Abstract
The Recombination Activating Genes 1 and 2 (Rag1 and Rag2) encode the key enzyme that is required for the generation of the highly diversified antigen receptor repertoire central to adaptive immunity. The longstanding model proposed that this gene pair was acquired by horizontal gene transfer to explain its abrupt appearance in the vertebrate lineage. The analyses of the enormous amount of sequence data created by many genome sequencing projects now provides the basis for a more refined model as to how this unique gene pair evolved from a selfish DNA transposon into a sophisticated DNA recombinase essential for immunity.
Keywords: RAG1, RAG2, transposase, evolution, adaptive immunity
One hallmark of the adaptive immune system shared by all jawed vertebrates is a highly diversified repertoire of antigen receptors. These recognition molecules are encoded by the immunoglobulin (Ig) and T cell receptor (TCR) genes which are assembled from individual gene segments in a process named V(D)J recombination. This site-specific DNA recombination process is catalyzed by the proteins encoded by the recombination activating genes 1 and 2 (RAG1 and RAG2). This review will focus on the evolutionary origin of the Rag1/2 proteins, and how they evolved from a mobile DNA element into a sophisticated and tightly controlled DNA recombinase that is only active in lymphocytes.
Cloning of the Rag genes – a historical overview
In their germline configuration, the Ig and TCR gene loci consist of an array of individual V, D, and J gene segments and thus do not encode functional proteins. When this unique gene structure was discovered in 1976 by the Tonegawa lab, it immediately suggested that their assembly into functional antigen receptor genes requires an ordered DNA rearrangement process [reviewed in 1]. Further analyses revealed that each V, D, and J gene segment was flanked by conserved DNA sequence tags which were thought to mark these segments as building blocks and which served as substrates for a site-specific DNA recombinase [2]. Hence these tags were named recombination signal sequences (RSS), and found to occur in two flavors: 12-RSS and 23-RSS, depending on the length of the spacer that separates conserved heptamer and nonamer sequences. The order and organization of the RSS within the Ig and TCR gene loci indicated that recombination should only occur between a 12-RSS and 23-RSS. The inverted repeat structure and the left/right asymmetry of the RSS resemble the terminal repeat ends of prokaryotic insertion sequences, mobile DNA elements encoding transposases. The most parsimonious model explaining these features was that V(D)J recombination is a variation of the basic principle of transposition and is likely mediated by descendants of a respective transposase [2].
Using a functional complementation approach, the Baltimore lab identified a gene pair that allowed for recombination of RSS-flanked DNA sequences, even when expressed in non-lymphoid cells [3, 4]. The two genes, tightly linked, were hence named Recombination Activating Genes 1 and 2 (RAG1 and RAG2). The encoded proteins were unique as they shared no significant sequence or functional homology with any other protein known at that point, and zoo blots and PCR with degenerate oligos suggested that the presence of these genes is restricted to vertebrates. Subsequent biochemical studies showed that this gene pair indeed encodes a DNA recombinase complex that can excise an RSS-flanked DNA fragment from a linear/circular piece of DNA [5, 6]. Later on, two landmark studies from the Gellert and Schatz labs filled the last missing gap in the model by demonstrating that the Rag1/Rag2 protein complex were able to catalyze a transposition reaction in vitro [7, 8]. More recent data suggested that his also happens at extremely low levels in vivo [9, 10]. Taken together, these observations provided increasing support for the commonly accepted model that we owe our diversified antigen receptor repertoire to a mobile DNA element that entered the genome of a common ancestor of all jawed vertebrates [reviewed in 11, reviewed in 12].
Rag1 and Rag1-like transposases
Mouse Rag1 is a protein 1040 amino acid in length, that when expressed together with Rag2 confers cells the ability to perform V(D)J recombination on integrated or episomal substrates [3]. Rag1 is thought to represent the catalytic subunit of the Rag1/Rag2 recombinase complex, while Rag2 acts as a regulatory subunit that is essential for all activities. At the time of its cloning, Rag1 showed no sequence homologies that would have helped identify important domains and residues. Thus it was initially operationally divided into a “core region” (384-1008) that is essential for all activity in vivo and in vitro, and the respective N- and C-terminal “non-core” regions (1-383, and 1009-1040, respectively) [13, 14]. The core region of Rag1 harbors all elements and domains (including the sequence specific DNA binding domain, the active site, and the Rag2 interaction domain) that are important for its enzymatic activities and shows striking similarities to many cut-and-paste transposases (discussed below) [reviewed in 12, reviewed in 15]. Even with an ever increasing number of vertebrate genomes being sequenced, the amino acid sequence of the Rag1 core remains truly unique. In contrast, the N-terminal non-core region of Rag1 (1-383) contains a RING domain fold that is found in a large number of eukaryotic proteins [16]. RING domains are an essential part of all ubiquitin ligases [reviewed in 17], and there is evidence that the RING domain of Rag1 also exhibits ubiquitin ligase activity [reviewed in 18].
DNA binding domain
All transposases contain a sequence specific DNA binding domain that recognizes and binds to the terminal inverted repeats (TIRs) at the end of the respective mobile DNA element. Similarly, Rag1 harbors a nonamer binding domain (NBD, 389-441), that confers the sequence specificity of the Rag1/Rag2 recombinase by recognizing the conserved nonamer sequence (ACAAAAACC) within the RSS [19, 20]. It had been suggested that NBD of Rag1 might be similar to the DNA binding domain of the prokaryotic Hin recombinase as both recognize highly similar DNA sequence motifs [19]. While this had suggested that at least this domain of Rag1 originated from a bacterial DNA recombinase, a recent report of the crystal structure of the NBD disproved this idea as its protein fold differs from that of the Hin DNA binding domain [21]. For DNA cleavage to occur, additional contacts with the RSS, in particular at the heptamer, are essential [reviewed in 22]. A systematic analysis of proteolytically stable fragments of Rag1 revealed to two subdomains (amino acids 528-760 and 761-960) that showed DNA binding activity [23, 24]. While amino acids 528-760 of Rag1 showed some specificity for single- and double-stranded DNA containing RSS heptamer sequences, the crystal structures of Tn5 and Hermes transposases indicated that the interaction of such transposases with the site of cleavage is not mediated by a typical DNA binding domain fold but rather by residues within their retroviral integrase fold [25, 26]. This suggests that Rag1 would also rely on several structural components for the interaction with the heptamer, but only an experimentally determined structure of the entire Rag1 core will shed light on details of how the Rag1 subdomains contribute to the heptamer-protein interaction surfaces.
Molecular mechanism
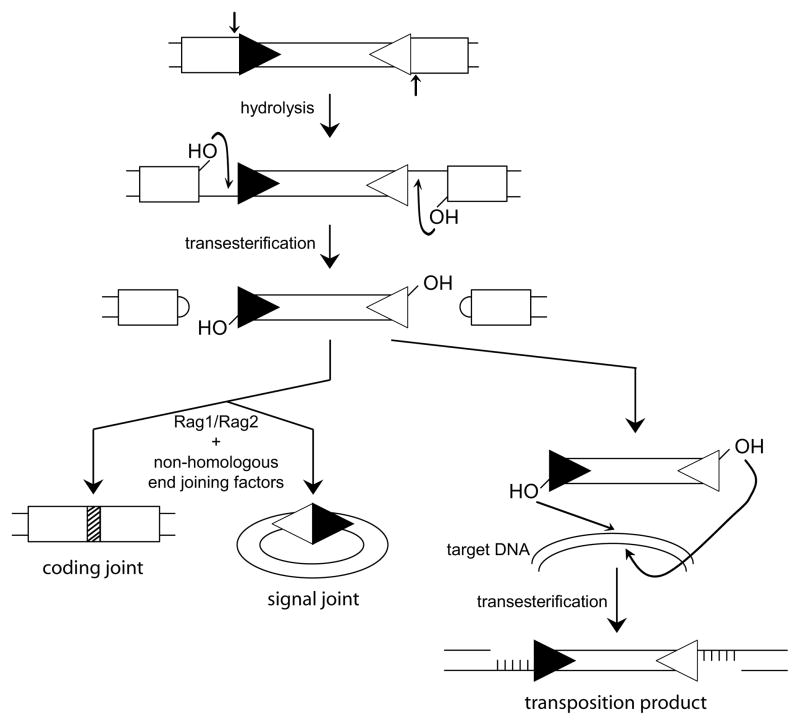
The Rag1/Rag2 complex uses the same reaction chemistry as many cut-and-paste transposases (including Tn5, Tn10, and Hermes), and cleaves the DNA at the RSS by a set of hydrolysis and transesterification reactions: an initial hydrolysis step creates a nick in the top strand of the DNA, followed by a transesterification using the 3′-OH group as the nucleophile (Fig. 1) [reviewed in 22, reviewed in 27]. The result is a hairpin DNA structure at the end of the gene segment, and a blunt DNA end at the RSS flanking the excised DNA (Fig. 1). Although most cut-and-paste transposition reactions that fall in this category and have been studied in detail, proceed with opposite polarity (i.e. the ends of excised DNA fragments carry the hairpin structure, instead of the ends of the chromosomal DNA break), recent biochemical studies of the hAT family transposase Hermes indicated that this enzyme utilizes the same reaction polarity as the Rag1/Rag2 enzyme [28].
Figure 1. The enzymatic activities of Rag1/Rag2.
This is a schematic representation of the reactions catalyzed by the Rag recombinase in vivo and in vitro. Gene segments are indicated as boxes on the double-stranded DNA, and the flanking RSSs are shown as triangles. The initial hydrolysis step, creating single strand breaks, occurs at the RSS-gene segment borders, and is followed by a direct transesterification process, giving rise to hairpin-sealed “coding ends” at the gene segments and “signal ends” at the RSSs. In vivo, the DNA ends get rejoined leading to precise “signal joints”, and imprecise “coding joints” (hatched box) as this ligation step requires additional processing of the DNA ends. Note that the joining steps involve the Rag1/Rag2 complex but also require non-homologous end joining DNA repair factors. The alternative fate of the RSS-flanked DNA segment in vitro (and to a limited extend in vivo) is the result of a second transesterification that integrates this fragment into a target DNA location. This pathway represents a bona fide transposition reaction, and does not require any additional enzymes besides the Rag1/Rag2 complex.
Catalytic center
Although there is almost no conservation between Rag1 and any of the well-studied prokaryotic transposases at the level of their primary amino acid sequences, secondary structure prediction allowed the identification of a triad of acidic residues within mouse Rag1 (D600, D708, and E962) forming the catalytic center of the Rag1/Rag2 complex [29–31]. These two aspartates and one glutamate form a so called DDE motif that chelates two divalent metal ions essential for catalysis [32]. Importantly, DDE (and the related DDD) motifs represent the common active site shared by many cut-and-paste transposase superfamilies [33, 34]. As all three residues reside within Rag1, this finding provides a strong argument that this protein is closely related to transposases and thus may share a common evolutionary origin.
Transib transposases
All the relationships between Rag1 and bona fide cut-and-paste transposases described above are limited solely at the functional level, and could easily be explained by convergent evolution, i.e. evolutionary forces generating very similar solutions to the same problem from very different starting points. The first direct evidence for a genetic relationship between Rag1 and transposases came from the systematic search for Rag1 related genes within the many hundreds of thousands of sequence traces and small contigs from numerous invertebrate genome sequencing projects [35]. The sequence of the retrieved elements from sea urchin (Strongylocentrotus purpuratus), hydra (Hydra magnipapillata), sea anemone (Nematostella vectensis), and lancelet (Branchiostoma floridae) showed a surprising level of similarity with the core region of Rag1. Although most of these elements lack flanking TIRs, they encoded proteins that that still showed a striking similarity with Transib transposases and hence can be considered to be Transib-like elements. The prototypical Transib transposons were originally identified in fruit flies (Drosophila melanogaster) and mosquitoes (Anopheles gambiae) [36], and are now known to also be present in the genomes of several other insects and nematodes. Thus far, no in vitro or in vivo studies have been performed to determine the reaction chemistry, active site motif, and DNA binding modules used by these cut-and-paste invertebrate transposases. There is, however, significant sequence similarity between Transib TIRs and Rag1/Rag2 RSSs, and both Transib and Rag1/Rag2 generate 5 bp target site duplications at transposon integration sites. These features suggest that Transib transposases might indeed catalyze reactions identical to those mediated by the Rag1/Rag2 complex [35]. It is intriguing that transposases with strongest similarities to Rag1 (including Hermes and in particular Transib) are only found in invertebrates, and not present in prokaryotes, suggesting that some of their properties might have evolved to cope with different “lifestyle” requirements compared to their closest relatives amongst the bacterial transposons, including Tn5 and Tn10.
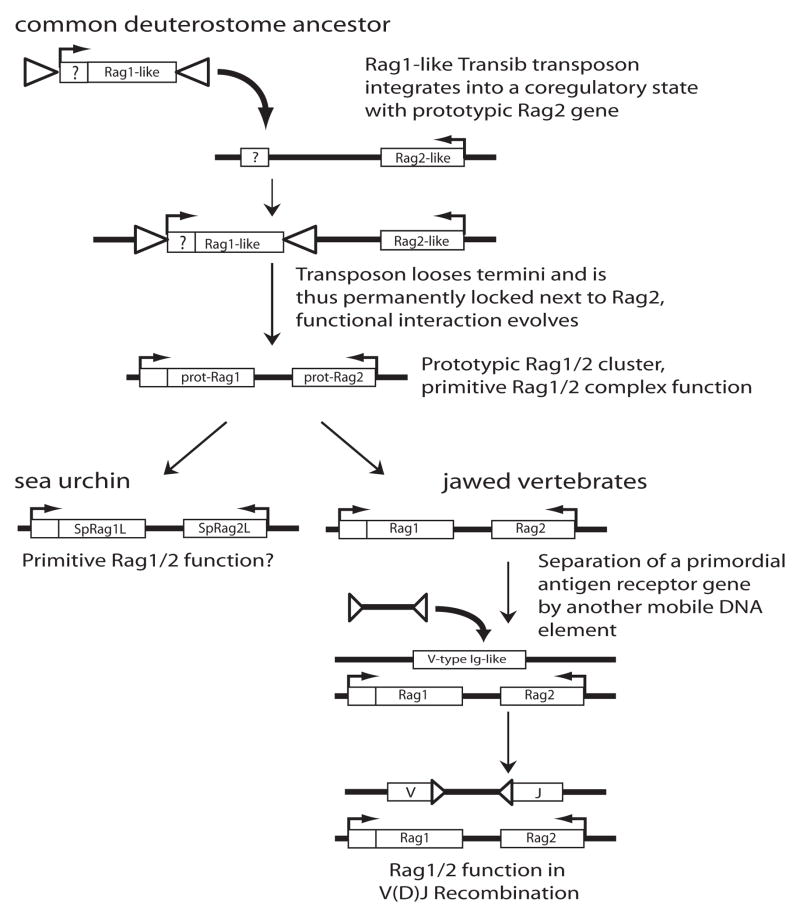
In contrast to the Rag1/Rag2 gene pair, all Transib and Transib-like elements identified thus far contain only a single open reading frame with similarity to Rag1 [35]. Thus there is no evidence for a Rag2-like gene being part of Transib in any of the instances, although some of the earlier studies of Transib elements might have been compromised by the rudimentary state of the genome assemblies at the point of analysis. Alternatively, many transposable elements in the genomes of vertebrates and invertebrates are inactivated by mutation and/or (partial) deletions of transposon ends or coding regions, though it is highly unlikely that every single Transib element would have lost its Rag2-like open reading frame to render them inactive. A much simpler explanation for the absence of Rag2-like open reading frames within these transposon remnants is that Rag2 was never a part of them, and an ancestral Rag1-like Transib sequence simply integrated next to a prototypical Rag2 gene that served a different cellular function at that time (Fig. 2).
Figure 2. Current model of the origins of the Rag1/Rag2 gene cluster.
This is a schematic representation of the “Rag1-only” transposon concept, and also illustrates the “two step” concept of the evolution of V(D)J recombination (see section Open Questions). Coding regions are drawn as open boxes with the name of the gene inside, RSS-like TIRs and RSSs of flanking mobile elements as triangles, and directions of transcription are shown by arrows. In a common ancestor of all living deuterostomes, a Rag1-like transposable element integrates next to a Rag2-like gene. The box with “?” indicates that the N-terminus of Rag1 may have either been part of the original transposon or of a gene into which it integrated. The TIRs flanking the transposon are lost, and a functional interaction of the prototypical Rag1 and Rag2 (prot-Rag1 and prot-Rag2) proteins emerged, serving an unknown primitive function. As the lineages that led to sea urchins and jawed vertebrates diverged, the sea urchin gene cluster (SpRag1L/SpRag2L) may have retained this original function, while the vertebrate genes (Rag1/Rag2) evolved into the V(D)J recombinase. The disruption of a V-type Ig-like receptor gene by an RSS-flanked mobile element, an event that is predicted to have occurred in a common ancestor of all jawed vertebrates, represents a critical step in this process.
The sequence similarity between vertebrate Rag1 and the Transib elements is limited to its core region [35]. Surprisingly, an analysis of transcripts and genomic fragments from the mollusk Aplysia californica revealed a new class of transposable element, named NRAG1-TP that shows a striking similarity with the N-terminal conserved non-core region of Rag1 [37]. This includes the RING domain, but all the remaining parts of NRAG1-TP, including the region thought to contain its active site, show no discernible similarities to Rag1. This finding raises the idea that the ancient Rag1 transposon might have been generated by a recombination event between a Transib and an NRAG1-TP element. Several sequence matches with limited similarity to the N-terminus of Rag1 (though lacking similarity to Rag1 core) were also found in the genomes of sea urchin, lancelet, sea anemone, and hydra [35], but whether they share a genetic origin with Rag1 and/or NRag1-TP elements is unclear. This, however, suggests another attractive model, namely that Rag1 could also have originated from the integration of a Transib element into the 3′end of a ubiquitin ligase gene that was located next to the ancestral Rag2 (Fig. 2).
Rag2 – origin and function
In contrast to Rag1, Rag2 lacks any apparent sequence similarity to any known transposon encoded protein. While there is also only little homology with any known invertebrate or vertebrate protein at the level of its primary amino acid sequence, secondary structure prediction indicates that Rag2 consists of two distinct domains that are found in various proteins throughout the eukaryotic kingdom: an N-terminal 6-bladed β-propeller, and a C-terminal plant homeo (PHD) domain [38, 39]. β-propeller domains and the related WD40 domains are compact globular folds that consist entirely of β sheets in the form of Kelch repeats [reviewed in 40]. There is no specific molecular function assigned to these domains, but in many cases they serve as docking platforms for proteins or small signaling molecules. In the case of mouse Rag2, the β-propeller represents the minimal core (residues 1-387) that is required for all V(D)J recombinase activities [14, 41]. While the actual molecular function of the Rag2 core remains elusive, all evidence so far suggest that it induces a conformational change within Rag1 that switches the catalytic center into its active conformation. In addition, it is also thought to contribute to the strong DNA contacts of the Rag1/Rag2 complex at the border of the RSS heptamer and the flanking gene segment [42, 43]. PHD domains are important recognition folds for posttranslationally modified histone tails, and are found in a diverse group of proteins that control transcription or regulate chromatin structure [44]. The PHD domain of mouse Rag2 recognizes trimethylated histone H3K4 [39, 45, 46]. This histone modification is thought to occur mostly within accessible chromatin, in particular areas that are actively transcribed. The C-terminus of Rag2 also contains a threonine residue (T490) that is a target of the Chk2 kinase and controls the degradation of Rag2 by the proteasome at the G1/S transition of the cell-cycle [47]. This is important to prevent the formation of Rag1/2-initiated DNA breaks during replication.
The currently prevailing concept is that Rag2 was an integral component of an ancestral Rag transposon, which would explain both, the abrupt appearance of Rag2 in the genomes of all jawed vertebrates and its tight genomic linkage to the transposase-derived Rag1 gene. Importantly, there has not been a case of a Rag2-like gene not flanked by Rag1, but Rag1-like transposons lacking Rag2 do exist (see section Rag1 and Rag1-like transposases). Along those lines, Rag2 shows clear features of eukaryotic proteins and all functional data is consistent with a model that Rag2 has originated from a simple domain shuffling event that connected a globular β-propeller fold with a histone tail recognition domain. This model also readily explains the presence of transcriptional control elements ensuring the lymphocyte-restricted expression pattern of Rag1/Rag2, as those could have already controlled transcription of the primordial Rag2 in primordial lymphocyte-like cells. This is in contrast to the transposon model, in which such enhancer sequences would have to be acquired gradually as none of the neighboring genes shares the distinctive expression pattern of Rag1/Rag2 (S.D.F. unpublished data). In summary, the model of a prototypic Rag2 gene, already preexisting in the genome of a common deuterostome ancestor (Fig. 2), explains many (but clearly not all) observations with respect to the origin of the Rag1/Rag2 recombinase.
Rag-like genes in the purple sea urchin
Outside the jawed vertebrate lineage, the purple sea urchin is single only other species in which a Rag1/Rag2-like gene pair has been identified thus far [48]. This appearance in echinoderms represents the earliest occurrence of a complete Rag1/Rag2 gene pair in evolution, considering that similarities with cut-and-paste transposases in lower organisms are limited to Rag1 by itself. In the context of the sea urchin genome sequencing project, an initial analysis readily identified sequences traces (and later on small contigs) showing significant similarity to Rag1; some of these sequence fragment were initially assigned to be fragments and remnants of Transib transposases by Kapitonov and Jurka (see section Rag1 and Rag1-like transposases) [35]. A subsequent thorough analysis of a first complete assembly of genome, however, revealed that one of the elements (parts of are present in contig 29068) encoded a protein with remarkable sequence similarity to Rag1 that extended beyond the core region into the conserved N-terminus [48]. Importantly, a gene with remarkable similarity to Rag2 that is transcribed in the opposite direction was found downstream of it (see below). For the purpose of this review, we will refer to the two genes and the encoded protein as Strongylocentrotus purpuratus Recombination Activating Gene 1/2-like (SpRag1L and SpRag2L, respectively). Note that parts of the SpRag1L gene are identical to the 29068_SP element described prior to completion and of the genome sequence assembly [35].
Within SpRag1L the many of important features of Rag1 are well conserved [48]: the three catalytic residues D548, D708, and E962, their immediate sequence environment, the zinc finger domain important for the interaction with Rag2, and parts of the N-terminus. The NBD that recognizes the nonamer motif within the RSS is less well conserved, and this raises the question whether the SpRag1L/SpRag2L protein complex recognizes and utilizes vertebrate RSSs as its DNA substrates. A recent crystal structure of the mouse Rag1 NBD bound to a consensus nonamer sequence identified several basic residues that are directly contacting the DNA strands [21]. Interestingly, many of these amino acids are conserved in the sea urchin protein, suggesting that SpRag1L might indeed bind to at a sequence similar to the RSS nonamer of jawed vertebrates.
Unlike the prototypical Transib elements in fruit flies and mosquitoes, the SpRag1L gene is located next to a Rag2-like gene (SpRag2L) [48]. While the similarity is quite low at the level of the primary amino acid sequence, structure prediction of the encoded protein indicated that it consists of two distinct structural domains: an N-terminal 6-bladed β propeller linked to a C-terminal PHD domain. The only other proteins with this particular cadence of domains are the Rag2s found in jawed vertebrates.
The similarity of SpRag1L and SpRag2L with vertebrate Rag1 and Rag2 extends to their transcriptional control, namely that both genes are always coexpressed. Transcripts were identified in developing sea urchin embryos as well as in distinct coelomocyte populations in adult sea urchins [48]. Every tissue examined showed either expression of both genes, or of neither of them. This finding is suggests that there exist shared regulatory DNA elements that control the coordinated transcription of both genes in the appropriate cell types at the appropriate developmental stage. As cis-regulatory sequences are frequently less well conserved than coding DNA, the identification of such elements and the promoters they act on will likely require wet-lab experiments rather than in silico approaches.
The similarity of the sea urchin Rag1/2 gene pair with their vertebrate counterparts suggests a common origin, and therefore it is likely that they serve a similar function although they diverged more than 400 million years ago. The exclusive function of vertebrate Rag1/2 is in adaptive immunity to catalyze V(D)J recombination: assemble functional antigen receptor genes during the development of B- and T-lymphocytes. Coelomocytes, the cells that express SpRag1L/SpRag2L in adult sea urchins, are immune cells, consistent with a role of these proteins in immunity. There is no evidence for Ig and TCR genes in the sea urchin genome, and thus far there is no compelling evidence for an adaptive arm of the immune system in sea urchins and many other invertebrates. Recently, however, a highly diversified family of receptor genes, named 185/333, has been identified that is strongly induced by innate immune stimuli in coelomocytes [49], suggesting that our view of invertebrate immunology being limited to fixed pattern recognition receptors might need to be reconsidered. There is, however, no evidence that SpRag1L and SpRag2L are involved in the generation of this diverse receptor repertoire, and hence the roles of these genes in sea urchin immunity remain to be elucidated. The molecular characterization of the SpRag1L/SpRag2L proteins yielded a number of observations that are consistent with the complex having DNA modifying activities, including a putative function as a transposase or DNA recombinase. Such molecular function would fit into the conceptual framework of vertebrate and sea urchin Rags sharing common ancestry. A key feature of the vertebrate Rag1/2 complex is that the formation of the complex itself is essential for any catalytic activity to occur. Importantly, SpRag1L and SpRag2L form complexes that are likely to be structurally closely related to those of bona fide Rag1/2, as it is possible to swap components between sea urchin Rag complexes and those of jawed vertebrates [48]. While the sea urchin Rag complex does not show activities in transient V(D)J recombination assays in human 293T cells (S.D.F. unpublished data), the recombinase activities of such hybrid complexes has not been determined yet. An important property of any protein that acts on chromosomal DNA is its ability to enter the nucleus of the cell, which is mediated by nuclear localization signals in vertebrate Rags. Similarly, both SpRag1L and SpRag2L are able to enter the nucleus when expressed in 293T cells [50].Lastly, the vertebrate Rag complex is not only able to utilize naked double-stranded DNA substrates (in vitro or as transiently transfected plasmids), but also on the endogenous antigen receptor gene loci in the context of chromatin. The PHD domain of vertebrate Rag2 plays an important role in this context as it binds to histone H3K4me3; this binding to histone tails is also a property of sea urchin Rag2 albeit with a slightly altered preference for the dimethylated H3K4me2 [50].
Overall, these observations point towards a role of the SpRag1L/2L complex in the maintenance/alteration of DNA in the context of chromatin. Based on the evolutionary conservation of the active site of SpRag1L it is tempting to speculate that this sea urchin protein might also be involved in a site-specific DNA recombination process. As the NBD is poorly conserved between SpRag1L and Rag1, the sequence of the SpRag1L DNA substrate is unclear. An extensive search for clusters of gene segments with similarity to the vertebrate V, D, and J gene segment from the Ig and TCR clusters did not turn up any candidate gene loci. Similarly, no apparent clusters of RSS-like sequences were found anywhere within the sea urchin genome. The identification of the genes SpRag1L and SpRag2L are acting upon remains a challenging goal for future studies.
Rag1 and Rag2 in jawed vertebrates
The Rag1/Rag2 cluster is a tightly linked gene pair in all jawed vertebrates in which the locus has been characterized. The Rag1 genes or fragments thereof have been cloned from far more than 100 different species, and reveal a strong conservation of the encoded amino acid sequence in particular of the core region; the evolutionarily most distant shark and human Rag1 share overall 65% identity and 77% similarity [51]. Only one vertebrate Rag1 protein has been reported to show a unique feature not shared by any other Rag1 sequenced thus far. The Rag1 from bull shark (Carcharhinus leucas) carries a unique C-terminal repeat (nine copies of a TILEDD consensus hexapeptide), but the biological function and significance of this sequence for Rag1 activity in shark is unknown [51]. Rag2 is overall less well conserved than Rag1 (54% identity and 68% similarity between shark and human Rag2), but the hydrophobic residues that are important for the structural integrity of the β-propeller and the PHD domain remain intact throughout all vertebrate Rag2 reported thus far.
As all jawed vertebrate species rely on the assembly of RSS flanked V, D, and J segments to generate functional Ig and TCR genes, it is likely that the V(D)J recombinase activity of Rag1/Rag2 is conserved. Importantly, the sequences of their DNA substrates, the RSSs, are highly conserved from shark to humans. Although almost all published experiments were performed using mouse or human Rag proteins or variants thereof, it is very likely that all conclusions are valid for the Rag1/Rag2 from all other species as well. It is tempting to propose that getting locked in its genomic location was an essential feature on the way to developing a tightly controlled DNA recombinase system. Therefore it was considered unlikely to find any RSS-like elements in the Rag1/Rag2 gene locus. A sophisticated computational analysis, however, revealed conserved sequences flanking mouse and human Rag1/Rag2 that theoretically could act as RSS [52], but their conservation in other species and whether they are indeed remnants of the ancestral TIRs remains to be determined.
Open questions
The recent identification of Transib transposons and the SpRag1L/SpRag2L gene pair in sea urchin has led to a revised conceptual framework regarding the origin of the vertebrate Rag1/Rag2 cluster (Fig. 2). While this model explains many previous observations, there are still a number of key questions that need to be resolved.
What was the ancient Rag transposon that gave rise to the Rag1/Rag2 genes?
Was it a mobile DNA element that integrated next to a primordial Rag2-like gene? Or did the mobile DNA element already contain both Rag1 and Rag2? As more and more drafts and complete genomes from a variety of invertebrates become available, evidence for one or both will emerge. Thus far no Rag2-like gene has been identified in a species in the absence of Rag1, but the presence of Transib and Transib-like elements (based on their sequence related to the vertebrate Rag1) in the genome of several invertebrates in the absence of a Rag2-like gene currently tips the balance in favor of a “Rag1-only” transposable element.
When did the Rag transposon enter the genome of higher eukaryotic organisms?
The presence of the Rag1/Rag2 gene pair in an echinoderm species and in jawed vertebrates suggest that the transposon either entered the genome of a common ancestor of all living deuterostomes (Fig. 2), or that two related Rag1-like elements independently entered the genome of an ancestral jawed vertebrate and an ancestral echinoderm. The “one event” model fits readily with the “Rag1-only” transposon idea, as it seems unlikely that the very same Rag2-like locus is targeted twice in independent events in distant species. The “two event” model, however, provides a straightforward explanation for the apparent absence of Rag2-like genes in species that are more closely related to vertebrates, such as the sea squirt (Ciona intestinalis) and the lancelet. It is however worth noting, that the sea squirt has a highly compacted genome lacking many genes present in both, echinoderms and vertebrates. Again, a careful systematic analysis of already completed invertebrate genomes and newly sequenced genomes is likely to provide more evidence to address this question.
3) How did the segmented antigen receptor gene loci arise?
To generate the V(D)J recombination system for antigen receptor diversity, two key events were required: the Rag1/Rag2 gene pair needed to be acquired and co-opted for recombinase activity, and a primordial V-type antigen receptor gene needed to be disrupted by an RSS-flanked DNA element. This raises the question whether these events occurred sequentially (as presented in Fig. 2), or at the very same moment, i.e. the Rag transposon itself disrupted the receptor gene, and only later on got separated from this gene locus. The presence of the SpRag1L/SpRag2L gene pair in sea urchins in the absence of any discernible Ig and TCR gene loci supports the sequential model, and suggests that the first event was the co-option of the Rag genes that subsequently served an as of yet unknown primitive function. There is, however, no evidence of non-autonomous RSS-flanked mobile DNA elements required for the second step in any prokaryotic or invertebrate genome analyzed thus far.
4) How did the Rag gene pair acquire its recombinase activity?
The purpose of a transposase is rather selfish in that it works to move its own genetic information from one position in the host genome into another. In the context of V(D)J recombination the Rag1/Rag2 complex present two properties that are unusual for a transposase: it cares about the chromosomal DNA break and participates in its resealing (Fig. 1), and it prefers to join the two open RSS elements instead of using them to integrate into a target location (Fig. 1). While it has been suggested that the C-terminus of Rag2 control the transposase activity [reviewed in 15], it is likely that several other changes in Rag1 also contributed to these altered properties.
Future experiments, ranging from sophisticated genome sequence analysis down to detailed biochemical analyses of the enzymatic properties of the Rag1/Rag2 complex will provide answers to at least some of these questions. Despite the enormous progress in the field over the last decades, the continuous efforts of many laboratories will reveal more of the mysteries of the Rag recombinase that evolved from a selfish DNA element to the master regulator of adaptive immunity.
Acknowledgments
I want to thank Jonathan Rast, Martin Flajnik and all members of my lab for stimulating discussions, and Shu Yuan Yang for helpful comments and suggestions on this manuscript. This work was supported by the Intramural Research Program of the National Institute on Aging/National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–94. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 3.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–23. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 4.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–48. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 5.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–34. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 6.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–95. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 7.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–70. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–51. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 9.Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. Embo J. 2003;22:1381–8. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26:1558–68. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–9. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 12.Schatz DG. Antigen receptor genes and the evolution of a recombinase. Semin Immunol. 2004;16:245–56. doi: 10.1016/j.smim.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–50. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci U S A. 1993;90:6100–4. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–48. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 16.Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 17.Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–39. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 18.Jones JM, Simkus C. The roles of the RAG1 and RAG2 "non-core" regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz) 2009;57:105–16. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- 19.Spanopoulou E, Zaitseva F, Wang FH, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–76. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 20.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–62. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 21.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 23.Arbuckle JL, Fauss LA, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276:37093–101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- 24.Peak MM, Arbuckle JL, Rodgers KK. The central domain of core RAG1 preferentially recognizes single-stranded recombination signal sequence heptamer. J Biol Chem. 2003;278:18235–40. doi: 10.1074/jbc.M302041200. [DOI] [PubMed] [Google Scholar]
- 25.Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, et al. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol. 2005;12:715–21. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 26.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 27.Oettinger MA. Molecular biology: hairpins at split ends in DNA. Nature. 2004;432:960–1. doi: 10.1038/432960b. [DOI] [PubMed] [Google Scholar]
- 28.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 29.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 30.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–69. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–80. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao W, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 2009;26:983–93. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–2. doi: 10.1038/nrg2165-c1. author reply 4. [DOI] [PubMed] [Google Scholar]
- 35.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–74. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panchin Y, Moroz LL. Molluscan mobile elements similar to the vertebrate Recombination-Activating Genes. Biochem Biophys Res Commun. 2008;369:818–23. doi: 10.1016/j.bbrc.2008.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–91. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 41.Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–9. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 43.Nagawa F, Hirose S, Nishizumi H, Nishihara T, Sakano H. Joining mutants of RAG1 and RAG2 that demonstrate impaired interactions with the coding-end DNA. J Biol Chem. 2004;279:38360–8. doi: 10.1074/jbc.M405485200. [DOI] [PubMed] [Google Scholar]
- 44.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–4. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A Plant Homeodomain in Rag-2 that Binds Hypermethylated Lysine 4 of Histone H3 Is Necessary for Efficient Antigen-Receptor-Gene Rearrangement. Immunity. 2007 doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–8. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, Desiderio S. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 1999;11:771–81. doi: 10.1016/s1074-7613(00)80151-x. [DOI] [PubMed] [Google Scholar]
- 48.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–33. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nair SV, Del Valle H, Gross PS, Terwilliger DP, Smith LC. Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. Identification of unexpected immune diversity in an invertebrate. Physiol Genomics. 2005;22:33–47. doi: 10.1152/physiolgenomics.00052.2005. [DOI] [PubMed] [Google Scholar]
- 50.Wilson DR, Norton DD, Fugmann SD. The PHD domain of the sea urchin RAG2 homolog, SpRAG2L, recognizes dimethylated lysine 4 in histone H3 tails. Dev Comp Immunol. 2008;32:1221–30. doi: 10.1016/j.dci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci U S A. 1996;93:9454–9. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cowell LG, Davila M, Ramsden D, Kelsoe G. Computational tools for understanding sequence variability in recombination signals. Immunol Rev. 2004;200:57–69. doi: 10.1111/j.0105-2896.2004.00171.x. [DOI] [PubMed] [Google Scholar]