll et al., 2006; Papugani et al., 2008), RBP-Jκ protein may act as a key mediator in cells for recruiting ORF50 protein to the target promoter DNA.
In addition to nucleotide sequence homology and the capacity to bind ORF50 or RBP-Jκ protein in vitro, classification of the ORF50 REs can be also achieved by measuring their response to DNA-binding deficient ORF50 mutants (Chang et al., 2005). Previously, we found that several DNA-binding deficient ORF50 mutants retained the capacity to activate indirect targets, such as the ORF57 and vMIP-1 promoters, but were markedly defective at activating direct targets, such as the PAN and K12 promoters (Chang et al., 2005). These DNA-binding deficient ORF50 mutants may be useful to classify new cellular or viral targets of ORF50.
In this report, we show that ORF50 can activate the promoter of the ORF47 gene that encodes glycoprotein L (gL). The minimal ORF50 RE in ORF47p was mapped to a 25-bp element that contains a consensus RBP-Jκ binding site. Although there is another consensus RBP-Jκ recognition sequence nearby in the ORF47 promoter, this element is bound weakly by RBP-Jκ protein in vitro and confers little ORF50 responsiveness. Thus a conserved RBP-Jκ sequence itself is not sufficient to confer an ORF50 response. By study of mutant ORF50 response elements from the ORF 47p and four homologous RBPJ-κ-binding elements from the human genome, we demonstrate that not all elements bound by RBP-Jκ protein mediate ORF50 responsiveness.
Results
ORF47 behaves with early gene kinetics during lytic replication
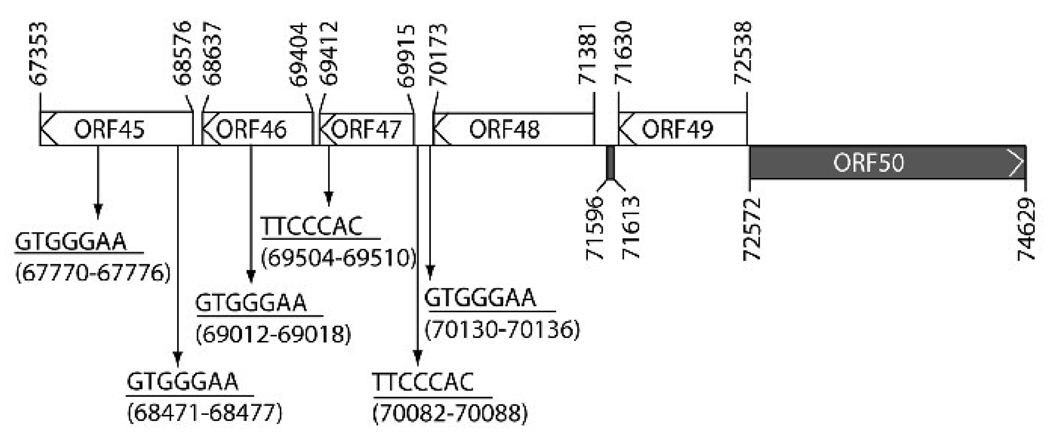
RBP-Jκ elements have been implicated to play a critical role in regulating ORF50 promoter activity (Chang et al., 2005; Liang and Ganem, 2003). Analysis of sequences upstream of the ORF50 gene revealed six consensus RBP-Jκ recognition sequences, GTGGGAA (Tun et al., 1994), which were dispersed in the region between the ORF45 and the ORF48 genes (Fig. 1). Two of these RBP-Jκ elements, located at nucleotides (nt) 70082 to 70088 and 70130 to 70136, were found in the promoter region of the ORF47 gene (Fig. 1). The ORF47 gene encodes a homolog of herpesvirus glycoprotein L (gL) (Russo et al., 1996), which was thought to be a late gene during lytic replication. Since several viral promoters containing the RBP-Jκ recognition sequence belong to the class of delayed-early genes that are regulated by ORF50 protein, we speculated that the ORF47 gene could be a target of ORF50 protein and behave as a delayed-early gene.
Fig. 1.
Six consensus RBP-Jκ recognition sequences are dispersed in the region encompassing ORFs 45, 46, 47 and 48 of the KSHV genome. Forward or reverse RBP-Jκ recognition sequences are indicated as GTGGGAA or TTCCCAC. Numbers in the diagram represent nucleotide positions of the KSHV genome (Russo et al., 1996). The back box indicates a rightward open reading frame, and white boxes indicate leftward open reading frames.
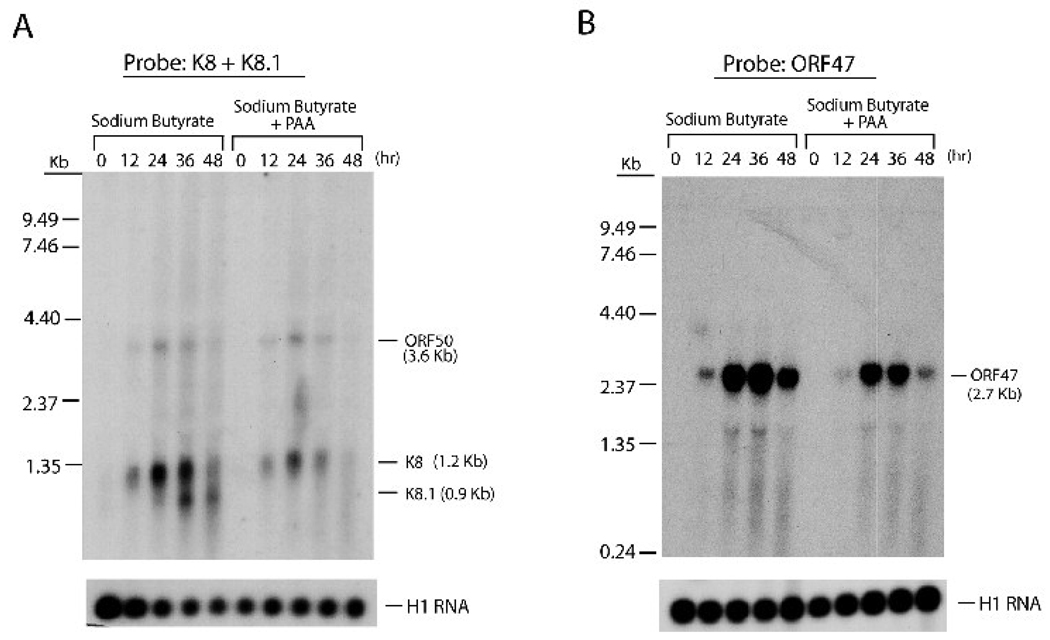
To analyze the expression kinetics of the ORF47 gene during the lytic cycle, northern analysis was carried out using RNA samples of HH-B2 cells treated with n-butyrate. In one set of cultured cells, phosphonoacetic acid (PAA), an inhibitor of KSHV lytic DNA synthesis, was added to block viral DNA synthesis and late gene expression (Sun et al., 1999). After treatment with n-butyrate, a 2.7-kb mRNA containing ORF47 was markedly induced (Fig. 2B). The 2.7-kb transcript was a polycistronic mRNA containing open reading frames 47, 46 and 45 (data not shown). PAA treatment only slightly affected ORF47 mRNA expression, indicating that the ORF47 gene was not a late gene. The same RNA samples were also used for detecting expression of ORF50 (immediate-early gene), K8 (delayed-early gene) and K8.1 (late gene), which are also part of a polycistronic transcript. Like the ORF47 mRNA, expression of the ORF50 and K8 mRNAs was resistant to PAA treatment (Fig. 2A). By contrast, treatment with PAA completely abolished expression of the K8.1 gene (Fig. 2A). These results indicated that the ORF47 gene belongs to the early gene family.
Fig. 2.
The ORF47 gene is expressed in the early stage of viral reactivation. Total RNA was prepared from HH-B2 cells that had been treated with sodium butyrate in the absence or presence of phosphonoacetic acid (PAA). At different times (12, 24, 36 and 48 hrs) after chemical treatment, expression of ORF50, K8, K8.1 (A) and ORF47 (B) was examined by northern blot analysis. A 2.7-kb ORF47 mRNA induced by sodium butyrate was resistant to PAA treatment. Hybridization with H1RNA of RNase P served as a loading control.
The ORF50 response element within the ORF47 promoter contains an RBP-Jκ binding site
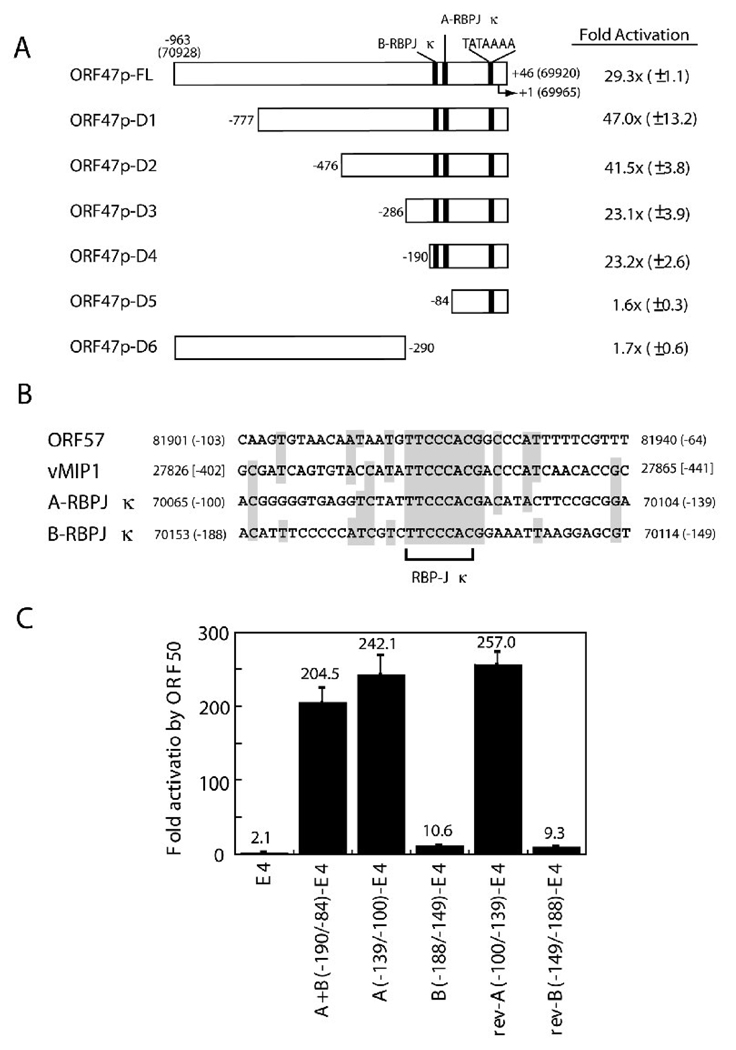
Using total RNAs of HH-B2 cells treated with n-butyrate, the transcriptional start site of the ORF47 promoter was mapped to nt 69,965 of the KSHV genome by RACE (rapid amplification of cDNA ends). A consensus TATA box, TATAAAA, was present between −30 and −24 relative to the transcription start site (Fig. 3A). To determine whether the ORF47 promoter was a target of ORF50 protein, a 1,008-bp DNA fragment corresponding to the promoter region from −963 to +46 was fused 5’ to a CAT reporter. Co-transfection of the ORF47p/CAT reporter plasmid with an ORF50 expression plasmid led to a 29-fold enhancement of CAT activity in BJAB cells (Fig. 3A). This result demonstrated that the ORF47 promoter was a target of ORF50 protein.
Fig. 3.
Defining the ORF50 response element (ORF50 RE) of the ORF47 promoter. (A) Transcriptional activation of deletion mutants of the ORF47 promoter by ORF50 protein. A series of deletions of the ORF47 promoter in pCAT-Basic are indicated. The numbers in the diagram represent nucleotide positions of the KSHV genome and the positions relative to the transcriptional start site (+1). Two RBP-Jκ binding elements in the ORF47 promoter are designated A-RBPJκ and B-RBPJκ, respectively. CAT activities of reporters co-transfected with pCMV or pCMV-ORF50 were measured in BJAB cells. (B) Sequence alignment of the RBP-Jκ-containing elements in the ORF47 promoter and in the ORF50 REs of the ORF57 and vMIP-1 promoters. (C) Response to ORF50 protein of the A-RBPJκ and B-RBPJκ elements and flanking sequence in pE4CAT. The region from −190 to −84 contains both A-RBPJκ and B-RBPJκ. Both forward and reverse (rev) orientations of the A-RBPJκ and B-RBPJκ elements in pE4CAT were examined for their response to ORF50 protein.
To define the location of the ORF50 response element (ORF50 RE) in the ORF47 promoter, a series of deletion mutants were constructed and reporter assays were performed. As shown in Fig. 3A, 5’ deletion to −190 relative to the ORF47 transcriptional start site still retained nearly maximal response to ORF50 protein. However, further deletion to −84, which removed the two putative RBP-Jκ binding elements, completely abolished the response of pORF47p/CAT to ORF50 protein (Fig. 3A, ORF47p-D5). These results indicated that the 107-bp region between −190 and −84 in the ORF47 promoter contained elements required for ORF50 responsiveness. Two consensus RBP-Jκ recognition sequences were present in this region; these were designated A-RBPJκ (−139 to −100) and B-RBPJκ (−188 to −149) (Fig. 3A and 3B). Both elements in the ORF47 promoter shared sequence homology with the ORF50 REs of the ORF57 and vMIP1 promoters (Fig. 3B). To verify the importance of the region between −190 and −84 in the ORF47 promoter, this region was fused to the heterologous adenovirus E4 promoter and its response to ORF50 protein was examined (Fig. 3C). When the reporter containing the entire −190/−84 region was co-transfected with an ORF50-expressing plasmid in cells, CAT activity was activated over 200-fold [Fig. 3C, A+B(−190/−84)-E4]. When either the A-RBPJκ(−139/−100) or B-RBPJκ (−188/−149) element in pE4CAT was co-transfected with the ORF50-expressing plasmid, only the reporter plasmid containing the A-RBPJκ(−139/−100) element conferred robust activation by ORF50 protein. Forward or reverse orientations of the A-RBPJκ or B-RBPJκ in pE4CAT did not significantly change their response to ORF50 protein (Fig. 3B and 3C). This experiment showed that not all elements that fit a consensus RBP-Jκ recognition sequence confer ORF50 responsiveness.
RBP-Jκ protein binds strongly to the A-RBPJκ element, but weakly to the B-RBPJκ element
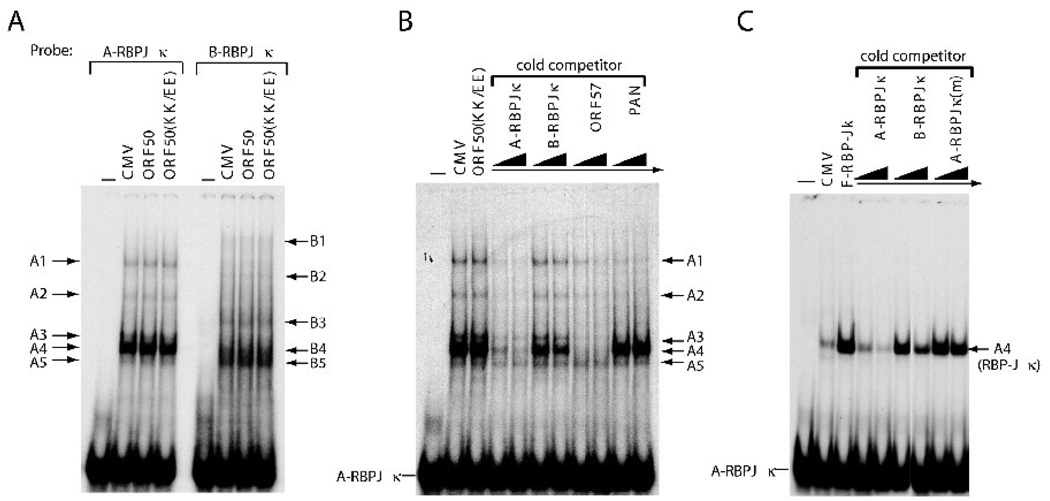
To understand the mechanism underlying the differential response of the A- and B-RBPJκ elements to ORF50 protein, binding of ORF50 and cellular proteins to the A-RBPJκ and B-RBPJκ elements was examined (Fig. 4). Total cell lysates of HKB5/B5 cells transfected with empty vector, pCMV-ORF50 or pCMV-ORF50(KK/EE) were used in electrophoretic mobility shift assays (EMSAs). The ORF50(KK/EE) protein contains two amino acid substitutions at positions K527 and K528 which markedly enhance DNA-binding capacity (Chang and Miller, 2004; Chang et al., 2008). As shown in Fig 4A, neither wild-type ORF50 nor the KK/EE mutant bound the ORF47 promoter elements in vitro. However, several cellular proteins bound to the A-RBPJκ and B-RBPJκ elements in EMSA experiments and the pattern of binding of cellular proteins to the two elements was different (Fig. 4A). The variation in cellular proteins binding to A-RBPJκ and B-RBPJκ was confirmed by cross competition with cold oligonucleotides (Fig. 4B). Cold A-RBPJκ competitor, but not B-RBPJκ cold competitor, significantly reduced binding of cellular proteins to the A-RBPJκ element (Fig. 4B). This result suggested that specific cellular proteins bound to the A-RBPJκ element may be critical for mediating the ORF50 transactivation.
Fig. 4.
RBP-Jκ protein specifically binds to the ORF50 RE of the ORF47 promoter. (A) EMSAs of A-RBPJκ (−139 to −100) and B-RBPJκ (−188 to −149). The radioactive probes (A-RBPJκ or B-RBPJκ) were incubated with cell extracts of HKB5/B5 transfected with pCMV, pCMV-ORF50, or pCMV-ORF50(KK/EE) and analyzed by EMSA. The different protein/DNA complexes are indicated. (B) Cross competition with various ORF50 REs from viral promoters (ORF57p and PANp) in EMSA. The indicated cold competitors used in EMSA were in 50- and 100-fold molar excesses over hot probe DNA. (C) Binding of the A-RBPJκ element by ectopically expressed RBP-Jκ protein. Extract of HH-B2 cells transfected with pCMV-FLAG-RBP-Jκ was used in EMSA; a cross-competition assay was performed to determine the binding specificity of RBP-Jκ protein to the probe.
Two distinct ORF50 REs from the ORF57p, an indirect target, and PANp, a direct target promoter, were also used as cold competitors in EMSA. The ORF57p element, but not the PANp element, completely disrupted the formation of all complexes in EMSA (Fig. 4B). These data suggest that the A-RBPJκ and ORF57 elements may possess binding sites for related cellular proteins, especially for the protein component in abundant complex “A4”. RBP-Jκ was a likely candidate protein component in complex A4.
To determine whether RBP-Jκ protein bound to the A-RBPJκ element, protein extracts of HKB5/B5 cells transfected with a plasmid expressing RBP-Jκ protein were used in EMSA. An abundant DNA/RBP-Jκ complex could be detected, which co-migrated with complex A4 (Fig. 4C). In cross-competition analysis, a mutant competitor with point mutations in the RBP-Jκ site (Fig. 7A) lost its competitive ability in EMSA [Fig 4C, A-RBPJκ(m)]. The B-RBPJκ cold competitor behaved more like A-RBPJκ(m) than the A-RBPJκ element. It did not significantly affect formation of DNA/RBP-Jκ complex. These results indicated that RBP-Jκ protein was the major component of the complex A4. Although the B-RBPJκ element contains a consensus RBP-Jκ binding site, it did not efficiently bind RBP-Jκ protein.
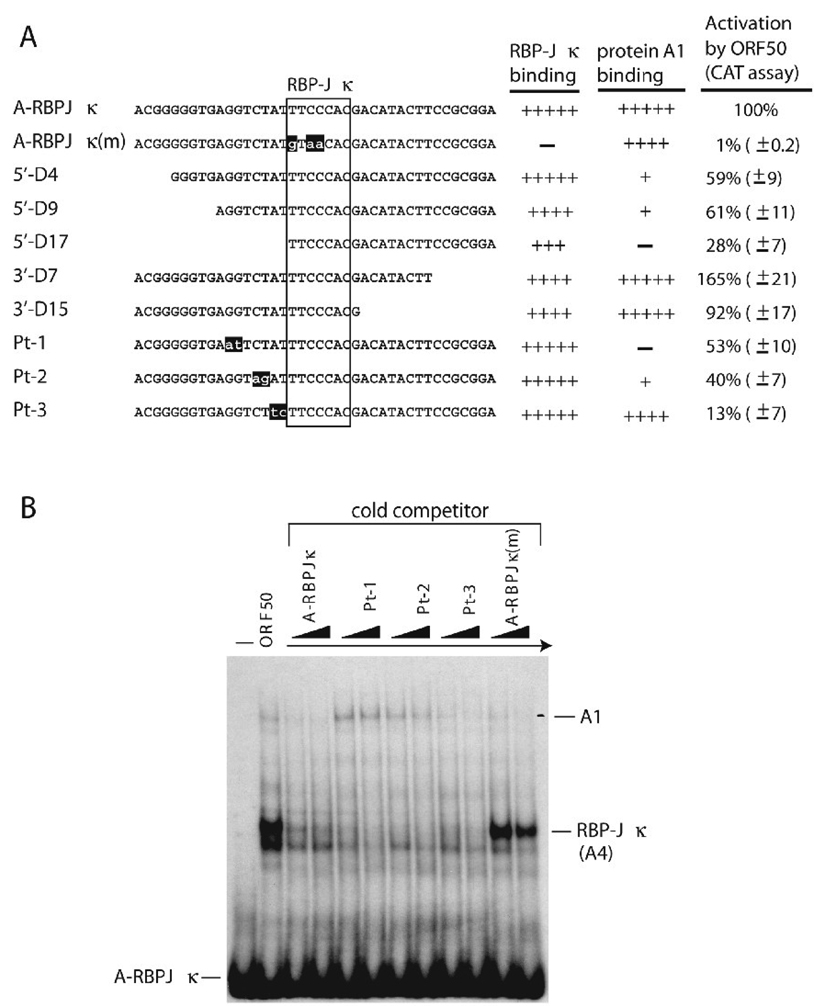
Fig. 7.
Optimal ORF50 responsiveness of the A-RBPJκ element requires both a consensus RBP-Jκ site and flanking sequences. (A) Effect of deletions or mutations in the A-RBPJκ element on capacity to bind RBP-Jκ protein and to respond to the ORF50 protein. The relative binding capacity of the mutant elements to RBP-Jκ (a component of complex A4) and to A1 protein (a component of complex A1) was measured by competition experiments in EMSA. ORF50 responsiveness was determined by CAT assays in BJAB cells as described in Fig. 3. (B) Cross competition analysis by the A-RBPJκ mutant elements in EMSA. The radioactive probe and an extract of HKB5/B5 cells transfected with ORF50 protein were used in EMSA. The cold competitors Pt-1, Pt-2 and Pt-3 as described in (A) were used for competition analysis. They were added in 50 and 100-fold molar excesses over the hot probe.
The A-RBPJκ element is responsive to a DNA-binding deficient ORF50 mutant
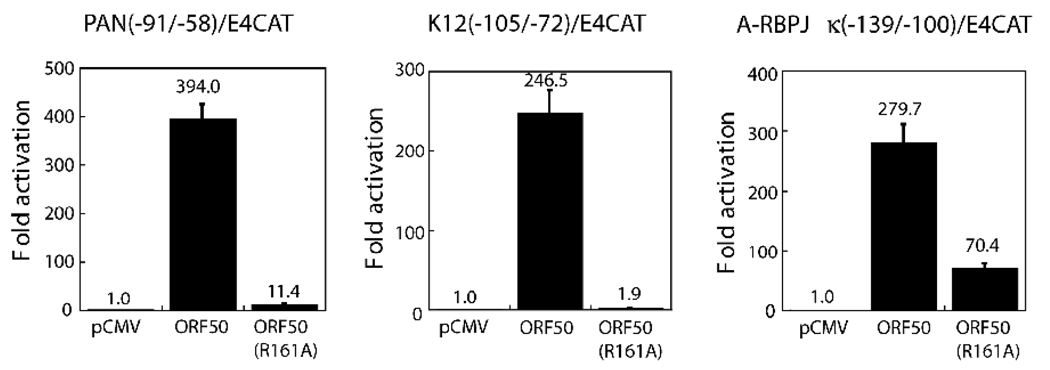
Previously, we showed that several DNA-binding deficient ORF50 mutants were capable of activating the indirect targets of ORF50 such as ORF57p and vMIP1p (Chang et al., 2005). Here, we determined whether ORF50(R161A), a DNA binding-deficient ORF50 mutant, could activate the ORF47 promoter. As shown in Fig. 5, ORF50(R161A) failed to activate the PAN and K12 promoters, two direct targets of ORF50, in a CAT reporter assay. By contrast, ORF50(R161A) retained the capacity to strongly activate the ORF47 promoter (70-fold), although the transactivation function of the R161A mutant was lower than that of wild-type ORF50 (Fig. 5). These results confirmed the experiment illustrated in Fig. 4, showing that ORF50 protein did not bind directly to the ORF47p. Activation of ORF47p by ORF50 protein is mediated, at least in part, through an indirect interaction with DNA (see Discussion, Fig. 10).
Fig. 5.
Transcriptional activation of the PANp, K12p and ORF47p elements by ORF50 protein and ORF50(R161A), a DNA-binding deficient point mutant of ORF50. The transcriptional capacities of wild-type ORF50 and ORF50(R161A) on the identified ORF50 REs from the PAN, K12 and ORF47 promoters were determined by CAT assay in BJAB cells.
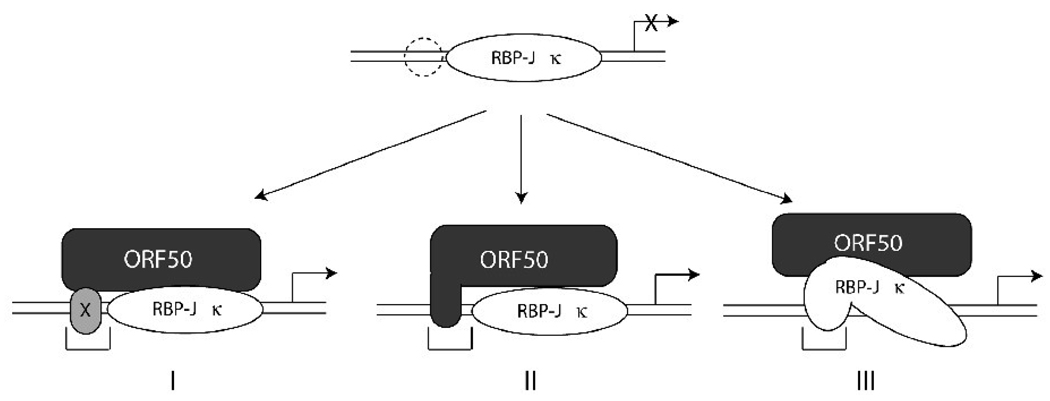
Fig. 10.
Models depicting possible mechanisms of ORF50-mediated transactivation through RBP-Jκ protein. In addition to the RBP-Jκ binding site, an auxiliary flanking DNA element is partly required for optimal ORF50-mediated transactivation. The specific cis-acting region may provide a platform for binding of other cellular protein(s) (designated × in model I) or for trapping the ORF50 protein (model II). This event may enhance the formation and stability of the active ORF50/RBPJκ/DNA ternary complex. The recognition sequence of RBP-Jκ protein in target DNA may be changed upon the recruitment of ORF50 protein (model III). During the period of the ORF50-mediated transcriptional activation, the specific flanking sequence may provide a new binding sequence for RBP-Jκ.
Activation of the ORF47 promoter by ORF50 protein is dependent on the expression of RBP-Jκ
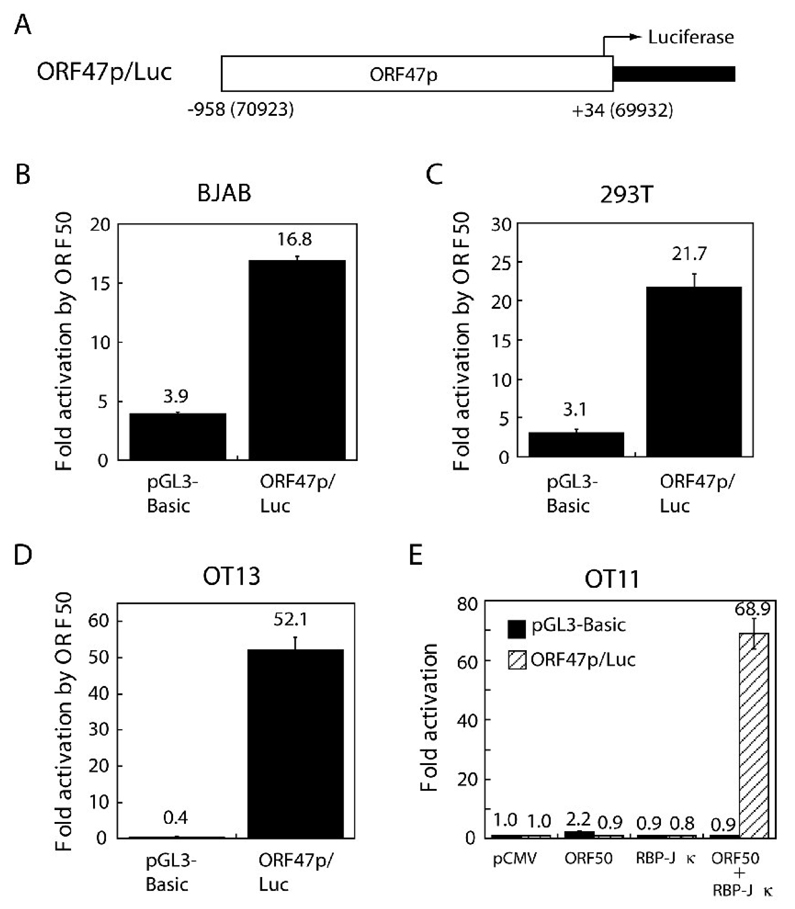
The experiment illustrated in Fig. 4 demonstrated that RBP-Jκ protein bound to the ORF50 RE of the ORF47 promoter, but did not address the critical functional role of RBP-Jκ protein in activation of the ORF47 promoter by ORF50. A reporter was constructed in which ORF47p was fused to luciferase (Fig. 6A). A reporter assay was conducted in 4 different cell backgrounds, including BJAB, 293T, OT11 (an RBP-Jκ-null cell line) and OT13 (a wild-type counterpart cell line of OT11). Co-tranfection of the ORF47p/ luciferase reporter with an expression vector for ORF50 revealed a 17-, 21- and 52-fold enhancement of luciferase expression in BJAB, 293T and OT13 cells. ORF50 protein activated ORF47p/luc at background levels in OT11 cells (Fig. 6). Transactivation of the ORF47p/Luc reporter by ORF50 in OT11 cells could be rescued by ectopic expression of RBP-Jκ (Fig. 6E). This result indicated that RBP-Jκ plays an essential role in activation of the ORF47 promoter by ORF50.
Fig. 6.
RBP-Jκ contributes to activation of the ORF47 promoter by ORF50. (A) A luciferase reporter construct containing the ORF47 promoter. The region from −958 to +34 of the ORF47 promoter was cloned 5’ to the luciferase gene of pGL3-Basic (Promega). The resulting plasmid pORF47p/luc and the control vector (pGL3-Basic) were assessed for ORF50-responsiveness in BJAB (B), 293T (C), OT13 (D) and OT11 cells (E). OT11 is an RBP-Jκ knockout mouse fibroblast cell line. A rescue experiment was carried out with the RBP-Jκ expression vector introduced into OT11. All cells were harvested 48 h post-transfection. Fold-activation was calculated by comparing luciferase activity stimulated by effectors to that stimulated by the control vector. The values of fold- activation represent at least three independent transfections with duplicate samples in each transfection.
The consensus RBP-Jκ recognition site and 5’ flanking sequence in the A-RBPJκ element are required for an optimal ORF50 response
To determine the minimal nucleotide requirement of the A-RBPJκ element for ORF50 responsiveness, deletions or point mutations in the A-RBPJκ element were created in the reporter plasmid (Fig. 7A). Mutation of the RBP-Jκ site in the A-RBPJκ element resulted in complete loss of ORF50 activation [Fig. 7A, A-RBPJκ(m)]. Point mutation in the RBP-Jκ recognition site also eliminated specific binding by RBP-Jκ protein (Fig. 4C and Fig 7B). These results confirmed that binding of RBP-Jκ protein to the response element correlated with trans-activation by ORF50. Deletions 5’ and 3’ to the consensus RBPJκ site in the A-RBPJκ element in pE4CAT were also examined for their responses to ORF50 protein. Deletions of all but one nucleotide of 3’ flanking sequences did not appreciably alter the response to ORF50; however, 5’ deletions reduced the response to ORF50 by 72% (Fig. 7A, 5’-D17). Thus, optimal ORF50 transactivation required both the RBP-Jκ binding site and 5’ flanking sequences in ORF47p. In EMSA experiments with constructs containing 5’deletions in ORF47p the formation of RBP-Jκ/DNA complex A4 and complex A1 were diminished (Fig. 7A; data not shown).
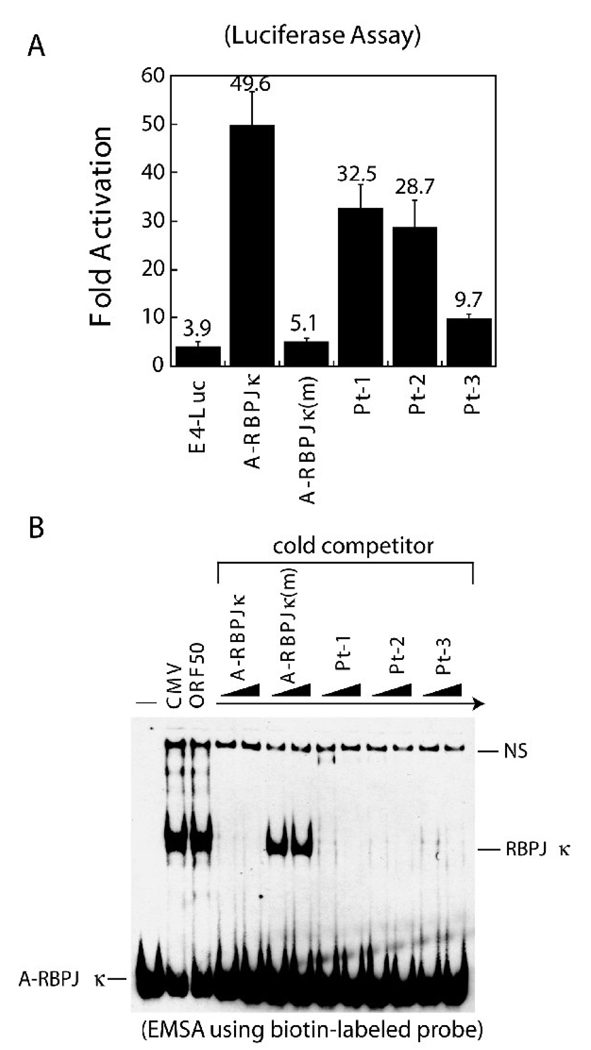
Point mutations in the 5’ flanking sequences were characterized for ORF50 responsiveness and for binding of cellular proteins. In CAT reporter assays, adjacent two nucleotide point mutations in the 5’ flanking region decreased ORF50 transactivation by 47%, 60% and 87% in mutants Pt-1, Pt-2 and Pt-3, respectively (Fig. 7B). ORF50-mediated transactivation was dramatically reduced in the Pt-3 mutant promoter; yet this mutant retained strong DNA binding affinity for RBP-Jκ protein (Fig. 7A and 7B). To confirm these results, the luciferase reporter system was used for further examining the importance of these mutations in ORF50-mediated transactivation (Fig. 8A). As shown in Fig. 8A, the luciferase assay also showed that mutations Pt-3, directly flanking the RBP-Jκ recognition site of the A-RBPJκ element, reduced ORF50 responsiveness by 80%. The DNA-binding assay with a radio-labeled probe was repeated in an EMSA experiment using a biotin-labeled probe (Fig. 8B). Identical results were obtained when compared to that using a radioactive probe (Fig. 7B). Mutants in A-RBPJκ, such as Pt-3, which were markedly reduced in transcriptional response to ORF50 still bound strongly to RBP-Jκ protein in EMSA. These results indicate that binding of RBP-Jκ protein to the target DNA is separable from trans-activation by ORF50.
Fig. 8.
Functional characterization of the 5’ flanking sequences in the A-RBPJκ (−139 to −100) element by luciferase assay and by non-isotopic EMSA. (A) Wild type A-RBPJκ (−139 to −100) element and the mutant elements (Pt-1, Pt-2 and Pt-3 as shown in Fig. 7A) were inserted in pE4-Luc, a plasmid containing the luciferase reporter gene. A luciferase assay was carried out in HKB5/B5 cells 48 hr post-transfection. Relative luciferase activities were measured as fold-activation by ORF50 from at least three independent transfections. (B) EMSA using a biotin-labeled probe. The annealed double-stranded oligonucleotide was labeled with biotin-11-UTP using terminal deoxynucleotidyl transferase. Extracts of HKB5/B5 cells transfected with pCMV or pCMV-ORF50 were prepared for EMSA. Cold competitors used in EMSA were in 50- and 100-fold molar excess over the hot probe. NS: nonspecific complex.
RBP-Jκ-binding elements from human DNA, homologous to those in ORF47p, confer ORF50 responsiveness
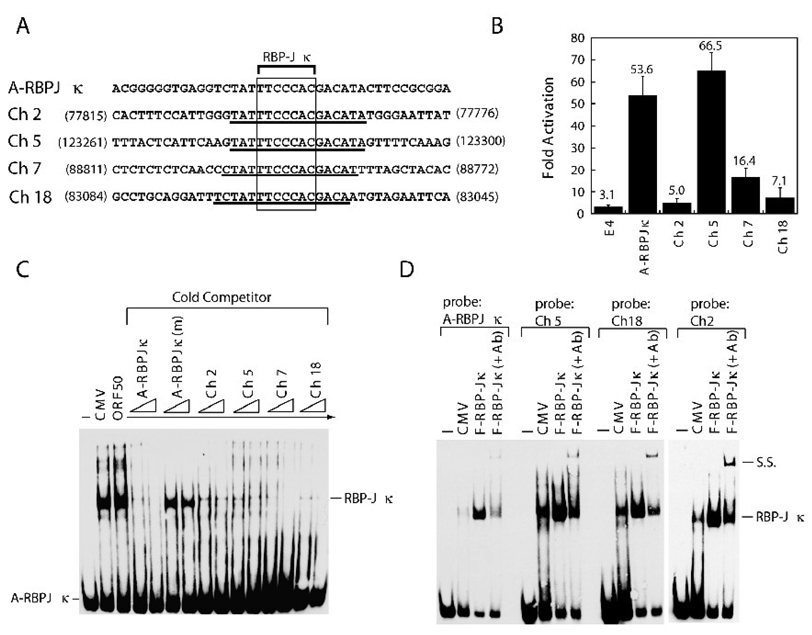
The results from Fig. 7 and 8 show that not all RBP-Jκ-binding elements conferred equivalent responsiveness to ORF50. This phenomenon was validated with four DNA sequences resembling the A-RBPJκ element of ORF47p that were identified in the human genome (Fig. 9). Each homologous element contained a consensus RBP-Jκ recognition site and shared at least 16 consecutive identical nucleotides with the A-RBPJκ element of ORF47p (Fig. 9A). The sequences from cellular DNA shared from 3 to 5 nucleotides in the 5’ flanking region and from 4 to 6 nucleotides in the 3’ flanking region with the A-RBPJκ element. In luciferase reporter assays, these cellular RBP-Jκ elements displayed heterogeneous responses to ORF50 protein. The elements from chromosome 2 and 18 exhibited little response to ORF50, while the element from chromosome 5 was strongly responsive to ORF50 protein (Fig. 9B). When these homologous elements were used as cold competitors in EMSA, all human DNA elements blocked formation of complex A4 as efficiently as did the A-RBPJκ element from ORF47p (Fig. 9C). Direct binding of RBP-Jκ protein to these elements was further demonstrated using ectopically expressed RBP-Jκ protein (Fig. 9D). Taken together, these results confirmed that not all DNA sequences that efficiently bind RBP-Jκ protein confer a transcriptional response to ORF50 protein.
Fig. 9.
ORF47p-related elements from human DNA confer ORF50-responsiveness. (A) Alignment of DNA sequences of the ORF47p-related elements. Four elements encompassing sequences homologous to the A-RBPJκ (−139 to −100) element were identified from human chromosome 2, 5, 7 and 18 by nucleotide-nucleotide BLAST (NCBI). Numbers indicate the nucleotide positions of the individual sequences in databank. Accession no: AC082651.6 (Ch 2); AC091907.4 (Ch5); AC002454.1 (Ch 7); AC091691.7 (Ch 18). (B) Response of the ORF47p-related elements in pE4-Luc to ORF50 protein. The luciferase assays were performed in HKB5/B5 cells. (C) Cross-competition analysis in EMSA. A biotin-labeled A-RBP-Jκ probe was used in EMSA. The indicated cold competitors used in EMSA were in 25- and 50-fold molar excess over probe DNA. (D) Binding of RBP-Jκ protein to the ORF47p-related human DNA elements. All probes were labeled with biotin. Cell extracts of HKB5/B5 transfected with pCMV or pCMV-FLAG-RBP-Jκ were used in EMSA. Antibody to FLAG was used to supershift the protein/DNA complex. SS: supershifted complex
Discussion
This report provides new information about regulation of expression of the ORF47 gene of KSHV and describes novel findings about the mechanisms of activation of the ORF47 promoter (ORF47p) through interactions of ORF50 protein and RBP-Jκ. Three novel findings about the role of RBP-Jκ in activation of ORF47p by ORF50 include the following: i) While RBP-Jκ is essential for activation of ORF47p by ORF50 protein, neither the presence of RBP-Jκ consensus sequences in the promoter nor binding of RBP-Jκ to a DNA sequence ensures a robust transcriptional response to ORF50 protein. ii) Sequences flanking the RBP-Jκ consensus site contribute to activation by ORF50 protein. iii) Cellular DNA contains RBP-Jκ consensus sequences, flanked by sequences that are homologous to those present in the ORF47 promoter. All of these cellular sequences bind RBP-Jκ, but only some of them respond strongly to ORF50 protein.
Regulation of Expression of ORF47
ORF47, which encodes a homologue of herpesvirus glycoprotein L, behaves with early gene kinetics (Fig. 2). In Herpes simplex virus and Pseudorabies virus, the equivalent gene UL1 is classified as a member of the βγ or early-late gene family (Dean and Cheung, 1993; Rajcani et al., 2004; Singh and Wagner, 1993), but little information is available about the kinetic class of gL in the gamma herpesviruses of which KSHV is a member. Although the mRNA that encodes ORF47 (gL) is expressed at an early kinetic stage, the pattern and kinetics of gL protein expression remains to be determined. Further experiments aimed at understanding the correlation between the kinetics of expression of gL mRNA and protein will be needed to elucidate whether gL is subject to post-transcriptional regulation.
The lytic 2.7-kb ORF47 transcript is a component of a polycistronic early leftward mRNA encompassing ORFs 47, 46, and 45 (Fig.1). (More details about the structure of this transcript will be provided separately). In simian varicella virus ORF47 and ORF46 (uracil DNA glycosylase) share a common 5’RNA start site (Ashburn and Gray, 2002). We map the start site of ORF47 transcription and we identify a sequence, located in the promoter of the ORF 47 gene, −139 to −100 relative to the start of transcription, that confers activation by ORF50 protein (Fig. 3). Neither wild-type ORF50 protein, nor a mutant of ORF50 protein (K527E/K528E) that is enhanced in binding to DNA (Chang and Miller, 2004; Chang et al., 2008) detectably binds in EMSA to the ORF50 response element in the ORF47 promoter (Fig. 4). Moreover, we find that the ORF50 response element in ORF47p can be activated by a mutant of ORF50 protein (R161A) that does not bind to DNA (Fig. 5). Therefore, we conclude that ORF47p is activated by an indirect mechanism, rather than by ORF50 binding directly to DNA.
RBP-Jκ plays an essential role in activation of ORF47p by ORF50 protein
The ORF50 response element from −139 to −100 in ORF47p contains an RBP-Jκ consensus sequence that is homologous to RBP-Jκ sites in the promoters of two other KSHV genes, ORF57 and vMIP-1, that are activated by ORF50 through an indirect mechanism (Fig. 3B). Therefore we designate the ORF50 response element, A-RBPJκ. Another RBP-Jκ consensus sequence, GTGGGAA was located upstream at −188 to −149. This element, that we designated B-RBPJκ, conferred only a minimal 5-fold response to ORF50 protein that was 4% of the response of A-RBPJκ (Fig. 3C). The B-RBPJκ element also bound RBP-Jκ protein with considerably weaker affinity than did A-RBPJκ (Fig. 4). The A- and B-RBPJκ elements also bound different cellular proteins. Therefore not all DNA elements that contain a consensus RBP-Jκ sequence confer an equivalent response to ORF50 protein and not all RBP-Jκ sequences are equally capable of binding RBP-Jκ protein. Binding of other cellular proteins may affect the binding of RBP-Jκ, ORF50 protein and the activation response.
Nonetheless, the capacity to bind RBP-Jκ is an important component of an indirect ORF50 response element. A mutant A-RBPJκ DNA element, with 3 nucleotide changes in the RBP-Jκ consensus, that has little or no capacity to bind RBP-Jκ, does not confer a transcriptional response to ORF50 protein (Fig.7 and 8). ORF50 protein does not activate ORF47p in mouse cells with a knock-out of RBP-Jκ (Fig.6). This deficiency can be rectified by providing RBP-Jκ in trans.
Neither an RBP-Jκ consensus site nor RBP-Jκ binding is sufficient to provide a robust response to ORF50 protein
The finding (Fig. 3) that the B-RBPJκ element in ORF47p was very weakly activated by ORF50 protein prompted us to search for adjacent elements that might influence the response. In preliminary experiments conducted by one of us (Joseph Boonsiri) we found, using chimeric reporter constructs with portions of A-RBPJκ fused to B-RBPJκ, that the 5’flanking sequences of A-RBPJκ were essential to confer a full response to ORF50 (data not shown). The resulting chimeric construct was similar to mutants 3’-D15 or 3’-D7 (Fig. 7A) in which sequence deleted from the promoter was replaced with sequence provided by vector DNA. These mutants also conferred a wild-type response to ORF50 protein. In further work (Fig. 7 and data not shown) we found that deletions of the flanking sequences 5’ to the A-RBPJκ site had a greater effect than those 3’ to the site in reducing activation by ORF50 protein. Moreover, point mutations in the 5’ flanking sequence of A-RBPJκ reduced the response (Fig. 7 and 8). However, a surprising finding was that these 5’ point mutations, even those such as Pt-3 that reduced the response to ORF50 by more than 85%, did not significantly affect binding of RBP-Jκ to the A-RBPJκ element (Fig. 7 and 8).
The two nucleotide substitutions in mutant Pt-3 were more deleterious to a response to ORF50 protein than was the entire 5’ deletion in mutant 5’-D15 (Fig. 7A). This paradoxical result can be explained by vector DNA sequences present in the 5’ deletion construct 5’-D15. In this mutant vector DNA has provided 4 nucleotides that are identical to those present in the ORF47 promoter. The sequence of the 5 nucleotides 5’ to the RBP-Jk site was TCTAT while the 5’flanking sequence of the 5’-D15 construct was TCTAG. The comparable flanking sequence of the two-residue substitution mutant Pt-3 was TCTTC. It is likely that the replaced vector sequence has corrected the 5’ deletion.
Since RBP-Jκ binding sites are not restricted to the viral genome, we extended the scope of our study by examining the relationship among ORF50, RBP-Jκ and cellular DNA. The correlation between binding of RBP-Jκ and response to activation by ORF50 protein was examined in studies of cellular DNA sequences that contained consensus RBP-Jκ sites flanked by short stretches of sequences identical to those in A-RBPJκ. All four cellular DNA sequences efficiently bound RBP-Jκ protein in EMSA, whether assessed as cold-competitors of A-RBPJκ binding or directly as probes for binding RBP-Jκ protein (Fig. 9). However, it is not yet known whether the interactions of ORF50 protein with these cellular RBP-Jκ sites control cellular gene expression or affect viral replication.
These experiments confirmed the conclusion that binding of RBP-Jκ to DNA is not the sole determinant of trans-activation by ORF50 protein. Two previous studies support this conclusion. In studies of the ORF57 promoter Lukac et al. (2001) and Carroll et al. (2006) described mutations flanking the consensus RBP-Jκ site that resulted in reduction of response to ORF50 protein. However, these studies provided limited information about the effect of these mutations on binding of RBP-Jκ protein. Papugani et al. (2008) showed that a DNA binding-defective mutant of ORF50 (K152E) was capable of forming a DNA/RBP-Jk/ORF50 ternary complex, but was not sufficient to mediate ORF50 responsiveness. Thus, after full assembly of the ternary complex recruitment of additional co-factors may be essential to promote activation.
Models for the role of sequences flanking the core RBP-Jκ element
We propose three models to explain the importance of sequences flanking the RBP-Jκ core element in mediating activation by ORF50 protein (Fig. 10). In the first model, one or more auxiliary proteins binds the flanking sequence and is required for enhancing the formation of the ORF50/RBP-Jκ/DNA ternary complex and for subsequent ORF50 activity. Since elimination of the core RBP-Jκ sequence destroys the response to ORF50, binding of the auxiliary factor(s) alone to target DNA is not sufficient to confer a response to ORF50. The only possible clue to the identity of such an auxiliary protein, in the data presented here, is that two of the three point mutants in flanking DNA sequence that reduce ORF50 activity also are deficient in binding to protein in complex A1 (Fig. 7). However, Pt-3, which is most deficient in activation by ORF50, appears to bind protein A1 at levels comparable to wild-type A-RBPJκ. There may be more than one auxiliary protein.
In the second model, the flanking sequence may provide a transient docking site for ORF50 protein. Such an interaction may favor and enhance the association between ORF50 and RBP-Jκ. Such a model would predict a direct interaction between ORF50 and cognate DNA. We could not detect any direct interaction between ORF50 protein and target DNA in our experiments using EMSA (Fig. 4A). However, we cannot rule out a weak association between ORF50 protein and DNA that cannot be detected using EMSA with cell extracts. Previously Liao et al. showed that partially purified ORF50 protein could bind to tandem arrays of A/T rich trinucleotide motifs in vitro (Liao et al., 2003). One such A/T trinucleotide motif is present immediately 5’ of the RBP-Jκ core element in the ORF47p (Fig. 3B). Pt-3 destroys this motif.
In the third model, recruitment of ORF50 protein to the target promoter by RBP-Jκ protein may induce a conformational change of RBP-Jκ. The new configuration of RBP-Jκ may be competent to interact with an expanded region of DNA. In effect, recruitment of ORF50 protein may create a new upstream binding site for RBP-Jκ. None of these models is mutually exclusive. To better distinguish among these models in future studies, DNase footprinting with purified proteins may elucidate the detailed interaction between RBP-Jκ protein and target DNA in the presence or absence of ORF50 protein.
In summary, our study shows that binding of RBP-Jκ to the ORF47p, while a critical component of activation by ORF50, by itself is not sufficient to confer a transcriptional response. Sequences flanking the RBP-Jκ core element are essential for regulating the magnitude of the response and presumably for selecting those viral and cellular gene targets whose expression is modulated by ORF50 protein.
Materials and Methods
Cell cultures and transfections
BJAB cells are derived from a B-cell lymphoma and grown in RPMI 1640 medium with 8% fetal bovine serum (FBS). 293T is human embryonic kidney cell line transformed with the E1 region of adenovirus and the simian virus 40 T antigen. The 293T cells were grown in high-glucose DMEM supplemented with 10% FBS. HKB5/B5 is an EBV-negative cell line formed by fusion of HH514-16 cells with 293T cells (Chang et al., 2002). HH-B2 cells are derived from a primary effusion lymphoma infected with KSHV (Gradoville et al., 2000). HKB5/B5 and HH-B2 cells were cultured in RPMI 1640 medium supplemented with 8% and 15% FBS, respectively. Mouse RBP-Jκ−/− (OT11) and wild type (OT13) embryonic fibroblast cell lines (Kato et al., 1997) were kindly provided by T. Honjo (Kyoto University, Kyoto, Japan) and were grown in high-glucose DMEM supplemented with 10% FBS and 100 U/ml of mouse interferon gamma. Transfection assays were performed in BJAB and HH-B2 cells using electroporation, in HKB5/B5 using DMRIE-C reagent (Invitrogen), and in 293T, OT11 and OT13 using Lipofetamine 2000.
Plasmid construction
For construction of CAT reporter plasmids, DNA fragments of the KSHV ORF47 promoter were amplified by PCR from pORF50p(−3870/+20)/CAT (Chang et al., 2005) and cloned into pCAT-Basic (Promega) digested with HindIII and PstI. The plasmids including pE4CAT, pPAN(−91/−58)/E4CAT, pK12(−105/−72)/E4CAT, pCMV-ORF50, pCMV-ORF50(KK/EE), pCMV-ORF50(R161A) and pCMV-FLAG-RBPJκ were described previously (Chang and Miller, 2004; Chang et al., 2002). The PCR-amplified DNA fragments or double-stranded oligonucleotides encompassing the RBP-Jκ recognition site of the ORF47 promoter were cloned into pE4CAT digested with HindIII and XbaI. For constructing the luciferase reporter plasmid, the promoter region from −958 to +34 of the ORF47 gene was cloned into pGL3-Basic (Promega). The reporter plasmid pE4-Luc was constructed by inserting a minimal E4 promoter of adenovirus into the pGL3-Basic. Various double-stranded oligonucleotides with a consensus RBP-Jκ site were cloned into pE4-Luc digested with NheI and XhoI.
CAT and luciferase assays
In CAT assays, BJAB cells (1.0 × 107) were transfected with 5 µg of pCMV-ORF50 or pCMV vector and 5 µg of reporter DNA. CAT activity was determined as described elsewhere (Serio et al., 1997). Activation was calculated as percent acetylation of chloramphenicol in the presence of activator divided by percent acetylation in the presence of control vector. In luciferase assays, 293T, HKB5/B5, OT11 and OT13 cells (7 × 105) were transfected with a fixed amount (1 µg) plasmid DNA, including effector plasmids and the reporter. The reporter assays were carried out according to manufacturer’s protocol for the luciferase reporter assay system (Promega).
Rapid amplification of 5’ cDNA ends
To map the transcriptional initiation site of the ORF47 gene, the GeneRacer kit (Invitrogen) was used. Briefly, HH-B2 cells were treated with 3 mM sodium butyrate for 40 hr and total RNA was prepared from the treated cells. Two µg of total RNA was treated with calf intestinal phosphatase to remove the 5’ phosphates of truncated mRNA and non-mRNA. After decapping of full-length mRNA by tobacco acid pyrophophatase, an RNA oligonucleotide was ligated to the mRNA by T4 RNA ligase. Reverse transcription was performed to obtain the first-strand cDNA by random primers. Subsequent PCR (or nested PCR) amplification was carried out using an ORF47 specific primer 5’CTTTTGACCTGCGTGCGC (or nested ORF47 specific primer 5’CTGCTTTTAGCCCGAGTCTGACTT) and the primers provided by kit. The amplified DNA fragments were cloned and sequenced. Nine out of the 10 clones contained the same initiation site at nucleotide 69965 of KSHV genome (Russo et al., 1996).
Electrophoretic mobility shift assays (EMSAs)
Cell extracts prepared from transfected HKB5/B5 cells were used for EMSAs. HKB5/B5 cells were suspended in a lysis buffer (0.42 M NaCl, 20 mM HEPES [pH 7.5], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 µg aprotinin per ml). Lysates were centrifuged at 90,000 rpm at 4 °C for 15 min in a benchtop ultracentrifuge and supernatants were harvested and stored at −80 °C. Annealed double-stranded oligonucleotides were end-labeled with 32P using T4 polynucleotide kinase or labeled with biotin-11-UTP using terminal deoxynucleotidyl transferase (PIERCE). Binding reactions contained 10 to 15 µg of protein extract in a solution containing 10 mM HEPES (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 2.5 µM ZnSO4, 0.5 mM EDTA, 1 mM dithiothreitol, 15% glycerol and 0.5 µg poly(dIdC) in a total volume of 20 µl. For competition assays, unlabeled competitor DNA was added to the initial reaction mix. Anti-FLAG antibody to the FLAG-RBPJκ protein was used for supershift analysis.
Northern blot analysis
HH-B2 cells were treated with 3 mM sodium butyrate in the presence or absence of 200 µg phosphonoacetic acid (PAA) per ml for 12, 24, 36 and 48 hrs. Total cellular RNAs from the treated HH-B2 cells (1.2 × 107) were prepared with an RNeasy kit (QIAGEN), fractionated on 1% formaldehyde-agarose gels, and transferred to nylon membranes (Hybond-N; Amershann Pharmacia Biotec). All probes were labeled by the random-primed method. Detection of the ORF50, K8 and K8.1 RNAs has been described previously (Chang et al., 2005). For detection of ORF47 RNA, DNA fragments corresponding to nucleotides 69421 to 69917 of KSHV genome was used as a probe. Hybridization was carried out in 6X SSC (1X SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5X Denhard’s solution, 0.5% SDS, and 100 µg of salmon DNA per ml at 60°C overnight. Membranes were washed in 2X SSC-0.5% SDS once for 10 min and in 0.1X SSC-0.5% SDS three times for 25 min at 60°C.
Acknowledgements
This work was supported by grants CA16038 and CA12055 from NIH to G.M. and by a medical research grant CMRPD650013 from Chang-Gung Memorial Hospital (Taiwan) and by a grant NSC95-2320-B-182-054-MY3 from National Science Council (Taiwan) to P-J C. We thank Dr. Tasuku Honjo (Kyoto University) for mouse RBP-Jκ−/− (OT11) and wild type (OT13) embryonic fibroblast cells. We thank Jianjiang Ye for critical reviews of the manuscript and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburn CV, Gray WL. Expression of the simian varicella virus glycoprotein L and H. Arch Virol. 2002;147(2):335–348. doi: 10.1007/s705-002-8323-6. [DOI] [PubMed] [Google Scholar]
- Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O'Leary JJ. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1(12):1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- Carroll KD, Bu W, Palmeri D, Spadavecchia S, Lynch SJ, Marras SA, Tyagi S, Lukac DM. Kaposi's Sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the Notch pathway. J Virol. 2006;80(19):9697–9709. doi: 10.1128/JVI.00746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS- related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chang PJ, Miller G. Autoregulation of DNA binding and protein stability of Kaposi's sarcoma-associated herpesvirus ORF50 protein. J Virol. 2004;78(19):10657–10673. doi: 10.1128/JVI.78.19.10657-10673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Gradoville L, Cho MS, Chen LW, Chang J, Miller G. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J Virol. 2002;76(7):3168–3178. doi: 10.1128/JVI.76.7.3168-3178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Miller G. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J Virol. 2005;79(14):8750–8763. doi: 10.1128/JVI.79.14.8750-8763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Miller G. A mobile functional region of Kaposi's sarcoma-associated herpesvirus ORF50 protein independently regulates DNA binding and protein abundance. J Virol. 2008;82(19):9700–9716. doi: 10.1128/JVI.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chen J, Ueda K, Sakakibara S, Okuno T, Yamanishi K. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J Virol. 2000;74(18):8623–8634. doi: 10.1128/jvi.74.18.8623-8634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B, Jeong JH, Bowser BS, DeWire SM, Staudt MR, Dittmer DP. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J Virol. 2004;78(10):5491–5499. doi: 10.1128/JVI.78.10.5491-5499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean HJ, Cheung AK. A 3' coterminal gene cluster in pseudorabies virus contains herpes simplex virus UL1, UL2, and UL3 gene homologs and a unique UL3.5 open reading frame. J Virol. 1993;67(10):5955–5961. doi: 10.1128/jvi.67.10.5955-5961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Song MJ, Chu JT, Sun R. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) J Virol. 2002;76(16):8252–8264. doi: 10.1128/JVI.76.16.8252-8264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Young A, Sun R. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J Gen Virol. 2000;81(Pt 12):3043–3048. doi: 10.1099/0022-1317-81-12-3043. [DOI] [PubMed] [Google Scholar]
- Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74(13):6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74(6):2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol. 2001;75(4):1798–1807. doi: 10.1128/JVI.75.4.1798-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124(20):4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3(9):840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002;16(15):1977–1989. doi: 10.1101/gad.996502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ganem D. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc Natl Acad Sci U S A. 2003;100(14):8490–8495. doi: 10.1073/pnas.1432843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ganem D. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J Virol. 2004;78(13):6818–6826. doi: 10.1128/JVI.78.13.6818-6826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Tang Y, Kuo YL, Liu BY, Xu CJ, Giam CZ. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J Virol. 2003;77(17):9399–9411. doi: 10.1128/JVI.77.17.9399-9411.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Y, Liang D, Gao Y, Xia T, Robertson ES, Lan K. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element. Virology. 2008;380(2):264–275. doi: 10.1016/j.virol.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. Dna binding by kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J Virol. 2001;75(15):6786–6799. doi: 10.1128/JVI.75.15.6786-6799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73(11):9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252(2):304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71(1):314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papugani A, Coleman T, Jones C, Zhang L. The interaction between KSHV RTA and cellular RBP-Jkappa and their subsequent DNA binding are not sufficient for activation of RBP-Jkappa. Virus Res. 2008;131(1):1–7. doi: 10.1016/j.virusres.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajcani J, Andrea V, Ingeborg R. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes. 2004;28(3):293–310. doi: 10.1023/b:viru.0000025777.62826.92. [DOI] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2(3):342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci U S A. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Kolman JL, Miller G. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol. 1997;71(11):8726–8734. doi: 10.1128/jvi.71.11.8726-8734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Wagner EK. Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology. 1993;196(1):220–231. doi: 10.1006/viro.1993.1470. [DOI] [PubMed] [Google Scholar]
- Song MJ, Brown HJ, Wu TT, Sun R. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75(7):3129–3140. doi: 10.1128/JVI.75.7.3129-3140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Deng H, Sun R. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 2003;77(17):9451–9462. doi: 10.1128/JVI.77.17.9451-9462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Li X, Brown HJ, Sun R. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 2002;76(10):5000–5013. doi: 10.1128/JVI.76.10.5000-5013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86(4):1276–1280. [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma- associated herpesvirus. Proc Natl Acad Sci U S A. 1998;95(18):10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73(3):2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22(6):965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Ishikawa K, Nishimura K, Sakakibara S, Do E, Yamanishi K. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J Virol. 2002;76(23):12044–12054. doi: 10.1128/JVI.76.23.12044-12054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu S, Wu M, Geng Y, Wood C. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch Virol. 2001;146(7):1415–1426. doi: 10.1007/s007050170102. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yuan Y. Essential role of RBP-Jkappa in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology. 2007;359(1):19–27. doi: 10.1016/j.virol.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chiu J, Lin JC. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 1998;17(9):735–742. doi: 10.1089/dna.1998.17.735. [DOI] [PubMed] [Google Scholar]
- Zhu FX, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma- associated herpesvirus. J Virol. 1999;73(7):5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer J, Grundhoff A, Ganem D. Exploring the DNA binding interactions of the Kaposi's sarcoma-associated herpesvirus lytic switch protein by selective amplification of bound sequences in vitro. J Virol. 2006;80(6):2958–2967. doi: 10.1128/JVI.80.6.2958-2967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]