Abstract
Measurement of brain change due to neurodegenerative disease and treatment is one of the fundamental tasks of neuroimaging. Deformation-based morphometry (DBM) has been long recognized as an effective and sensitive tool for estimating the change in the volume of brain regions over time. This paper demonstrates that a straightforward application of DBM to estimate the change in the volume of the hippocampus can result in substantial bias, i.e., an overestimation of the rate of change in hippocampal volume. In ADNI data, this bias is manifested as a non-zero intercept of the regression line fitted to the 6 and 12 month rates of hippocampal atrophy. The bias is further confirmed by applying DBM to repeat scans of subjects acquired on the same day. This bias appears to be the result of asymmetry in the interpolation of baseline and followup images during longitudinal image registration. Correcting this asymmetry leads to bias-free atrophy estimation.
Keywords: Deformation-Based Morphometry, Longitudinal Image Registration, Neurodegenerative Disorders, Alzheimer’s Disease Neuroimaging Initiative, Unbiased Estimation, Neuroimaging Biomarkers
1 Introduction
Neuroimaging will play an important role in future clinical trials of disease-modifying treatments for Alzheimer’s disease (AD) and other neurodegenerative disorders. One of the great promises of neuroimaging is that it will allow shorter and smaller clinical trials, thus reducing the costs of developing a successful treatment. Macroscopic changes in brain anatomy, detected and quantified by magnetic resonance imaging (MRI), consistently have been shown to be highly predictive of AD pathology and highly sensitive to AD progression (Scahill et al., 2002; de Leon et al., 2006; Jack et al., 2008b; Schuff et al., 2009). Compared to clinical measures and neuropsychological testing, MRI-derived biomarkers require an order of magnitude smaller cohort size to detect disease-related changes over time. Theoretically, such biomarkers will be equally effective in detecting the effects of disease-modifying treatments, and will allow smaller and shorter clinical trials.
Deformation-based morphometry (DBM) is a widely used and cost-effective technique for estimating longitudinal brain atrophy (Chung et al., 2001; Studholme et al., 2004; Leow et al., 2006). To measure atrophy in a given anatomical structure across two time points with DBM, one must (1) label the structure of interest in the baseline image; (2) perform deformable image registration between the baseline image and the followup image; (3) measure the change in volume induced by the deformation on the structure of interest. With many automatic segmentation and registration algorithms available as free software, DBM has become a very accessible and low-cost technique for longitudinal image analysis. DBM also offers advantages in terms of statistical power, particularly when compared with the frequently used alternative (e.g., recent work on hippocampal atrophy by Schuff et al. (2009)) of segmenting the structure of interest in each time point, and taking the difference in the volumes of the segmentations. This alternative is subject to repeat measurement errors, whereas DBM measures the difference between time points more directly.
However, one of the drawbacks of DBM for atrophy estimation is its susceptibility to bias. In general, bias can occur when a system of measurement is not blinded to the independent variables. In the context of a study like (Schuff et al., 2009), the segmentation of the structure of interest in different time points is performed independently; it may even be randomized, and the individuals performing the segmentation may be blinded to avoid bias completely. However, in the context of DBM, it is not as straightforward to blind the method to which image is the baseline image, and which images are followup images. Specific aspects of underlying registration methodology, usually obscured from the user, can cause atrophy to be systematically overestimated or underestimated.
Such bias strongly undermines the utility of DBM in neuroimaging biomarker research. Overestimation of atrophy in a pilot study can cause the subsequent clinical trial to be underpowered, leading to a waste of resources and an unnecessary burden on the patients. Presence of bias also makes it difficult to compare the statistical power of different atrophy estimation methods.
In this paper, we examine the bias associated with DBM in the context of measuring hippocampal atrophy in mild cognitive impairment (MCI) and healthy aging. The data for this study come from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005; Jack et al., 2008a), a large multi-center MRI imaging study. We propose two techniques for measuring bias in estimation of hippocampal atrophy. The first technique examines the intercept of the regression line fitted to atrophy estimates from 6-month and 12-month longitudinal data. The second technique uses repeat scans from a single time point, where we expect to find zero atrophy in the absence of DBM-related bias. With both techniques, we find substantial, statistically significant bias when using “routine” DBM with no built-in bias correction. The bias is of the same order of magnitude as the known rate of hippocampal atrophy in MCI. Bias of this magnitude would lead to severe underpowering of a subsequent clinical trial. 1 In subsequent analysis, we find that DBM-associated bias can be eliminated if the global transformation between the baseline image and the followup image is applied symmetrically. Symmetric application of the deformable transformation between baseline and followup images does not affect the bias significantly in our experiments.
This paper is organized as follows. Section 2 discusses the subset of ADNI data used in this study and the DBM methodology that we employ. Section 3 describes the results of atrophy measurement experiments with and without bias correction. Section 4 discusses how the findings relate to other work on longitudinal brain atrophy estimation, including previous work on unbiased techniques. The conclusions of this paper are in Section 5.
2 Materials and Methods
2.1 Subjects and Imaging Data
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research – approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years.
ADNI MRI data includes 1.5 Tesla structural MRI from all 800 subjects and 3 Tesla structural MRI from 200 subjects. Our study is conducted using only 3 Tesla MRI, and it only includes data from MCI patients and controls. We also use only a subset of the imaging time points in ADNI: baseline, 6 months and 12 months. The demographic characteristics of the subjects whose data are included in this study are given in Table 1.
Table 1.
Summary of the relevant characteristics of the subset of ADNI subjects included in this study.
| Group | N | MMSE | Age (y.) | LHV (mm3) | RHV (mm3) |
|---|---|---|---|---|---|
| MCI | 80 | 26.7 ± 1.9 | 74.3 ± 7.8 | 1455 ± 323 | 1406 ± 348 |
| Control | 57 | 29.4 ± 0.8 | 75.4 ± 4.8 | 1782 ± 277 | 1704 ± 290 |
The MRI imaging protocol for ADNI is described by Jack et al. (2008a). Each session includes a T1-weighted high-resolution MP-RAGE scan, a repeat MP-RAGE scan, a pair of low-resolution B1 calibration scans, and a TSE scan weighted for proton density and T2 contrast. Phantoms are used to ensure scanner parameters and performance remain consistent across imaging sessions. ADNI performs some post-processing of the imaging data. Researchers at the Mayo clinic compare the two MP-RAGE scans acquired in every imaging session and designate one of the scans as having superior quality. The superior scan is then post-processed by ADNI researchers. The specific postprocessing procedures are MRI scanner specific. At the most, they include “corrections in image geometry for gradient nonlinearity, i.e., 3D gradwarp (Hajnal et al., 2001; Jovicich et al., 2006); corrections for intensity nonuniformity due to nonuniform receiver coil sensitivity (Narayana et al., 1988); and correction of image intensity nonuniformity due to other causes such as wave effects at 3 T.” (Jack et al., 2008a). The raw, unprocessed MP-RAGE scans are also available in the ADNI database. We use all three of these images in this study. We refer to the post-processed image as Ipp, the raw superior image as Irs and the raw inferior image Iri.
2.2 Hippocampal Atrophy Estimation with Deformation-Based Morphometry
We begin by describing what we consider the “established” DBM pipeline. Later, we discuss the modifications to the pipeline used to remove bias. The standard DBM pipeline for hippocampal atrophy estimation includes four basic steps:
Segmentation. The left and right hippocampus is labeled in each subject’s baseline image.
Global Registration. The followup image at time t is aligned to the baseline image using a linear global coordinate transformation.
Deformable Registration. A locally varying, high-dimensional, smooth and invertible (i.e., diffeomorphic) transformation is computed between the baseline image and the aligned followup image.
Atrophy Estimation. The change in volume induced by the local transformation is computed throughout the hippocampus ROI and integrated over the ROI to calculate total atrophy.
The sections below describe each of these four steps in slightly more detail. Each step is implemented using freely available open-source tools. Later in the paper, we repeat some of the analysis with alternative tools, and find that the findings largely transcend the choice of tool.
2.2.1 Segmentation
The left and right hippocampal regions of interest (ROI), consisting of the hippocampus proper, dentate gyrus, a small medial portion of the subiculum, and including alveus and some intra-hippocampal cerebrospinal fluid, is segmented in each baseline image. We use a hybrid segmentation approach, where an initial segmentation is computed automatically using landmark-guided registration to a labeled brain atlas. This segmentation is then edited by a trained human operator to produce the final segmentation. This approach saves a great deal of time over fully manual segmentation, without compromising segmentation quality. Our approach is similar to the one used by ADNI researchers at UCSF to segment 1.5 Tesla MRI ADNI data (Schuff et al., 2009; Hsu et al., 2002; Haller et al., 1997). The details of our approach are given in (Pluta et al., 2009).
2.2.2 Global Registration
Global (six or nine-parameter) registration is used to bring the baseline image and the followup image of each subject into global alignment. Global registration is performed using the FLIRT software from the FSL suite (Smith et al., 2004). The algorithm in FLIRT searches for the linear transformation that minimizes the correlation ratio metric between the two images. We specify the baseline image as the reference image and the followup image as the moving image.
In this paper, we primarily use the six-parameter rigid transformation model, because the baseline and followup images are from the same subject. However, following Scahill et al. (2002), Paling et al. (2004), and Leow et al. (2009), we also conduct experiments with a 9-parameter (rigid plus anisotropic scaling) model. Paling et al. (2004) argued that variation in voxel size over time in MRI scanners can account for errors in annual atrophy rates as large as 0.5%, and suggested that global registration with nine degrees of freedom (rigid transformation plus anisotropic scaling) may correct for such changes. However, the authors did not find statistically significant differences between 9 and 6-parameter global transformations. Leow et al. (2009) adopted 9-parameter global transformation in their longitudinal analysis of ADNI data. In one of our experiments below, we compare six and nine-parameter global registration in terms of atrophy estimation bias and power. However, in all other experiments, we use the six-parameter rigid model.
2.2.3 Deformable Registration
Deformable registration computes a spatially varying mapping between a pair of images, such that the similarity between points linked by the mapping is maximized. There are many deformable registration approaches available in the literature: (Christensen et al., 1997; Rueckert et al., 1999; Ashburner and Friston, 1999; Crum et al., 2005; Beg et al., 2005), just to name a few. This paper employs the Symmetric Normalization (SyN) approach by Avants et al. (2008) because of several desirable properties: (1) the algorithm is symmetric with respect to the two input images; changing the order of the images does not affect the mapping computed by SyN; (2) the algorithm guarantees that the mapping is smooth and invertible (i.e., diffeomorphic), and generates an inverse mapping; (3) the algorithm admits a wide range of similarity metrics; (4) the implementation can be used on single-processor computer hardware. In a recent comparison of 14 publicly available software implementations of deformable registration algorithms, SyN was one of the top two performers (Klein et al., 2009).
We give only a brief summary of SyN in this section, referring the reader to (Avants et al., 2008) for a full description of the method. The theoretical foundations of SyN are closely linked to large deformation diffeomorphic metric mapping (Dupuis et al., 1998; Beg et al., 2005). The main distinction is that SyN optimizes an energy function that is defined symmetrically with respect to the input images I and J. This optimization has the form:
| (1) |
subject to
| (2) |
In this formulation, φ1(x, t) and φ2(x, t) are time-dependent mappings of the image domain Ω onto itself, with t ∈ [0, 1] the time variable; v1(x, t) and v2(x, t) are time-dependent vector fields defined on Ω, over which the objective function is minimized; || · ||L denotes the Sobolev norm of a vector field under the differential operator L (see (Dupuis et al., 1998; Beg et al., 2005)); I(x) and J(x) are a pair of images defined on the domain Ω; and Π is an operator that measures dissimilarity between images. Since φi(x, t) are defined as the solutions of the flow ordinary differential equation (2), they are guaranteed to be diffeomorphic if the vector fields v1(x, t) and v2(x, t) are smooth. SyN employs a greedy optimization strategy to find φ1(x, t) and φ2(x, t). Greedy optimization is an alternative to direct optimization over the space of time-varying vector fields vi(x, t), as in (Beg et al., 2005). The greedy approach offers improved computational performance and requires less memory, albeit at the cost of lacking certain attractive theoretical properties of optima computed by direct optimization.
In our experiments, we use SyN with the normalized cross-correlation image match metric, with the radius of four voxels in each dimension. Registration is performed using only one resolution level, at the native resolution of the input images. We do not use the multi-resolution features of SyN because the deformations between the baseline and followup images are very local. The maximum number of iterations allowed in SyN registration is 60. The smoothing applied to the deformation field at each iteration uses a Gaussian kernel with σ = 2.0 mm in each dimension. The baseline and followup images themselves are not smoothed. The step size in the time dimension is 0.2. SyN normalization is performed using the open-source Advanced Normalization Tools (ANTS) software implementation (http://picsl.upenn.edu/ants).
2.2.4 Estimation of Atrophy
To estimate atrophy in the hippocampus between the baseline image and the followup image, we use the following simple approach. We place a volumetric tetrahedral mesh inside of the hippocampus segmentation, and apply the deformation field computed by the registration algorithm to each vertex of the mesh. We measure the volume of each tetrahedron in the mesh before and after the deformation and add up the volumes. We define atrophy as the ratio
where Vbl and Vfu are the volumes of the mesh in the baseline and followup images, respectively.
The mesh-based approach to estimate atrophy is more direct than computing the Jacobian determinant of the deformation field at each voxel of the baseline image and integrating over the hippocampus segmentation. In the context of non-parametric registration methods like SyN, the latter requires finite difference approximation, which requires deformation fields to be very smooth in order to avoid numerical errors.
2.3 Composition of Rigid and Deformable Transformations
A subtle, but very important detail is the way in which global and deformable transformations are combined in this approach. In fact it is this detail that affects whether bias is present in the results of the longitudinal study.
Before we proceed, let us define a notation for image resampling. Given an image I, i.e. a set of values {Ij} defined on a lattice of points {xj}, we define the resampling of image I under transformation ψ as a new image I′= R(I, ψ) given by
where ℒ is the interpolation kernel, e.g., a box function for nearest neighbor interpolation, or a tent function for linear interpolation. Recall that repeated application of interpolation and resampling to an image results in smoothing and/or aliasing, depending on which kernel is used. In this paper we use linear interpolation.
Arguably, the most straightforward strategy to combine global and deformable registration in DBM would be to apply the global transformation T to the followup image, producing a new resampled image R(Ifu, T). Then, the metric computation in SyN would take the form:
This formulation is clearly non-symmetric, since the baseline image would be sampled only once, and the second image would be sampled twice. An alternative is to have SyN compose the global and deformable transformations applied to the followup image, resulting in the following form:
This form is symmetric in the number of resampling operations applied to each image. However, at the beginning of the deformable registration iteration, the baseline image is not really resampled because φ1 is identity, while the followup image undergoes global transformation by T. So some asymmetry remains, and as we see below, this asymmetry contributes to bias.
To eliminate asymmetry, we adopt a simple solution motivated by the work of Guimond et al. (2000), Joshi et al. (2004) and others on unbiased population-specific atlases for image registration. This solution involves splitting the global transformation T into two equal global transformations T1/2, such that T = T1/2 ◦ T1/2. To find T1/2, we write T(x) = Qx + b, where Q is a 3 × 3 matrix of rotation and, for the 9-parameter global transformation, scaling; and b is a translation vector. Then it is easy to verify that the desired transform is given by
| (3) |
where I is the identity matrix and Q1/2 is the matrix square root of Q. The square root of Q can be computed effciently using the Denman and Beavers (1976) iterative algorithm (see Appendix).
By applying T−1/2 to the baseline image and T1/2 to the followup image, and passing the resampled images to SyN, we can make the metric computation truly symmetric:
| (4) |
Lastly, to avoid resampling each image twice, we can have SyN compose the global and non-global transformations during computation, leading to the following symmetric formulation:
| (5) |
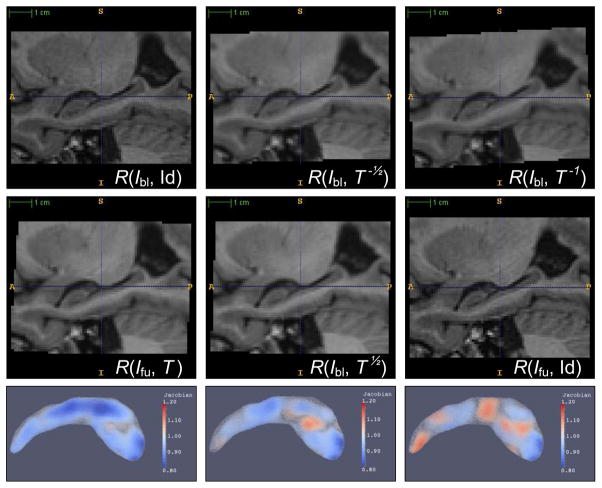
Fig 1 illustrates the effects of applying global rigid registration symmetrically and asymmetrically. Asymmetry in the sampling of image data causes images passed in to SyN to have different intensity characteristics, which leads to different atrophy estimates.
Fig. 1.
Example of DBM configurations with different resampling of the baseline image Ibl and followup image Ifu. In the first column, the global transformation is applied only to Ifu. In the second column, the transformation is split equally between Ibl and Ifu. In the last column, the transformation is applied only to Ibl. The two images in the first column have different degree of smoothing and aliasing, as do the two images in the last column. This leads to bias when registration is used to compute atrophy between these pairs of images. The two images in the middle column have roughly the same degree of smoothing and aliasing. In the bottom row, the Jacobian map resulting from applying SyN to the resampled images is shown. The Jacobian map is computed using a tetrahedral mesh, and is plotted here using volume rendering. Overall, there is most volume reduction in the leftmost column, and least volume reduction in the rightmost column. These columns correspond to FU/HW, HW/HW and BL/HW in Tables 2, 3 and 4.
For completeness, this paper also examines the effect of symmetry in the diffeomorphic registration method on bias. With a small modification to the SyN algorithm, we can implement an asymmetric diffeomorphic registration approach. We simply enforce either v1(x, t) = 0 or v2(x, t) = 0 in (1), which in turn causes either φ1 or φ2 to become identity. Let us call this approach asymmetric normalization (aSyN).
With SyN and aSyN, there are nine different ways in which we can split the transformation between the baseline image and the followup image. The diffeomorphic transformation can be applied to either of the images only (with aSyN) or to both images (SyN). Likewise, the global transformation can be applied to either image, or split into equal half-transformations using (3). The metric computations corresponding to these nine different approaches all have the form
where the transformations ψbl and ψfu can be summarized in a table:
| (6) |
Notice that in all these computations, the global and deformable transformations are composed, and at most one image interpolation is applied to each image. In Sec. 3 we examine the bias associated with each of these nine formulations of registration.
2.4 Alternative DBM Approach
To show that the bias related to asymmetry in image resampling is not unique to SyN, we repeat a subset of the experiments with a different deformable image registration technique. We chose to use the Image Registration Toolkit (IRTK) from IXICO, Inc., which is the official implementation of the B-spline based Free-Form Deformation (FFD) deformable image registration algorithm by Rueckert et al. (1999). The reasons for selecting this particular algorithm included its wide use in the literature, the high rating that it received in the recent evaluation study by Klein et al. (2009), the availability of a free software implementation, and ease of interfacing between IRTK and other tools used in this study.
FFD differs from SyN in several aspects. In FFD, the deformable registration is formulated asymmetrically, i.e., the deformation is applied to one of the images only. The deformation in FFD is parametric and smooth by construction. Smoothness is controlled by the spacing of B-spline control points. The parameters of the FFD algorithm were largely set to their defaults, with the following exceptions. As in SyN, registration was performed at the native image resolution; i.e., the multi-resolution registration scheme was not employed. This is due to the very local nature of the anatomical changes that the registration is intended to measure. The B-spline control point spacing was set to 4.8 mm in all three dimensions, allowing for a smooth deformation. The Gaussian blurring parameter for the baseline and followup images was set to 0.6 mm. The normalized mutual information metric (Studholme et al., 1997) was used. We purposely used a different metric from SyN experiments. It is by no means our intention to compare FFD to SyN in terms of registration accuracy or sensitivity to atrophy in MCI. Rather, we aim to demonstrate that the issues of bias in DBM of longitudinal data are not limited to a particular method or a particular metric.
2.5 Direct Estimation of Bias
The ADNI dataset provides a unique opportunity to estimate registration bias in a controlled experiment. Recall from Sec. 2.1 that each ADNI imaging session includes a pair of MPRAGE images, one ranked superior (Irs) and one ranked inferior (Iri). Since no longitudinal changes have taken place between these scans, we would expect the average atrophy detected by the registration to be zero. However, since some systematic differences may be present between the images acquired earlier and later in an MRI scan, or between inferior and superior MRI images, we randomly assign each of two images labels “baseline” and “followup,” and then perform the DBM longitudinal analysis on these data. The only difference between this bias estimation experiment and the actual longitudinal study is that we do not repeat the hippocampus segmentation effort for the former. Hippocampi were segmented in the post-processed “superior” MRI images. We map these segmentations into the images Irs and Iri by global registration to the post-processed image.
3 Experimental Results
3.1 Metrics Used to Compare Transformation Models
We use several metrics to analyze the bias and statistical power of different DBM-based atrophy estimation configurations. Bias can be estimated in two distinct ways. The first way is the direct estimation of bias from the randomized experiment described in Sec. 2.5. We report the mean and standard deviation of the atrophy rate estimated in this experiment for each flavor of DBM discussed above. For a DBM configuration to be unbiased, mean atrophy must not be significantly different from zero.
A complementary way to measure bias uses data from the “real” longitudinal experiment, where atrophy is computed between the baseline image of each subject and the 6 and 12 month followup images. For each group, the intercept of the regression line fitted to the atrophy estimated at the two time points should be zero.2 We report the mean and standard deviation of the intercept value for the MCI group and control group. We also plot the empirical cumulative distribution functions of 6 and 12-month atrophy for the two groups.
To measure the power of DBM-based atrophy estimation, we compare the atrophy rates between control and MCI groups using data from baseline and 12 months. We report the mean and standard deviation of 12-month atrophy for each group, as well as the p-value of the Student t-test with the null hypothesis that the two means are equal, and the alternative hypothesis that atrophy is greater in MCI. We also perform power analysis and report the sample size required to detect a 25% reduction in MCI atrophy relative to the control atrophy with statistical power β = 0.8, significance level α = 0.05 and two-sided alternative hypothesis. The sample size calculation is given by the formula:
| (7) |
where zt is the t-th quantile of the normal distribution, μMCI and μCTL are the estimates of the mean atrophy in MCI and control populations, and σMCI is the estimate of the standard deviation of atrophy in the MCI population. Smaller sample size indicates greater power of the DBM-based atrophy estimation method.
3.2 Asymmetry in Rigid and Deformable Transformations
In Sec. 2.3 we described nine DBM configurations in which global and deformable transformations are divided differently between the baseline image and the followup image. Specifically, each type of transformation can be applied only to the baseline image, only to the followup image, or split equally between the two images. The direct bias estimated for each of these nine con-figurations is shown in Table 2. For each configuration, the table lists the mean atrophy estimated in the bias experiment, the standard deviation, and the p-value from a Student t-test with null hypothesis of no atrophy (i.e., no bias). The results show a clear effect of asymmetry in the global component on the bias. When the global component is applied to the baseline image, there is significant negative bias, and when the global component is applied to the followup image, the bias is significantly positive. When the global component is split equally between the two images, the bias is not significant, except in one configuration, where it reaches significance with p = 0.01.
Table 2.
Direct estimation of bias for nine DBM configurations summarized in Equation (6). The configurations are arranged in a 3 × 3 table. The columns correspond to three different ways to divide the global transformation between the baseline and followup image during registration. Column “BL” indicates that the transformation is applied to the baseline image only; “HW” means the transformation is split halfway between the two images; “FU” means that the transformation is applied to the followup image only. Likewise, the rows correspond to different ways to split the non-global transformation between the images. The cell “HW”,“HW” corresponds to fully symmetric registration. For each cell in the table, the sample mean and standard deviation of atrophy estimated in the bias experiment are given. Additionally, the p-value for the null hypothesis that atrophy is zero (i.e., no bias) is given.
| RIGID |
|||||
|---|---|---|---|---|---|
| BL | HW | FU | |||
| NON-RIGID | BL | μ: | −3.30% | 0.25% | 2.11% |
| σ: | 1.33% | 1.11% | 1.72% | ||
| p -value: | 0.000 | 0.010 | 0.000 | ||
| HW | μ: | −3.04% | 0.08% | 2.87% | |
| σ: | 1.55% | 0.99% | 1.45% | ||
| p -value: | 0.000 | 0.341 | 0.000 | ||
| FU | μ: | −2.34% | 0.03% | 3.12% | |
| σ: | 1.62% | 1.07% | 1.39% | ||
| p -value: | 0.000 | 0.742 | 0.000 | ||
Asymmetry in the deformable component of the transformation does not have as obvious an effect on the bias. When the global transformation is applied asymmetrically, the bias is increased slightly when the deformable transformation is applied on the same side as the global one (cells BL/BL and FU/FU in Table 2), and decreased when the two transformations are applied on opposite sides (cells BL/FU and FU/BL). This effect may be explained by the fact that in configurations BL/BL and FU/FU one of the images is assigned the identity transformation and is not interpolated at all, while in BL/FU and FU/BL both images are interpolated, although asymmetrically.
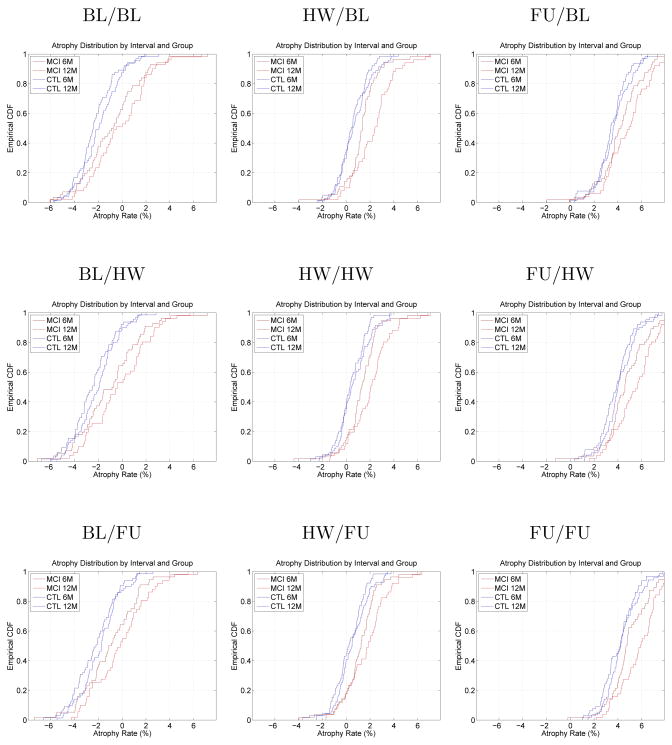
Fig. 2 shows the cumulative distribution plots for 6-month and 12-month atrophy in MCI and control groups. One plot is shown for each of the nine configurations. The plots clearly indicate that in experiments where the global registration is applied asymmetrically, bias is present. This visually confirms the findings from the direct bias estimation experiment in real longitudinal data. Table 3 further confirms this by listing for each cohort the average intercept of the regression line fitted to each subject’s 6-month and 12-month atrophy values. This intercept is an alternative way of estimating bias, and the general sense of the results from the direct bias estimation experiment is maintained. Asymmetrical application of the global transformation results in 2 – 3% bias, while asymmetry in the deformable registration has little e3ect on bias. The bias is of the same order of magnitude for control subjects and MCI patients. A t-test comparing bias between these two cohorts in each of the nine configurations yields two-sided p-values that range from 0.32 to 0.95, indicating that in neither of these configurations the difference in bias between cohorts is significant. This suggests that atrophy comparisons between cohorts should not be significantly affected by the presence of DBM-related bias.
Fig. 2.
Empirical cumulative distribution plots (CDF) of hippocampal atrophy over 6 and 12 months in MCI and control groups. The nine plots correspond to the nine DBM configurations in Equation (6). In absence of bias, we expect the four curves to be centered slightly to the right of the origin. However, in many DBM configurations, the curves are shifted either to the left or to the right, indicating negative or positive bias. The order of the CDF curves (6 month control, 12 month control, 6 month MCI, 12 month MCI) and the separation between them is roughly preserved in all DBM configurations.
Table 3.
Intercept-based atrophy bias estimation in nine configurations of DBM. See caption to Table 2 for the meaning of the rows and columns. Each cell in the table gives the mean intercept value of the regression line fitted to the 6-month and 12-month atrophy values, as well as the standard deviation of the intercept and the p-value of the t-test with the null hypothesis of zero intercept (i.e., no bias).
| MCI GROUP | |||||
|---|---|---|---|---|---|
| RIGID |
|||||
| BL | HW | FU | |||
| NON-RIGID | BL | μ: | −2.19% | 0.49% | 2.39% |
| σ: | 3.62% | 2.50% | 3.58% | ||
| p -value: | 0.000 | 0.122 | 0.000 | ||
| HW | μ: | −2.14% | 0.50% | 3.26% | |
| σ: | 3.63% | 2.49% | 3.41% | ||
| p -value: | 0.000 | 0.115 | 0.000 | ||
| FU | μ: | −1.50% | 0.25% | 3.81% | |
| σ: | 3.25% | 2.54% | 2.99% | ||
| p -value: | 0.001 | 0.439 | 0.000 | ||
| CONTROL GROUP | |||||
|---|---|---|---|---|---|
| RIGID |
|||||
| BL | HW | FU | |||
| NON-RIGID | BL | μ: | −2.78% | 0.13% | 1.91% |
| σ: | 2.57% | 2.19% | 2.72% | ||
| p -value: | 0.000 | 0.704 | 0.000 | ||
| HW | μ: | −2.23% | 0.06% | 2.89% | |
| σ: | 3.13% | 2.18% | 2.62% | ||
| p -value: | 0.000 | 0.850 | 0.000 | ||
| FU | μ: | −1.54% | 0.17% | 3.97% | |
| σ: | 3.16% | 2.38% | 2.18% | ||
| p -value: | 0.002 | 0.632 | 0.000 | ||
The effect of asymmetry in global and deformable transformations on the power of the MCI–control group difference comparison is summarized in Table 4. For each of the nine symmetry/asymmetry configurations, the table lists the mean and standard deviation of atrophy in each group, the one-sided p-value for the Student t-test, and the sample size for the power analysis described in Sec. 3.1. Lastly, the 90% confidence interval for the sample size is given, which is computed using the bias-corrected and accelerated (BCα) bootstrap method (Efron, 1987). There is substantial overlap between the confidence intervals for all nine configurations. The results in Table 4 confirm the results of intercept analysis: asymmetry appears to have no significant effect on the power of MCI–control group difference comparison.
Table 4.
Summary of results of 12-month longitudinal atrophy estimation experiments using nine DBM configurations summarized in Equation (6). See caption to Table 2 for the meaning of the rows and columns. For each cell, the mean atrophy and standard deviation are given in the MCI and control groups, as well as the p-value of the MCI-control group comparison, and the sample size needed to detect 25% reduction in MCI atrophy relative to control atrophy with 80% power and significance level 0.05. Lower sample size indicates more powerful atrophy estimation.
| RIGID |
|||||
|---|---|---|---|---|---|
| BL | HW | FU | |||
| NON-RIGID | BL | μ MCI: | −0.07% | 2.31% | 3.56% |
| σ MCI: | 2.50% | 1.92% | 1.84% | ||
| μ CTL: | −1.88% | 0.91% | 2.58% | ||
| σ CTL: | 1.64% | 1.13% | 1.55% | ||
| p -value: | 4.52E-06 | 1.76E-06 | 1.89E-03 | ||
| N: | 482 | 474 | 892 | ||
| CI 0.9 (N): | 263–1230 | 265–1183 | 386–4287 | ||
| HW | μ MCI: | 0.36% | 2.04% | 4.78% | |
| σ MCI: | 2.55% | 1.91% | 1.85% | ||
| μ CTL: | −1.57% | 0.69% | 3.50% | ||
| σ CTL: | 1.58% | 1.10% | 1.36% | ||
| p -value: | 1.06E-06 | 3.18E-06 | 2.1E-05 | ||
| N: | 438 | 508 | 519 | ||
| CI 0.9 (N): | 240–1146 | 283–1280 | 272–1459 | ||
| FU | μ MCI: | 0.93% | 1.69% | 5.80% | |
| σ MCI: | 2.35% | 1.91% | 1.80% | ||
| μ CTL: | −0.96% | 0.34% | 4.42% | ||
| σ CTL: | 1.48% | 1.11% | 1.19% | ||
| p -value: | 3.05E-07 | 2.93E-06 | 1.32E-06 | ||
| N: | 391 | 499 | 426 | ||
| CI 0.9 (N): | 218–1025 | 279–1214 | 232–1047 | ||
3.3 Repeated Interpolation
In the nine configurations presented above, the deformable and global components of the deformation are always composed, so that no image undergoes interpolation more than once. This is not always done in practice in DBM studies. Rigid and deformable registration may be performed using different tools, and there might not be a way to pass the global transformation to the deformable registration method as the initialization. The alternative is to resample images after global transformation and then perform deformable registration on resampled images. In this section we examine the effect of this extra level of interpolation on the bias and power of DBM-based atrophy estimation.
For simplicity, we only consider two of the nine configurations in the previous experiment: the fully symmetric configuration (HW/HW in Table 2) and the configuration where the baseline image is fixed and all transformation is applied to the followup image (FU/FU). In the HW/HW case, the metric computation with one level of resampling is given in equation (5), and the computation with two levels of resampling is in equation (4). The results of the comparison are in Table 5. Overall, repeated interpolation affects the asymmetric DBM configuration much more than the symmetric configuration. Curiously, in the symmetric DBM configuration with repeated interpolation, statistically significant bias is detected in the direct bias estimation experiment (p = 0.03). In the asymmetric DBM configuration, adding a second level of interpolation increases the bias detected in both direct and intercept-based experiments by approximately 2%.
Table 5.
Results of comparison between DBM configurations that perform resampling once (by composing global and deformable transformations) and DBM configurations that perform resampling twice (by applying the global and deformable transformations in sequence). The rows in the table correspond to the rows in Tables 2, 3 and 4.
| HW/HW | FU/FU | |||||
|---|---|---|---|---|---|---|
| resample once | resample twice | resample once | resample twice | |||
| Direct bias estimation | μ: | 0.08% | 0.22% | 3.12% | 5.15% | |
| σ: | 0.99% | 1.12% | 1.39% | 1.81% | ||
| p -value: | 0.34 | 0.03 | 0.0000 | 0.0000 | ||
| Intercept-based bias estimation | MCI | μ: | 0.50% | 0.40% | 3.81% | 6.04% |
| σ: | 2.49% | 2.67% | 2.99% | 3.45% | ||
| p -value: | 0.12 | 0.23 | 0.0000 | 0.0000 | ||
| CTL | μ: | 0.06% | 0.33% | 3.97% | 6.19% | |
| σ: | 2.18% | 2.12% | 2.18% | 2.56% | ||
| p -value: | 0.85 | 0.31 | 0.0000 | 0.0000 | ||
| 12-month longitudinal experiment | μ MCI: | 2.04% | 2.16% | 5.80% | 8.25% | |
| σ MCI: | 1.91% | 2.11% | 1.80% | 2.12% | ||
| μ CTL: | 0.69% | 0.60% | 4.42% | 6.78% | ||
| σ CTL: | 1.10% | 1.21% | 1.19% | 1.71% | ||
| p -value: | 3.2E-06 | 1.2E-06 | 1.3E-06 | 4.9E-05 | ||
| N: | 508 | 465 | 426 | 529 | ||
3.4 Alternative Deformable Registration Approach
Table 6 summarizes the findings of the experiments using the alternative DBM pipeline, which uses the Rueckert et al. (1999) free-form deformation (FFD) registration approach. In the FFD approach, it is not possible to make registration fully symmetric, because the deformable transformation in FFD registration is always applied to just one image. Of the three columns in Table 6, columns BL/FU and HW/FU are both “more symmetric” than the column “FU/FU”. In configuration BL/FU, all of the global transformation is assigned to the baseline image, and all of the deformable transformation is assigned to the followup image. In configuration HW/FU, the global transformation is split between the two images. In FU/FU all the transformation is applied to the followup image; the baseline image is sampled in its native space. As we would expect from the SyN results, the two “more symmetric” configurations result in less bias than the “less symmetric” configuration FU/FU. Indeed, in intercept experiments, the configuration HW/FU is the only one to yield insignificant bias. On the other hand, the direct bias estimation experiment finds significant bias in both “more symmetric” configurations, although the sign of the bias is negative for BL/FU and positive for HW/FU. In the 12-month longitudinal experiment, the BL/FU configuration of FFD yields the best statistical power of all experiments in this paper (N = 289).
Table 6.
Results of bias analysis using the Rueckert et al. (1999) free-form deformation approach. The three columns of numbers correspond to three DBM configurations. In configuration “BL”, the global transformation is applied to the baseline image only. In configuration “HW”, the global transformation is split between the baseline and followup images. In configuration “FU” the global transformation is applied to the followup image. In all three configurations, global and deformable transformations are composed whenever possible, so each image is resampled only once. The rows in the table correspond to the rows in Tables 2, 3 and 4.
| RIGID/DEFORMABLE config-n |
|||||
|---|---|---|---|---|---|
| BL/FU | HW/FU | FU/FU | |||
| Direct bias estimation | μ: | −0.65% | 0.59% | 1.41% | |
| σ: | 1.25% | 1.12% | 1.19% | ||
| p -value: | 0.0000 | 0.0000 | 0.0000 | ||
| Intercept-based bias estimation | MCI | μ: | −1.62% | −0.42% | 0.92% |
| σ: | 3.98% | 3.70% | 3.72% | ||
| p -value: | 0.0027 | 0.39 | 0.054 | ||
| CTL | μ: | −1.05% | −0.04% | 1.50% | |
| σ: | 2.89% | 3.42% | 3.27% | ||
| p -value: | 0.021 | 0.94 | 0.0039 | ||
| 12-month longitudinal experiment | μ MCI: | 2.23% | 2.49% | 4.15% | |
| σ MCI: | 2.47% | 2.65% | 2.61% | ||
| μ CTL: | −0.07% | 0.79% | 2.35% | ||
| σ CTL: | 1.52% | 1.89% | 1.88% | ||
| p -value: | 1.01E-08 | 9.18E-05 | 2.02E-05 | ||
| N: | 289 | 612 | 527 | ||
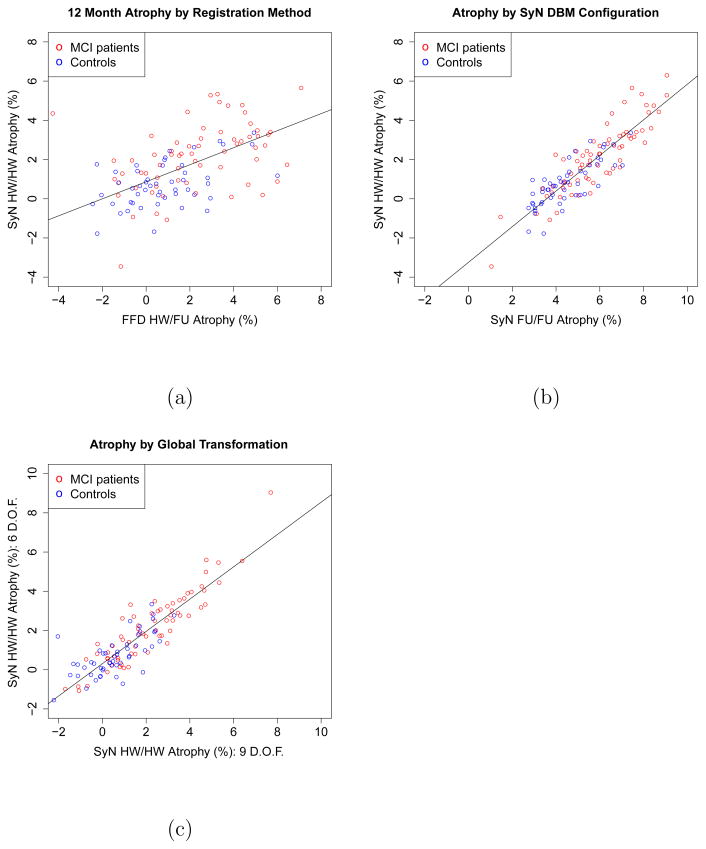
Fig. 3a shows a scatter plot of SyN-based atrophy values in the HW/HW configuration and FFD-based atrophy values with symmetric application of the global transformation. The atrophy values are significantly correlated, R2 = 0.38, F (1, 115) = 70, p ≪ 0.0001, although much of the variance in the data is not described by the correlation. By contrast, the correlation between atrophy values computed by different SyN configurations (HW/HW vs FU/FU), plotted in Fig. 3b, is much greater, R2 = 0.79, F (1, 120) = 446.3, p ≪ 0.0001);
Fig. 3.
(a). Correlation between hippocampal atrophy values computed by SyN-based DBM (HW/HW configuration) and FFD-based DBM (HW/FU) configuration. Each circle in the scatter plot represents a subject. Atrophy values are averaged for the left and right hippocampi. A regression line is fitted to the atrophy values. (b). Correlation between hippocampal atrophy values computed by two SyN-based DBM configurations: a fully symmetric HW/HW configuration, and the fully asymmetric FU/FU configuration. (c) Correlation between atrophy estimated using 6-parameter rigid global registration and 9-parameter (rigid + anisotropic scaling) global registration. This experiment uses SyN-based DBM in the HW/HW configuration.
3.5 Alternative Global Registration Approaches
Table 7 compares atrophy values and intercept-based bias statistics for DBM performed with six and nine-parameter global registration. Results are shown for two SyN-based DBM configurations: HW/HW and FU/FU. The results are remarkably similar for six and nine-parameter registration. Fig. 3c plots the correlation between atrophy values estimated using the HW/HW configuration with 6-parameter global transformation and atrophy values estimated by the same configuration with 9-parameter global transformation. The atrophy values are very highly correlated, R2 = 0.80, F (1, 120) = 494, p ≪ 0.001. This suggests that in ADNI data, the effect of changing voxel size is largely negligible, at least from the point of view of hippocampal atrophy analysis.
Table 7.
Comparison of three global registration approaches: FLIRT tool with six–parameter rigid registration, FLIRT with nine-parameter linear registration (rigid plus anisotropic scaling), and the RREG tool from IRTK with six-parameter rigid registration. The table lists intercept-based atrophy bias estimates and 12-month atrophy estimates for two configurations of DBM (symmetric HW/HW configuration and asymmetric FU/FU configuration), each implemented with 6 or 9 degree-of-freedom global registration. The rows in the table correspond to the rows in Tables 3 and 4.
| HW/HW | FU/FU | |||||||
|---|---|---|---|---|---|---|---|---|
| 6 d.o.f. FLIRT | 9 d.o.f. FLIRT | 6 d.o.f. RREG | 6 d.o.f. FLIRT | 9 d.o.f. FLIRT | 6 d.o.f. RREG | |||
| Intercept-based bias estimation | MCI | μ: | 0.50% | 0.47% | 0.45% | 3.81% | 3.89% | 3.93% |
| σ: | 2.49% | 2.69% | 2.47% | 2.99% | 3.14% | 2.83% | ||
| p -value: | 0.12 | 0.17 | 0.16 | 0.0000 | 0.0000 | 0.0000 | ||
| CTL | μ: | 0.06% | −0.01% | 0.13% | 3.97% | 3.92% | 4.02% | |
| σ: | 2.18% | 2.03% | 2.09% | 2.18% | 2.21% | 2.13% | ||
| p -value: | 0.85 | 0.97 | 0.68 | 0.0000 | 0.0000 | 0.0000 | ||
| 12-month longitudinal experiment | μ MCI: | 2.04% | 2.05% | 2.10% | 5.80% | 5.81% | 5.86% | |
| σ MCI: | 1.91% | 2.01% | 1.90% | 1.80% | 1.97% | 1.83% | ||
| μ CTL: | 0.69% | 0.54% | 0.71% | 4.42% | 4.30% | 4.44% | ||
| σ CTL: | 1.10% | 1.32% | 1.11% | 1.19% | 1.24% | 1.23% | ||
| p -value: | 3.2E-06 | 1.8E-06 | 1.6E-06 | 1.3E-06 | 9.6E-07 | 1.2E-06 | ||
| N: | 508 | 441 | 470 | 426 | 430 | 420 | ||
In addition, Table 7 provides a comparison between rigid registration in FLIRT global and the RREG rigid registration tool that is part of the IRTK software package. The measures of atrophy in each DBM configuration are remarkably similar. This indicates that the bias discussed in this paper is not endemic to a specific global registration tool.
4 Discussion
The most important finding of this paper is that the bias in DBM-based longitudinal analysis of hippocampal atrophy can largely be attributed to the asymmetry in the application of global transformations. This finding is important because it implies that the step of bias elimination can be introduced into researchers’ data processing pipelines in a fairly transparent manner, without requiring changes to the underlying complex image registration software. In particular, it suggests that specialized metrics that account for bias (Leow et al., 2007) may not be required in the context of atrophy estimation in the hippocampus.
Why does asymmetry in global transformation affect the bias in SyN experiments when other factors (asymmetry in deformable transformation, number of interpolations, the registration method) seem to have so little effect on it? One plausible explanation is that the deformable transformation between the baseline image and the followup image is largely determined by the initial gradient of the image match metric. In greedy diffeomorphic registration, the overall deformation is computed by repeatedly taking this gradient, smoothing it and composing the resulting smooth elastic deformations over multiple iterations. However, since the deformation between the baseline image and followup image is small to begin with, the initial gradient may account for much of the total deformation. Now, if the global transformation is applied asymmetrically, at the time the initial gradient is computed, one of the images has undergone a resampling/interpolation operation (which smooths the image) and the other has not. Thus, much of the initial gradient may be driven by differences in sampling and interpolation, rather than anatomical differences. When the global transformation is symmetric, the same kind of resampling/interpolation is applied to both images. So the initial gradient of the metric reflects anatomical differences, as well as noise. Whether the deformable registration is symmetric or not does not matter, because it is primarily driven by the initial gradient.
The idea of splitting the global transformation via the matrix square root operation is not new. It falls within the unbiased atlas framework proposed by Guimond et al. (2000); Davis et al. (2004); Joshi et al. (2004) and adopted by many studies. This framework finds the Frechét mean of the input anatomies in the space of image transformations. The Frechét mean of the baseline image and the followup image, within the space of global transformations, is precisely the matrix square root of the global transformation estimated between these two images by global registration. Of course, the unbiased atlas formulation also applies the Frechét mean to the diffeomorphic transformations. However, based on our findings, this step may not be required, at least in the context of hippocampal atrophy.
The power of the MCI vs. control comparison did not substantially change under different DBM configurations. This suggests that the effect of longitudinal bias may be altogether negligible when reporting group differences in atrophy. In the context of designing clinical trials, this suggests that sample size should be calculated relative to the control atrophy rate. In other words, when we ask, “how many subjects are needed in each cohort to detect an x% reduction in atrophy in the treatment group with given statistical power and given alpha level,” the term “reduction” should refer to the relative change from the MCI rate of atrophy to the control rate of atrophy, rather than absolute reduction in the MCI rate of atrophy. However, when absolute atrophy rate is used for power calculations, severe underpowering can occur.
4.1 Relationship to Prior Work
Bias in longitudinal image registration has been the subject of several papers in the recent years. Leow et al. (2007) introduced an unbiased DBM approach based on an additional regularization term that penalizes the logarithm of the Jacobian determinant in the non-rigid transformation. Yanovsky et al. (2009) further refined this method by introducing a symmetric unbiased DBM technique. The authors evaluated the technique in data from 10 ADNI AD subjects and 10 controls. As in the present study, Yanovsky et al. (2009) use scans acquired at short intervals to assess DBM-related bias in absence of real atrophy. They find that the symmetric unbiased and asymmetric unbiased DBM substantially reduce bias vis-a-vis methods that do not control for bias. However, the unbiased approaches from these authors do not examine the effects of asymmetry in global registration on bias. Hua et al. (2009) compared atrophy estimation in a large ADNI cohort using different configurations of the Leow et al. unbiased registration framework, including 6-parameter and 9-parameter global registration. However, the effect of symmetry in global transformation was not considered. As such, our paper arrives at a different set of conclusions regarding bias. Our results suggest that symmetry in the application of global transformation is sufficient to eliminate significant bias. By contrast, the papers discussed above suggest that bias reduction should be enveloped into the regularization prior of deformable registration. It is important to note that our results are constrained to a small anatomical region (the hippocampus) and may not extrapolate to other brain regions.
Camara et al. (2008) used a synthetic dataset with known gold standard atrophy to compare the accuracy of atrophy estimation by two global atrophy estimation techniques (Freeborough and Fox, 1997; Smith et al., 2002) and two DBM techniques. The two DBM techniques were the FFD method (Rueckert et al., 1999) and a fluid-based image registration method (Crum et al., 2005). The authors found statistically significant differences in atrophy rates reported by DBM techniques and the gold standard in presence of simulated deformations consistent with AD pathology (DBM techniques underestimated atrophy), but did not find significant differences when simulated atrophy was consistent with healthy aging. The paper did not discuss the specifics of how global transformations were applied to the data, nor the amount of smoothing applied to the images. Nevertheless, it is curious that the bias detected on simulated data was in the opposite direction of the results presented in this paper.
One of the explanations for this difference lies in the way that the volume change induced on the hippocampus by a given deformation is calculated. We use a mesh-based calculation, where the deformation field is applied to each vertex of a volumetric tetrahedral mesh and the change in mesh volume is calculated exactly. Camara et al. (2008) and many other authors integrate the determinant of the Jacobian matrix of the deformation over the region of interest. When used in the context of non-parametric registration (e.g., SyN), the latter calculation uses deformation field values from voxels adjacent to the region of interest, since to calculate the Jacobian discretely, a finite difference approximation is used. Many of the voxels adjacent to the hippocampus are in the cerebrospinal fluid, which expands when the hippocampus shrinks. Thus mixing deformation field values across hippocampus boundaries can reduce atrophy estimates, and cause underestimation of atrophy.
Other authors have argued against direct application of DBM for longitudinal atrophy estimation. Davatzikos et al. (2001) proposed RAVENS maps, which avoid Jacobian computations, and instead preserve tissue density under deformable transformations. Studholme et al. (2003) argued that the Jacobian map should be spatially filtered using a measure of normalization uncertainty derived from the normalization procedure. Rohlfing (2006) examined the Jacobian fields yielded by different DBM approaches and found them to be strikingly different despite similar region-wise normalization accuracy performance. Despite these widely cited limitations, DBM remains widely used for longitudinal atrophy analysis.
4.2 Utility for Clinical Studies
The DBM-based atrophy estimation approach, both in absence and presence of bias, finds statistically significant differences between 1-year hippocampal atrophy in MCI patients and atrophy in controls. Particularly, the statistical power of DBM-based analysis is substantially greater than in the analysis of ADNI data that uses independent semi-automatic segmentation of the hippocampus in multiple timepoints (Schuff et al., 2009). Based on 1.5 Tesla MRI data from 127 controls and 226 MCI patients, Schuff et al. (2009) report annual percent change of −0.8 ± 5.6 in controls and −2.6 ± 4.5 in MCI patients. 3 In our analysis of 3 Tesla MRI, we report annual percent change of −0.7±1.1 in controls and −2.0±1.9 in MCI patients (these are the results for the symmetric HW/HW comparison in Table 4). Our results detect a change in MCI that is less in magnitude than in (Schuff et al., 2009), although the 95% confidence intervals for our study (1.6 – 2.5) and Schuff et al. study (2.0 – 3.2) overlap. On the other hand, the variance in the DBM-based approach is significantly reduced. In terms of sample size calculation, our calculation (see Sec. 3.1) yields N = 1570 for the Schuff et al. (2009) study 4 and N = 508 for DBM-based estimation. It is unlikely that these findings are due to differences in MRI modality, as it was recently reported that field strength in ADNI does not significantly affect atrophy estimates (Ho et al., 2009). This indicates that DBM-based atrophy estimation is more sensitive than comparison of hippocampal volumes extracted using semi-automatic segmentation.
4.3 Limitations
One of the limitations of the current study is that it only assesses additive bias in atrophy estimation. There are other types of bias that our methods are not capable of detecting. For example, certain DBM configurations may introduce multiplicative bias that can not be detected by the two experiments used in this study. In the direct bias estimation experiment, true atrophy is zero, so multiplicative effect can not be seen. In the intercept-based experiment, multiplicative bias can not be detected if the factor by which true atrophy is multiplied is the same at 6 months and 12 months. Multiplicative bias may explain why the average MCI atrophy detected by the symmetric DBM configuration is lower than the atrophy reported by Schuff et al. (2009).
Intercept-based atrophy estimation makes an underlying assumption that atrophy is linear over time. This assumption is not uncommon in the evaluation of atrophy estimation techniques (Fox and Freeborough, 1997). The fact that in the unbiased configuration on DBM we observe intercept values not significantly different from zero substantiates this assumption. Additional experiments on ADNI data from all available time points would allow this assumption to be evaluated more extensively.
In the SyN experiments, the results of direct bias estimation and intercept-based bias estimation experiments are overall very consistent. But in the FFD experiment (Table 6), there was some inconsistency between these two ways of estimating bias. Direct estimation finds significant bias in the BL/FU and HW/FU configurations whereas intercept-based estimation finds significant bias in BL/FU but not in HW/FU. However, we do not expect bias to be zero in either of these experiments because the deformable registration (FFD) is not fully symmetric. Both configurations are less asymmetric than FU/FU, in which substantial bias is detected using both measures. So overall, the FFD results fit the pattern of SyN results. Nevertheless, a more extensive evaluation of bias in parametric registration methods is warranted.
Our analysis does not take into consideration the heterogeneity of the clinical groups, particularly the MCI subjects. The only accurate way of determining AD pathology is through autopsy, and many of the MCI patients likely do not have AD pathology. CSF biomarkers are available for a subset of ADNI subjects and may have been used to identify MCI subjects with an AD-like chemical biomarker profile. Reducing heterogeneity in the cohorts would probably reduce the variance in atrophy in each cohort as well as the sample size for the MCI-control comparisons. However, there would not be an obvious effect on the bias of DBM methodology. Hence, we felt that for the purpose of evaluating bias in DBM methodology, such partitioning of the subjects was not necessary.
The experiments in this paper can not detect spatial biases in atrophy estimation. It is entirely possible that atrophy detected in the hippocampus is partially attributable to atrophy in other surrounding structures. DBM, by design, can not estimate change in the volume of a particular small region independently of surrounding image regions. Deformation fields in DBM are smoothed, which causes propagation of information across voxels. Our study can not detect and measure this type of bias.
5 Conclusions
In summary, we presented a study of hippocampal atrophy in patients with mild cognitive impairment using 3 Tesla MRI data from ADNI. Our atrophy estimation used deformation-based morphometry, with some specific choices of parameters tuned for fine-scale longitudinal change detection. These included minimal smoothing of image data; relatively small amount of regularization of deformation fields; precise segmentation of the region of interest in baseline MRI scans; and volume change computation using volumetric meshes rather than Jacobian determinant integration. We found that “naive” application of these methods to ADNI MRI produced excellent statistical power, but also led to unwanted additive bias in atrophy estimates. Examining the possible causes of bias, we discovered that asymmetry in the application of the global transformation between serial MRI images is the leading contributor to bias, whereas the asymmetry in the high-dimensional deformable transformation is less implicated in the bias. This finding appears to transcend the choice of deformable image registration algorithm used, although only two methods were compared in the present study. This finding appears to transcend the choice of deformable image registration algorithm used, although only two methods were compared in the present study. Symmetric application of global transformations requires only a simple modification to existing image analysis protocols, and we are hopeful that other longitudinal studies may benefit from our findings.
Acknowledgments
This work was supported by the Penn-Pfizer Alliance grant 10295 and the NIH grant K25 AG027785.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol–Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
6 Appendix
The square root of a matrix Q can be computed using the iterative algorithm proposed by Denman and Beavers (1976):
where
Footnotes
Underpowering occurs when the absolute rate of atrophy in MCI patients is used as the basis for sample size calculations. Our results show that if relative atrophy (i.e, MCI vs. control) is used, the effect of bias on sample size becomes insignificant.
The term “regression” is an overstatement here, as the regression line is simply the line passing through the two time points; however, the concept generalizes to more time points.
Schuff et al. (2009) report standard errors; we convert to sample standard deviation to be consistent with the rest of the paper and allow comparison across different sample sizes.
This is an approximation obtained by applying (7) to the values reported in (Schuff et al., 2009).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J, Friston K. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7(4):254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vision. 2005;61(2):139–157. [Google Scholar]
- Camara O, Schnabel JA, Ridgway GR, Crum WR, Douiri A, Scahill RI, Hill DLG, Fox NC. Accuracy assessment of global and local atrophy measurement techniques with realistic simulated longitudinal Alzheimer’s disease images. Neuroimage. 2008;42(2):696–709. doi: 10.1016/j.neuroimage.2008.04.259. [DOI] [PubMed] [Google Scholar]
- Christensen G, Joshi S, Miller M. Volumetric transformation of brain anatomy. IEEE Transactions on Medical Imaging. 1997;16:864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage. 2001;14(3):595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Crum WR, Tanner C, Hawkes DJ. Anisotropic multi-scale fluid registration: evaluation in magnetic resonance breast imaging. Phys Med Biol. 2005;50(21):5153–5174. doi: 10.1088/0031-9155/50/21/014. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the ravens maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14(6):1361–9. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Davis B, Lorenzen P, Joshi SC. Large deformation minimum mean squared error template estimation for computational anatomy. Proc IEEE Int Symp Biomed Imaging; 2004. pp. 173–176. [Google Scholar]
- de Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, Rusinek H, Li J, Tsui W, Louis LAS, Clark CM, Tarshish C, Li Y, Lair L, Javier E, Rich K, Lesbre P, Mosconi L, Reisberg B, Sadowski M, DeBernadis JF, Kerkman DJ, Hampel H, Wahlund LO, Davies P. Longitudinal CSF and MRI biomarkers improve the diagnosis of mild cognitive impairment. Neurobiol Aging. 2006;27(3):394–401. doi: 10.1016/j.neurobiolaging.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Denman E, Beavers A. The matrix sign function and computations in systems. Appl Math Comput. 1976;2(1):63–94. [Google Scholar]
- Dupuis P, Grenander U, Miller M. Variational problems on flows of diffeomorphisms for image matching. Quarterly of Applied Mathematics. 1998;56(3):587. [Google Scholar]
- Efron B. Better bootstrap confidence intervals. Journal of the American Statistical Association. 1987;82(397):171–185. [Google Scholar]
- Fox NC, Freeborough PA. Brain atrophy progression measured from registered serial mri: validation and application to alzheimer’s disease. J Magn Reson Imaging. 1997;7(6):1069–1075. doi: 10.1002/jmri.1880070620. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging. 1997;16(5):623–629. doi: 10.1109/42.640753. [DOI] [PubMed] [Google Scholar]
- Guimond A, Meunier J, Thirion J-P. Average brain models: a convergence study. Comput Vis Image Underst. 2000;77(9):192–210. [Google Scholar]
- Hajnal J, Hill D, Hawkes D. Medical image registration. CRC Press; New York: 2001. [Google Scholar]
- Haller J, Banerjee A, Christensen G, Gado M, Joshi S, Miller M, Sheline Y, Vannier M, Csernansky J. Three-dimensional hippocampal MR morphometry by high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Ho AJ, Hua X, Lee S, Leow AD, Yanovsky I, Gutman B, Dinov ID, Lepor N, Stein JL, Toga AW, Jack CR, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM, and the Alzheimer’s Disease Neuroimaging Initiative Comparing 3 T and 1.5 T MRI for tracking Alzheimer’s disease progression with tensor-based morphometry. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, Weiner MW. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J Magn Reson Imaging. 2002;16(3):305–10. doi: 10.1002/jmri.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, Gutman B, Toga AW, Jack CR, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM, Initiative ADN. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009;48(4):668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008a;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Grundman M, Jin S, Gamst A, Ward CP, Sencakova D, Doody RS, Thal LJ the Alzheimer’s Disease Cooperative Study (ADCS), M. Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol Aging. 2008b;29(9):1285–1295. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23 (Suppl 1):S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient nonlinearity correction on phantom and human data. Neuroimage. 2006;30(2):436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM, Study ADNIPP. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31(2):627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chiang MC, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007;26(6):822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM, Initiative ADN. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009;45(3):645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana PA, Brey WW, Kulkarni MV, Sievenpiper CL. Compensation for surface coil sensitivity variation in magnetic resonance imaging. Magn Reson Imaging. 1988;6(3):271–274. doi: 10.1016/0730-725x(88)90401-8. [DOI] [PubMed] [Google Scholar]
- Paling SM, Williams ED, Barber R, Burton EJ, Crum WR, Fox NC, O’Brien JT. The application of serial MRI analysis techniques to the study of cerebral atrophy in late-onset dementia. Med Image Anal. 2004;8(1):69–79. doi: 10.1016/j.media.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Pluta J, Avants BB, Glynn S, Awate S, Gee JC, Detre JA. Appearance and incomplete label matching for diffeomorphic template based hippocampus segmentation. Hippocampus. 2009;19(6):565–571. doi: 10.1002/hipo.20619. [DOI] [PubMed] [Google Scholar]
- Rohlfing T. Transformation model and constraints cause bias in statistics on deformation fields. In: Larsen R, Nielsen M, Sporring J, editors. Medical Image Computing and Computer-Assisted Intervention — MIC-CAI 2006: 9th International Conference; Copenhagen, Denmark. October 1–5, 2006; [DOI] [PubMed] [Google Scholar]; Proceedings, volume 4190 of Lecture Notes in Computer Science. Springer-Verlag; Berlin/Heidelberg: 2006. pp. 207–214. [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A. 2002;99(7):4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Weiner MW, Initiative ADN. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, Luca MD, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Stefano ND, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23 (Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, Stefano ND. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17(1):479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Studholme C, Cardenas V, Blumenfeld R, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of semantic dementia with quantitative validation. Neuroimage. 2004;21(4):1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Studholme C, Cardenas V, Maudsley A, Weiner M. An intensity consistent filtering approach to the analysis of deformation tensor derived maps of brain shape. Neuroimage. 2003;19(4):1638–1649. doi: 10.1016/s1053-8119(03)00183-6. [DOI] [PubMed] [Google Scholar]
- Studholme C, Hill DL, Hawkes DJ. Automated three-dimensional registration of magnetic resonance and positron emission tomography brain images by multiresolution optimization of voxel similarity measures. Med Phys. 1997;24(1):25–35. doi: 10.1118/1.598130. [DOI] [PubMed] [Google Scholar]
- Yanovsky I, Leow AD, Lee S, Osher SJ, Thompson PM. Comparing registration methods for mapping brain change using tensor-based morphometry. Med Image Anal. 2009;13(5):679–700. doi: 10.1016/j.media.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]