Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease that predominantly affects women and presents with manifestations derived from the involvement of multiple organs including the kidneys, joints, nervous system, and hematopoietic organs. Immune system aberrations, as well as heritable, hormonal, and environmental factors interplay in the expression of organ damage. Recent contributions from different fields have developed our understanding of SLE and reshaped current pathogenic models. Here, we review novel information that deals with 1) genes associated with disease expression, 2) immune cell molecular abnormalities that lead to autoimmune pathology, 3) the role of hormones and sex chromosomes in the development of disease, 4) environmental and epigenetic factors thought to contribute to the expression of SLE. Finally, we emphasize molecular defects intimately associated with the disease process of SLE that represent ideal therapeutic targets and disease biomarkers.
Introduction
Significant progress has been made in the decade since the pathogenesis of systemic lupus erythematosus (SLE) (Box 1) was last reviewed in this journal [1]. New loci and genes have been associated with the expression of SLE. Extensive studies have shed light on the aberrant biochemistry that governs T cell function and cytokine production in SLE [1]. Exploitation of these abnormalities has been proposed for development of therapeutic targets and biomarkers. While studies have classically focused on the dysregulation of adaptive immunity in SLE, elements of the innate immune system have been identified as major contributors of disease pathogenesis. Furthermore, recent studies highlight the significance of epigenetic alterations in the aberrant expression or function of immune factors. Yet, despite these significant advances, the scarcity of novel therapies continues.
In this review, we discuss molecular and cellular aberrations of the immune system in patients with SLE. We address these alterations according to the main cell involved. However, one of the important messages that this article intends to communicate is that the immune system is broadly compromised in patients with SLE and that deregulation of single elements lead to altered behavior of the whole system.
1. Genes and Genetics in SLE
Genetic predisposition influences the development of SLE in major ways [2]. Although in rare cases this is accomplished by deficiency of a single gene (e.g. C1q) [1,2], it commonly results from the combined effect of a large number of genes. Each allele contributes only mildly (odds ratio ~1.5) and the accumulation of several genes is presumed necessary to significantly increase the risk of SLE. The combinations of risk alleles that lead to predisposition and the mechanisms through which they contribute to autoimmunity are poorly known. In fact, most single nucleotide polymorphisms (SNPs) associated to SLE fall within non-coding DNA regions and represent markers of co-segregated alleles. Notwithstanding, most of them are associated with genes presumed involved in the immune response.
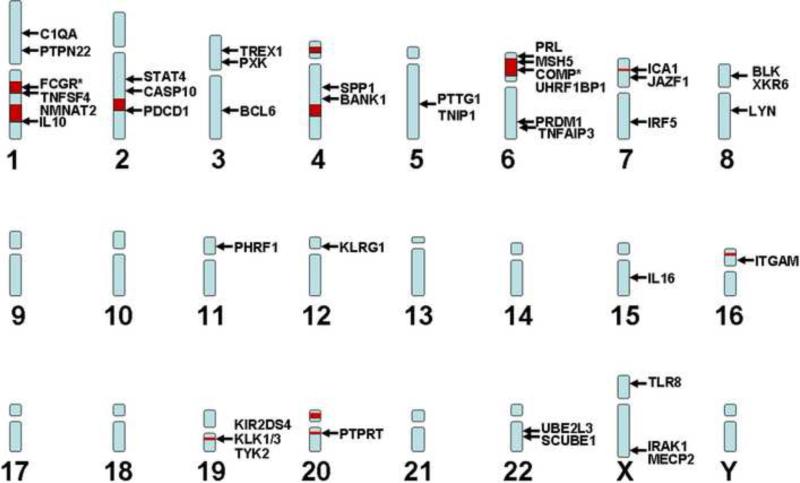
During the past few years, genome-wide analyses have substantially increased the number of candidate genes associated with SLE [3,4,5,6] (Figure 1). Their function is quite variable. Some, such as IRF5 [7], STAT4 [8], osteopontin [9], IRAK1 [10], TREX1 [11], and TLR8 [12], are involved in nucleic acid sensing and IFN production, whereas others are involved in T cell (PTPN22 [13], TNFSF4 [14], PDCD1) or B cell (BANK1, BLK [5], LYN [15]) signaling pathways (e.g. PTPN22 regulates lymphocyte activation [13]). BCL6 is the lineage-specific transcription factor of follicular helper T cells (TFH), a T cell subset that provides help to B cells in germinal centers [16]. Interestingly, IRF5 and STAT4 increase additively the risk of SLE [8]. Some genes have been associated with several autoimmune diseases (e.g. STAT4 with rheumatoid arthritis; PTPN22 with rheumatoid arthritis and diabetes), yet others appear to increase specifically the risk for SLE.
Figure 1. SLE-associated loci and genes.
The approximate position of SLE-associated loci (red squares) and genes (arrows) in the human genome is shown. Additional studies will identify the risk alleles responsible for these associations. This will allow a more comprehensive understanding of disease pathogenesis and the selection of better biomarkers and therapeutic targets. *FCGR stands for FCGR2A, FCGR3B, and FCGR3A; COMP stands for C2, and C4A, and C4B.
A recent large-scale replication study confirmed some of the above-mentioned associations and identified TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for SLE [17]. Although promising, the loci identified so far can account only for ~15% of the heritability of SLE [18]. Thus, although the identification of candidate genes and alleles represents an important step in our understanding of the pathogenesis, the relative importance of each gene in the overall disease process and its particular contribution to phenotype and severity remain to be defined. The pathways affected by the genes associated to SLE and their relationship with T and B cell functional aberrations described in this review will need to be addressed in future studies.
2. T Lymphocytes and cytokines in lupus
In SLE, T cells provide excessive help to B cells and mount inflammatory responses while failing to produce sufficient interleukin-2 (IL-2). Biochemical and gene expression defects have been identified that account for their aberrant function [19].
T cell activation and signal transduction
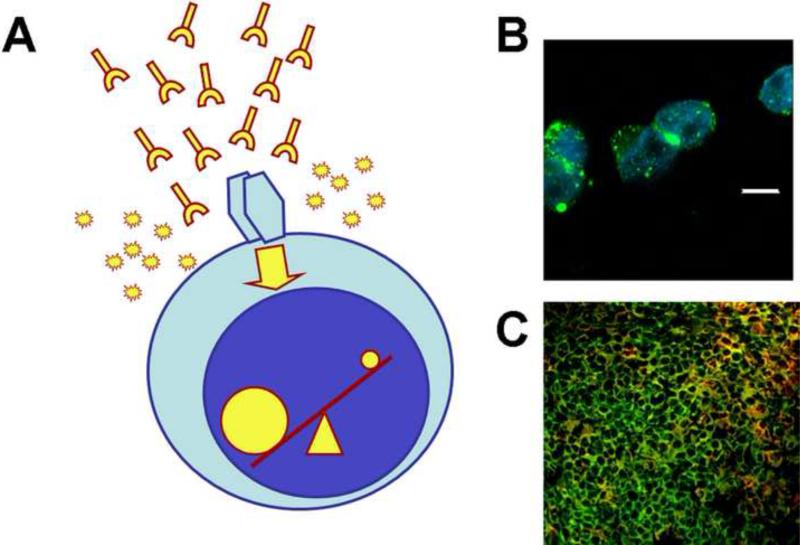
Cellular activation is altered in T cells from SLE patients. Engagement of the T-cell receptor (TCR)/CD3 leads to an early and enhanced signaling response manifested by increased intracytoplasmic calcium flux and cytosolic protein tyrosine phosphorylation. The CD3 complex in SLE T cells is rewired whereby the CD3ζ is replaced by the FcRγ common chain [19] (Figure 2). When FcRγ is present, the signal relies on the spleen tyrosine kinase (Syk) and not on the canonical ζ-associated protein (ZAP-70) [20]. Multiple molecular mechanisms have been described in SLE T cells that contribute to the diminished expression of CD3ζ. These include decreased transcription, abnormal mRNA splicing, decreased mRNA stability, and increased caspase 3-mediated protein degradation [21].
Figure 2. Altered signaling and gene transcription in T cells.
(A) Factors present in SLE sera (e.g. auto-antibodies, cytokines) and constitutive defects in signaling molecules alter T cell signaling events. This results in profound changes in the activation of transcription factors (i.e. more NFAT and CREM activation; less CREB phosphorylation and AP-1 activation) and subsequently in the function of T cells. (B) T cells from SLE patients produce increased amounts of pro-inflammatory cytokines such as IL-17. Shown are T cells from a patient with SLE, stimulated with anti-CD3 and anti-CD28, and stained with anti-IL-17 (green) and a nuclear dye (DAPI; blue) after permeabilization (white bar represents 5 μm). (C) CD3+ CD4- CD8- (double negative) T cells are expanded in patients with SLE and infiltrate affected kidneys. Shown is a dense T cell infiltrate from a renal biopsy of a patient with SLE stained with anti-CD3 (green) and anti-CD4 and CD8 (red). Green cells are double negative T cells and yellow cells are CD4 and CD8 T cells.
Another factor contributing to the enhanced T cell activation is the aggregation of lipid rafts on the cell surface [22]. These high-cholesterol membrane zones rich in signaling molecules polarize upon cell activation. Evidence for the in vivo relevance of these findings was provided by a study that showed that administration of an agent that enhances lipid raft clustering accelerates disease onset in a murine model of lupus (MRl/lpr), whereas injection of a drug that disrupts lipid raft clustering has the opposite effect [23]. Additional signaling abnormalities noted in T cells from SLE patients are listed in Table 1.
Table 1.
Signaling defects reported in SLE T cells
| Molecular defect |
|---|
| Decreased activity of PKC |
| Decreased PKA levels and activity |
| Increased calcium response |
| Deficient CD45 phosphatase activity |
| Increased phosphorylation of tyrosine residues |
| Decreased expression of CD3ζ |
| Decreased activity of Lck |
| Increased expression of FcRγ |
| Decreased MAP kinase activity |
| Increased activity of PI3K |
| Increased lipid raft clustering |
| Increased expression and activity of Syk |
Transcription factors and gene expression
The abnormal gene transcription profile observed in lupus T cells is complex. This altered pattern produces a characteristic phenotype that in some aspects resembles that of activated T cells, and in others shares characteristics with anergic cells.
NFAT and AP1
Nuclear factor of activated T cells (NFAT) is a transcription factor regulated by calcium influx. Upregulation of NFAT in SLE T cells may account for the increased expression of the costimulatory molecule CD40L. However, it fails to upregulate IL-2, because, in certain gene promoters it requires the concurrent binding of AP1 and AP1 is decreased in SLE T cells [24].
CREM and CREB balance
At position –180 of the IL2 promoter, a cyclic AMP response element (CRE) – a DNA region able to bind to the transcription factors CRE-binding protein (CREB) and CRE-modulator (CREM) – has been shown to be of significant pathogenic importance [25]. In patients with SLE, CREM binds preferably over pCREB at the –180 site because 1) nuclear levels of CaMKIV are elevated in response to increased signaling through TCR/CD3 and enhance CREM binding to the IL2 promoter [26], and 2) SLE T cells express increased amounts of PP2A, which dephosphorylates pCREB and limits its binding to the IL2 promoter [27].
Elf-1
Active Elf-1 binds to the promoter of the CD3ζ gene and enhances its activity. Interestingly, when Elf-1 binds to the promoter of the FcRγ gene, it represses its activity. The decreased levels of DNA-binding Elf-1 found in SLE T cells may thus explain decreased expression of CD3ζ and increased expression of FcRγ [28]. It appears though that PP2A is upstream of Elf-1 and indirectly determines the expression of CD3ζ and FcRγ [29].
CD4 helper T cells and cytokine production
After exposure to antigen, naïve CD4 T cells develop into effector subsets defined by the expression of distinct transcription factors that induce a particular phenotype and cytokine production profile. SLE patients have aberrant cytokine production and therefore, their effector capacities are compromised.
IL-2
The production of IL-2, a cytokine centrally involved in the process of T cell activation and proliferation, is defective in T cells from patients with SLE [19]. This may account for the known decreased cytotoxic activity, defective Treg function, and decreased activation-induced cell death in SLE patients.
IL-17
IL-17A and F are produced mainly by activated T cells and play important roles in the immune response against certain bacteria and fungi [30]. IL-17-producing cells have been implicated in the pathogenesis of several autoimmune diseases including multiple sclerosis and SLE [30,31]. Sera of patients with lupus contain abnormally high levels of IL-17 [32] and a high fraction of CD4+ and CD4–CD8– (double negative) T cells in these patients produce IL-17 [33]. Furthermore, IL-17-producing T cells have been found within kidney infiltrates of patients with lupus nephritis [33]. Increased production of IL-17 in patients with SLE correlates with disease activity [32,34]. Release of IL-17 amplifies the inflammatory response by recruiting effector cells to target organs. Furthermore, IL-17 contributes to the formation of germinal centers and, acting in concert with a B-cell-activating factor, increases the survival and proliferation of B cells and their transformation into antibody-secreting cells [32]. Support for the role of IL-17 in the pathogenesis of SLE has been lent by studies in lupus-prone mice [35], and mice deficient in Ro52 [36] and IBP [37,38] which develop SLE.
Differentiation of T cells into the TH17 subset is induced when priming occurs in the presence of TGF-β and certain inflammatory cytokines such as IL-1β, IL-6 or IL-21 [30]. Interestingly, the differentiation of naïve cells into this pro-inflammatory subset has been proposed to occur in a reciprocal fashion with the development of regulatory T cells (Treg) and the presence of an inflammatory signal seems to be the factor that determines whether pro-inflammatory or suppressive cells are generated [30]. Increased levels of the TH17-determining cytokines IL-6 [39], which may be linked to the increased expression of STAT3 [40] and IL-21 [41] are present in patients with SLE.
T follicular helper cells
Follicular helper T cells (TFH) comprise a recently described CD4 helper subset. Like TH17 cells, their differentiation is induced by IL-6 and IL-21, but depends on the costimulatory molecule ICOS and the absence of TGF [16]. TFH localize in the B cell zones of lymph nodes and produce IL-21 and express CD40L. Their main function is to provide B cells with signals for immunoglobulin production, isotype switching, and somatic hypermutation. In murine lupus, IL-21 and TFH are necessary elements for disease development [42,43] and treatment with IL-21R.Fc reduced disease progression [44]. Deficiency of ICOS protects MRL/lpr mice from lupus. This effect depends on the absence of a particular CD4 T cell type that promotes immunoglobulin production in extrafollicular compartments. This cell, observed in autoimmune-prone mice, is analogous to TFH [42].
Regulatory T cells
Most reports agree that CD4+FoxP3+ regulatory T cells (Treg) are low and functionally abnormal in patients with SLE [45]. However, the magnitude of the contribution of these defects to SLE course and pathology is still unknown. Moreover, other unconventional regulatory cell subsets have recently been reported to associate with SLE clinical remission, namely CD4+NKG2D+ [46] and CD8+FoxP3+ cells [47].
CD8 T cells and cytotoxic responses
Several reports point out deficient CD8 cytotoxic capacity [48]. SLE CD8 cells cannot suppress self B cells infected with EBV [49] and perforin-deficient mice have accelerated humoral autoimmunity and lupus-like disease in the MRL background [50]. Further, under certain conditions, CD8 T cells give rise to double negative T cells [51]. The differentiation of functional subsets of CD8 T cells and its contribution to SLE pathogenesis must be clarified in future work.
Double-negative T cells
T lymphocytes that lack the CD4 and CD8 co-receptors are called double-negative (DN) T cells. These cells comprise a scarce population in normal individuals (<5% of T lymphocytes), but are significantly expanded in patients with SLE and induce anti-DNA antibody production by autoreactive B cells. Recent work has shown that they also secrete other cytokines such as IL-1β and IL-17, and are found within cellular infiltrates in kidney biopsies of patients with lupus nephritis [33]. DN cells represent a fraction of CD8+ T cells that, upon activation, acquires a distinct gene expression profile that induces the loss of CD8 expression and the capacity to produce pro-inflammatory cytokines [51].
Adhesion molecules
The expression of the adhesion molecule CD44 is abnormally increased on T cells from patients with SLE [22]. Also, SLE T cells migrate at increased rates in response to the chemokine CXCL12; this is most likely because they express more CXCR4 receptors, which grants them an increased capacity to migrate into inflamed organs [22,52]. The expression of two of the many CD44 variants, CD44v3 and v6, is increased on T cells from lupus patients. This correlates with disease activity, presence of renal disease, and antibodies against double-stranded DNA [53]. The importance of these findings is further supported by the fact that T cells that infiltrate kidneys from SLE patients express CD44v3 and CD44v6 [54].
3. B lymphocytes
As T lymphocytes, B cells are commonly affected in patients with SLE. B cell lymphopenia and overactivity are among the most striking abnormalities encountered in SLE. B lymphocytes in SLE produce an array of autoantibodies against soluble and cellular constituents, but most commonly against intranuclear antigens (ANAs).
Analysis of B cell subpopulations in patients with juvenile-onset SLE disclosed that autoreactive B cells in SLE arise early in B cell ontogeny and that the usual tolerance checkpoints imposed during B cell development are violated [55]. Despite this overactive state, immunization of patients with SLE with tetanus toxoid results in decreased amounts of specific antibody production [56].
B cell subpopulations
In patients with active SLE, there is marked reduction in the numbers of naïve (CD19+CD27–) B cells and enhanced numbers of CD27highCD38+CD19dimsIglowCD20– CD138+ plasma cells in the periphery [57]. Additionally, increased numbers of CD27high plasma cells correlate with increased disease activity, suggesting that this may be a valuable marker of disease-activity [58].
B cell signaling
Stimulation of circulating B cells from patients with SLE through their surface IgM or IgD B-cell receptor (BCR) produces significantly higher fluxes of intracytoplasmic Ca2 and cytosolic protein tyrosine phosphorylation [59]. CR2 expression is decreased in SLE B cells, yet its engagement leads to increased BCR-mediated responses. Conversely, expression of the inhibitory receptor FcγRIIB on memory B cells and plasma cells is decreased in patients with SLE [60] and its ligation provides limited inhibition, suggesting that defects in costimulatory molecule function may account for the increased BCR signaling [61]. A SNP in the promoter of CR2 with functional repercussions may regulate its expression and function in SLE [62]. Likewise, a SNP in the human FCGR2B promoter (particularly found in European–American patients with SLE) results in decreased transcription, providing thus a molecular explanation for the dysregulation of FcγRIIB in some patients with SLE [63]. Protein tyrosine kinase Lyn has been reported to be decreased in the cytoplasm of B cells from 2/3 of patients with SLE [64]. Lyn is crucial for the function of several other signaling inhibitory molecules of B cells such as the surface receptors CD22 and FcgRIIBI, suggesting that decreased Lyn may contribute to lupus B cell overactivity [65]. Studies performed in mice suggest that the presence of a risk-conferring allele of the Ly108 gene (a member of the SLAM family) may facilitate autoimmunity by lowering the intensity of B cell signaling. This effect is explained by its consequences on B cell ontogeny. Auto-reactive B cells can transit through tolerance checkpoints without being deleted [66].
4. Dendritic cells (DCs) in SLE
Phenotypic characteristics of DCs in SLE
While earlier reports described normal and/or decreased numbers and function of SLE DCs (as compared with cells from normal individuals) [67], recent studies have reported an over-stimulated phenotype and function of SLE monocytes and DCs [68]. These discrepancies probably relate to the fact that most studies use monocyte-derived dendritic cells whose phenotype varies according to the environment in which they differentiate. Factors present in SLE sera, including IFN-α, CD40L, free nucleosomes, and autoantibody–DNA complexes cause differentiation and activation of normal DCs [69,70,71] and stimulate their production of cytokines, including IFN-α.
DC and apoptotic cell clearance in SLE
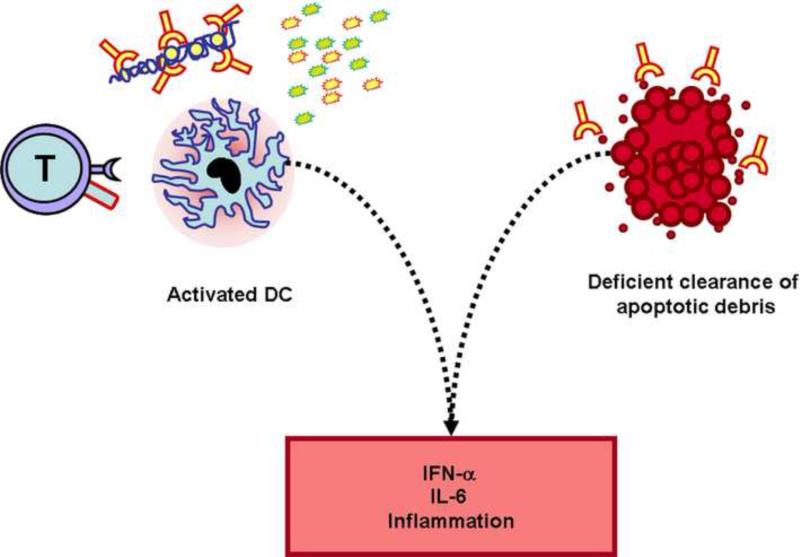
Under physiological conditions, the presence of apoptotic cells is interpreted by the immune system as an anti-inflammatory signal. Thus, when DCs pick up apoptotic cell fragments, autoantigens are presented in a fashion that leads to the inactivation of possible autoreactive T cells [72]. Whereas ingested necrotic cells are able to induce DC maturation, apoptotic cells fail to activate DCs under normal circumstances [73]. Disturbances in apoptosis and/or clearance of apoptotic cells can play an important role in the pathogenesis of SLE [74]. Moreover, SLE macrophages have an impaired capacity for taking up apoptotic cell material in vitro [75]. It has been shown that the interaction with iC3bopsonized apoptotic cells results in tolerizing DCs (with low expression of CD86 and MHC class II) [76] as well as DC interaction with thrombospondin-1 produced by apoptotic cells [77]. By contrast, high mobility group box protein 1 (HMGB1), which binds and stabilizes the structure of nucleosomes inside the nucleus, acts as a pro-inflammatory mediator when released from dying cells [78]. In SLE patients who are known to have a defect in apoptotic cell clearance, nonphagocytosed apoptotic cells undergo secondary necrosis [75]. HMGB1–nucleosome complexes are released from secondary necrotic cells and can be found in the blood of SLE patients [79] and induce cytokine expression in macrophages and maturation of DCs [80] (Figure 3).
Figure 3. Activated dendritic cells induce inflammation upon exposure to apoptotic and necrotic debris.
Dendritic cells from patients with SLE are activated by T cell co-stimulatory molecules (i.e. CD40L), inflammatory cytokines, and nucleic acid-containing immune complexes. Activated DCs produce increased amounts of pro-inflammatory cytokines (e.g. IL-6, IFN-α) that amplify the immune response. Apoptotic material is not recognized as anti-inflammatory and contributes to DC activation.
DC and nucleic acid sensors in SLE
Recent findings suggest that exogenous, pathogen-associated nucleic acids can exacerbate SLE pathology through stimulation of Toll-like receptors (TLRs) [81]. Moreover, the discrimination by TLRs between pathogen and self nucleic acids is not perfect and endogenous nucleic acids may also activate TLRs [82]. The apoptotic cells and necrotic debris may function as an endogenous source of nucleic acids. Bone marrow-derived myeloid DCs are activated by anti-nucleosome antibodies bound to chromatin [83] or by U1snRNP bound with anti-Sm antibodies [84]. However, these and other results indicate that DCs have an additional, TLR-independent, mechanism of nucleic acid sensing in the cytosol that triggers the secretion of IFN-β and IL-1β [85]. One of the new candidate molecules is the DNA-dependent activator of IFN-regulatory factors (DAI or DLM-1/ZBP1), which is able to trigger a strong type I IFN response upon activation [86]. Another candidate is the interferon-inducible HIN-200 family member Absent in Melanoma 2 (AIM2). This molecule binds directly to cytoplasmic DNA and triggers the assembly of an AIM2 inflammasome, resulting in caspase-1 activation and the maturation of IL-1β [87]. Recent reports have claimed an association between this gene family and SLE [88,89,90]. However, there is no information about the exact role of DAI and AIM2 in SLE and DCs so far.
DC and complement system deficiencies in SLE
Complement factors have been shown to mediate efficient clearance of apoptotic cells by means of macrophages [91,92] and the generation of tolerizing DCs [76]. C1q is a recognition protein of the classical pathway of the complement system. Immature DCs produce high amounts of C1q and are known to induce tolerogenic responses [93]. DC activation leads to maturation associated with a reduced capacity to produce C1q and the induction of immunogenic responses [94]. It appears that this may represent another mechanism that links C1q deficiency to the development of SLE. Complement has both beneficial and deleterious roles in the pathogenesis of SLE [95]. The complex role of C1q and other complement proteins in the pathogenesis of SLE is explained by their role in the clearance of apoptotic cells as opsonins (‘waste disposal’ hypothesis). The differentiation of DCs in the presence of C1q gives rise to CD1a+, DC-SIGN+ cells with high phagocytic capacity, impaired ability to stimulate alloreactive T cells and reduced production of IFN-γ [96]. By contrast, immobilized C1q is able to activate DCs and induce DC maturation, cytokine production, expression of co-stimulatory molecules and T cell stimulatory capacity [97]. Thus, C1q plays a protective role against the development and amplification of autoimmune responses by its participation in the clearance of apoptotic cells and immune complexes and perhaps by regulating DC activation.
DCs and the interferon signature in SLE
The expression levels of IFN-inducible genes are upregulated in SLE patients compared with normal controls or patients with other rheumatic diseases [98]. Plasmacytoid DCs secrete large amounts of type I IFN upon viral infection due to the activation of TLR7 and TLR9 [99], and probably represent the main source of IFN-α in SLE patients. Plasmacytoid DC numbers are reduced in SLE peripheral blood, perhaps due to their accelerated migration to inflammatory sites [69]. Indeed, plasmacytoid DCs massively infiltrate lupus skin and renal lesions [100].
DCs play a fundamental role in the development of immune responses. However, their broad capacity to detect immune complexes and nuclei acids grant them the power to amplify the autoimmune response in patients with SLE.
5. Environmental contributions to the expression of SLE
The contribution of the environment to the expression of SLE is unquestionable, as exemplified by the fact that clinical concordance of SLE in identical twins is limited to less than half of the pairs. Epigenetic changes such as DNA methylation have also been attributed to environmental factors associated with SLE. Exposure to ultraviolet light is a known risk factor in clinical disease, and various environmental toxins, including smoking, have been implicated in epidemiological studies [101].
Viral infections, including parvovirus B19 and cytomegalovirus (CMV), are common in patients with SLE [102]. Notably, much discussion has centered around the idea that a viral infection may trigger SLE [103]. The high prevalence of EBV in the adult population makes it difficult to draw any definitive conclusions about causality, but convincing evidence that EBV precedes SLE development was presented in a study in which serum samples were examined in patients before and after lupus development and it was observed that all patients developed antibodies to the EBV protein EBNA-1 prior to developing the hallmark SLE autoantibodies (e.g. anti-Ro) and SLE disease [104]. A significantly higher level of EBV seropositivity has been described in pediatric SLE patients in comparison with healthy children [105] and a high EBV viral titer is found in adult SLE patients, likely due to a T cell defect [106]. While chronic viral infection can lead to T cell exhaustion, viruses have also been implicated in contributing to autoimmunity through molecular mimicry. Some viral proteins are similar to self-antigens and therefore illicit specific immune responses that can cross-react with self-antigens. For instance, the EBV protein EBNA-1 crossreacts with the self-antigen Ro, a common target of autoantibodies [107]. Molecular mimicry is also observed with bacterial and parasitic epitopes.
6. The influence of sex hormones on SLE
Although men can develop lupus, the disease is much more prevalent in women of childbearing age. It has been shown in murine models that addition of estrogen or prolactin can lead to an autoimmune phenotype with an increase in mature high-affinity autoreactive B cells that can out-compete low-affinity autoreactive B cells [108]. Prolactin has also been shown to hasten disease development in lupus-prone mice [108]. While hormones influence SLE development in mice, it has recently been shown that the sex chromosomes themselves influence the expression of SLE. In gonadectomized female and male mice, that have been genetically manipulated to express XX, XO (female), XY or XXY (male), the presence of two X chromosomes increases the severity of SLE disease [109]. While it is clear that hormones can influence autoimmune development in murine models, the use of oral contraceptives does not influence SLE disease flares [110]. Pregnancy in general aggravates SLE, but this is not due to an increase in estradiol or progesterone – in fact the levels of these hormones are lower in the second and third trimester for SLE patients in comparison with healthy pregnant women [111]. Interestingly, a significant number of men suffering from SLE have higher estradiol levels and lower testosterone levels in comparison with healthy individuals [112]. Finally, treatment of patients with dihydroepiandrosterone conferred some clinical benefit [113].
7 Drugs and epigenetic regulation of gene expression
DNA accessibility and thus gene expression is regulated by DNA methylation and histone modifications (acetylation and methylation). SLE patients exhibit DNA hypomethylation in CD4 T cells. This defect has been shown to underlie the overexpression of several genes. While it occurs spontaneously in SLE patients, some commonly used drugs, such as hydralazine and procainamide, inhibit DNA methylation and can induce lupus in healthy individuals [114]. DNA demethylation causes increased CD4 T cell cytokine production and IgG hyperproduction by B cells that overexpress the co-stimulatory molecules CD70 and CD40L. Furthermore, hypomethylated T cells respond to APCs with empty MHC class II pockets, and overexpression of LFA-1 on these T cells can lead to killing of antigen-free macrophages [115]. The mechanisms involved in DNA hypomethylation are not clearly defined as conflicting studies have reported a decrease or no change in the transcription levels of various DNA methyltransferases [116,117], and one study demonstrated increased expression of the catalytic subunit of PP2A in the presence of a methylation inhibitor [118]. Defective activity of PKCδ, also reported in SLE T cells, has been associated with hampered DNA methyltransferase 1 activity through a pathway that depends on the faulty activity of mitogen-activated protein (MAP) kinases [119]. Elevated levels of IL-6, a pro-inflammatory cytokine produced in excess by mononuclear cells from lupus patients, have been reported to abrogate DNA methylation in B cells [120].
Abnormal histone acetylation has also been proposed to alter gene expression in SLE T cells. Some of the effects of CREM, particularly its effect on the IL2 promoter, depend on its capacity to recruit histone deacetylase 1 (HDAC1) [121]. PP2A also regulates the activity of HDAC, and some of its effects are known to be mediated through histone acetylation [122]. Treatment of SLE T cells with trichostatin A, a HDAC inhibitor, diminished the expression of CD40L and the production of IL-10, suggesting that histone acetylation plays a role in the overexpression of these molecules in lupus [123].
8. Tissue injury in SLE
The role of immune complexes in the expression of tissue injury in SLE cannot be overestimated. Immune complexes are cleared through FcR and complement receptors, and the fact that a strong genetic association has been noted between polymorphisms in the FcR genes [124] and the C3bi receptor gene (ITGAM) [5] and SLE further emphasizes their importance.
While the spectrum of autoantibody specificity in SLE may seem unrestricted, it is only a handful of these specificities that have been shown to contribute convincingly to disease-related tissue injury. The latter are best represented by the anti-blood-cell antibodies that activate complement and cause cytopenias and the cationic anti-dsDNA auto-antibodies that are thought to contribute to the expression of nephritis. Anti-phospholipid antibodies have been shown to cause fetal resorption in mice through complement activation [125]. Anti-T cell antibodies, as discussed, may cause decreased IL-2 production and associated T cell defects [26]. Anti-DNA antibodies have been shown to crossreact with NMDA receptors in the brain and cause hippocampal cell death and maybe other subtler neuropsychological manifestations [126]. Patients with lupus suffer ischemic injury of various forms (vasospasm, vasculitis or vasculopathy), and it is possible that circulating autoantibodies gain a tissue-damaging ability under these conditions. Lupus-prone mice develop more intense ischemia reperfusion injury than wild-type mice [127], and injection of anti-DNA, anti-histone or antiphospholipid antibodies to injury-resistant mice restores tissue damage [128].
It has been suggested that different genes dictate the development of acute and chronic glomerulonephritis in lupus-prone mice and that local responses may dictate the development of kidney damage [129]. Kallikrein production seems to mitigate murine and human lupus nephritis and kallikrein gene polymorphisms and promoter gene SNPs are associated with the development of nephritis in patients with SLE [130]. The genome-wide approaches in the study of human lupus may reveal loci and genes linked to the expression of SLE, as was the case with the identification of genes encoding kallikreins, but these discoveries should be followed by functional studies testing the contribution of these genes to disease. Accordingly, it is to be hoped that this line of investigation, that is the search for local factors that contribute, control or even instigate autoimmune tissue injury, may lead to the elucidation of completely different therapeutic approaches.
9. Prospects for the development of novel therapeutics
Patients with SLE are currently treated with non-specific immunosuppressive drugs. Although morbidity and mortality have been improved significantly over the last few decades, treatment-related morbidity remains a significant problem [131,132]. Advances in our understanding of the autoimmune-mediated organ damage has led to the introduction of successful biologics in the treatment of rheumatoid arthritis, spondyloarthropathies, and Crohn's disease, but the development of a new disease specific therapeutic agent for SLE is elusive. The clinical heterogeneity of the disease, the multiplicity of the involved pathogenic mechanisms, the lack of reliable biomarkers and the failure to design proper clinical trials have been frequently blamed for the failure. In Table 2, we have listed clinical trials which have either being concluded or are still in progress.
Table 2.
Biologies in the treatment of SLE
| Target | Treatment | Mode of action | Clinical Trials |
|---|---|---|---|
| BLyS APRIL | Belimumab (anti BLyS antibody) TACI-Ig | Blocks BLyS (and/or APRIL) effect on B cells | Efficacious in animal models. Positive large trial, albeit with moderate effects in patients with SLE (Phase III) |
| Interleukin-6 receptor (IL-6R) | Monoclonal antibody | Blocks IL-6 effect on lymphocytes such as immunoglobulin production. | Successful in mice. Phase I in patients |
| Interferon (IFN) | rhuMAbIFN alpha | Blocks IFN effects | Successful anti-IFN-γ treatment in murine lupus. Phase I of anti-IFN-alpha in patients with SLE |
| C5 | Monoclonal antibody | Blocks membrane attack complex (MAC) formation | Successful in mice. Effective and safe in patients with PNH. |
| CD20 | Chimeric Antibody | B cell depletion | Phase II/III trial in patients with SLE is ongoing. |
| CD22 | Humanized antibody | Modulation of B cell signaling. | Safe in phase I in SLE patients. Phase II is ongoing. |
| CD40L | Monoclonal antibody | Blocks T–B cell crosstalk. | Effective in mice and patients with SLE. Significant side-effects lead to discontinuation of the trials. |
| dsDNA B cell receptor | Abetimus | Blocks the production of anti-dsDNA antibodies. | Some effects on quality of life; better effect on patients with high levels of anti-dsDNA antibodies. |
| CD28–CD80/86 | CTLA-4.Ig | Prevents stimulation of T cells | Proven effectiveness in mice (with and without cyclophosphamide). Phase II/III in SLE patients. |
| ICOS-B7RP-1 | Antibody against B7RP-1 | Prevents T–B cell interaction | Effective in animals. Phase I trial in SLE patients |
BLyS is a cytokine that is involved in the survival of immature B cells especially at the transition stage, a major checkpoint for elimination of autoreactive B cells. Blockade of BLyS [133] with an anti-BLyS antibody (belimumab) has gathered sufficient attention. In the largest clinical trial ever done in SLE patients (865 patients), high dose anti-BLyS was found superior to placebo (57.6% vs. 43.6% patients reached the primary endpoint at 52 weeks) [134]. Although moderate, this effect is significant enough to make anti-BLyS a good therapeutic option for patients with mild to moderate SLE. IL-6 promotes antibody production in humans and mice with lupus [135] and is present in the urine of patients with lupus nephritis [136]. An anti-IL-6 receptor monoclonal antibody was tested in phase I clinical trials in patients with SLE; it led to a decrease in acute phase reactants but at the same time caused neutropenia in two patients. Given the role of IFN in the development of murine lupus and the established upregulation of IFN-α-inducible genes, the use of anti-IFN-α antibodies to treat SLE is currently under consideration. Phase I trials showed safety and a phase II trial is underway. Complement activation is profoundly increased in SLE patients and inhibition of C5 with an antibody, which proved efficacious in the treatment of patients with paroxysmal nocturnal hemoglobinemia, should be considered.
B cell depletion in the treatment of autoimmune and rheumatic diseases showed clinical efficacy. A chimeric anti-human CD20 antibody (rituximab) showed initially promise in small studies and case series in SLE [137] but a trial of rituximab with mycophenolate mofetil in SLE nephritis failed to reach its primary endpoint of remission at 52 weeks of treatment. Another phase II/III trial of rituximab in moderate to severe non-renal SLE is currently underway. CD22 is a molecule that is expressed on the surface of mature B cells but not plasma cells or memory B cells. A humanized anti-CD22 antibody (epratuzumab) is currently undergoing a phase II trial in patients with moderately active SLE. Given the initial failure of anti-CD20 treatment in lupus nephritis, the exact role, if any, of B cell depletion in SLE is unclear. Successful B cell depletion treatment may not come to fruition in SLE until we understand which subset of B cells (autoreactive vs. non-autoreactive) repopulate the empty B cell space and whether the production of B cell-tropic cytokines is altered following depletion. For example, increased production of BLyS following B cell depletion may negate the expected clinical benefit.
Attempts to reestablish B tolerance in patients with SLE have sufficient rationale. LJP-394 (abetimus sodium) is an artificial compound made of four deoxynucleotide-like molecules bound together and thus resembles ds-DNA. It can theoretically bind to autoreactive B cell receptors that recognize dsDNA. In clinical trials of SLE patients, abetimus led to a decrease in dsDNA antibody levels. However clinical trials in humans have not been successful aside from improving quality of life measures [138].
Considerations to block the cognate interaction between T and B cells have led to the use of a fusion molecule of CTLA4 with immunoglobulin (Abatacept). Abatacept with background treatment with mycophenolate mofetil or cyclophosphamide is in phase II/III clinical trial in SLE patients with nephritis. The inducible co-stimulator (ICOS) interaction with its ligand B7-related peptide-1 presents another costimulatory pair and blockade of this cognate interation with a human antibody is currently in a phase I clinical trial in SLE patients. Finally, the costimulatory pair CD40-CD40 ligand has been demonstrated important in the production of autoantibodies. Clinical trials have been conducted with two different types of anti-CD40L (BG9588 and IDEC-1) and although some clinical efficacy was noted (only by BG9588) they were both suspended because of unexpected thomboembolic events [139].
Failure to develop new drugs has not quenched effort and enthusiasm. The gained experience elucidates the absolute need of biomarkers, the design of better trials and consideration of the heterogeneous nature of the disease and should meet success in the near future.
10. Conclusion and future directions
The main question in the field of human SLE is why there has been no new drug for SLE in the last 5 decades. Is this because of a lack of proper biologics, poorly designed trials, lack of proper biomarkers, or the clinical and pathogenic heterogeneity of the disease? (Box 2).
There is no doubt that the heterogeneity of the disease and lack of adequate tools to diagnose and follow disease progress present major impediments. Serum levels of autoantibodies, complement levels and the clinical tools available for characterizing the symptoms certainly have some value but they have proved incomplete in capturing the spectrum of the disease. Several laboratories have identified molecular abnormalities that appear to contribute to the molecular pathogenesis of the disease and, if used properly, these could prove useful in defining subsets of patients with SLE.
Similarly, the available tools have proven not sufficiently powerful to serve as biomarkers of disease activity. Proper disease biomarkers are needed to conduct clinical trials. It is proposed that a good biomarker should be easily evaluated and should somehow contribute to the expression of disease pathology. Herein we have discussed a number of potential biomarkers, including the expression of CD44. This molecule, along with phosphorylated ERM (pERM), was shown to be expressed by T cells in the peripheral blood and kidneys of patients with lupus nephritis [22]. Assuming that CD44+ pERM+ cells appear first in the peripheral blood prior to entering the kidney, and, assuming that, after entering the kidney, they contribute to tissue damage, their detection in the peripheral blood might represent a proper biomarker.
Emerging evidence at this point suggests that local factors may contribute, in parallel with the immune abnormalities, to the expression of end-organ damage in patients with SLE. It thus becomes apparent that we should understand better the mechanisms whereby local factors contribute to disease pathology. It is expected that understanding of the immune and non-immune mechanisms involved in the damage of specific organs will allow us to treat patients more effectively.
The available biologics (for example anti-CD20 antibodies) accomplish what is expected of them with certain limitations. If anti-CD20 antibodies destroy B cells through antibody-mediated cell cytotoxicity, then patients with FcR isoforms that do not bind with sufficient avidity the Fc portion of the antibody, should display limited B cell depletion. If the biologic is an antibody that destroys its target by activating complement, then SLE patients with decreased levels of complement will respond poorly. Herein we have discussed several molecular abnormalities expressed by SLE immune cells. It is expected that correction of these abnormalities will result in normalization of the immune cell function and abrogate their ability to contribute to organ damage.
Box 1. A brief introduction to systemic lupus erythematosus (SLE).
SLE is a systemic autoimmune disorder driven by an immune response directed against ubiquitous, mostly intranuclear, self-antigens. Patients with SLE may develop a heterogeneous array of clinical manifestations including skin rash, photosensitivity, oral ulcers, arthritis, serositis (pleuritis or pericarditis), glomerulonephritis, neurological symptoms (e.g. seizures), hemolytic anemia, leukopenia, and thrombocytopenia. Antibodies against nuclear antigens (ANA), particularly anti-double stranded DNA are commonly detected. Diagnosis is based on the finding of these clinical and laboratory abnormalities.
SLE principally affects women during the child-bearing years of life (nine out of ten patients are women). African-American, Asian, and Hispanic persons are more commonly and more severely affected than Caucasians. Organ damage (e.g. renal failure) occurs as a consequence of the chronic, uncontrolled auto-immune response and the foundation of therapy is immunosuppression. This is accomplished by corticosteroids and immunosuppressive drugs.
Treatment is aimed at avoiding end-organ damage and the lack of reliable biomarkers impedes the evaluation of disease activity in the absence of conspicuous signs. The importance of this is that, along with the toxic therapeutic agents currently used, low-grade chronic inflammation predisposes patients to infections and cardiovascular disease, the major causes of death and morbidity in patients with SLE.
Box 2. Questions that remain to be addressed.
Identify the gene alleles that predispose to SLE and define the biologic mechanisms that underlie such predisposing effects.
Identify the functional relationships between predisposing genes that produce autoimmune proclivity.
Define if the biochemical defects of the SLE T cell represent a generalized T cell phenomenon or the signature of a pathogenic subset.
Determine the relationship between the abnormal transcription factor pattern and the skewed effector cell differentiation (e.g. abnormal Treg function, increased DN T cell generation).
Understand better the underlying biology that accounts for the failure of biologics, especially the B cell-depleting ones, to produce therapeutic efficacy.
Identify the organ-specific factors that determine susceptibility or resistance to immune-mediated tissue injury.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsokos GC, Kammer GM. Molecular aberrations in human systemic lupus erythematosus. Mol. Med Today. 2000;6:418–424. doi: 10.1016/s1357-4310(00)01798-6. [DOI] [PubMed] [Google Scholar]
- 2.Moser KL, et al. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozyrev SV, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 5.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 6.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham RR, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abelson AK, et al. STAT4 Associates with SLE through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariuki SN, et al. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009;10:487–494. doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob CO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 12.Armstrong DL, et al. Identification of new SLE-associated genes with a two-step Bayesian study design. Genes Immun. 2009;10:446–456. doi: 10.1038/gene.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zikherman J, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunninghame Graham DS, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40:83–89. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, et al. Genetic associations of LYN with systemic lupus erythematosus. Genes Immun. 2009;10:397–403. doi: 10.1038/gene.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009 doi: 10.1038/ng.468. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crispin JC, et al. How signaling and gene transcription aberrations dictate the systemic lupus erythematosus T cell phenotype. Trends Immunol. 2008;29:110–115. doi: 10.1016/j.it.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan S, et al. Differential expression and molecular associations of Syk in systemic lupus erythematosus T cells. J Immunol. 2008;181:8145–8152. doi: 10.4049/jimmunol.181.11.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsokos GC, et al. Rewiring the T-cell: signaling defects and novel prospects for the treatment of SLE. Trends Immunol. 2003;24:259–263. doi: 10.1016/s1471-4906(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, et al. Phosphorylated ERM is responsible for increased T cell polarization, adhesion, and migration in patients with systemic lupus erythematosus. J Immunol. 2007;178:1938–1947. doi: 10.4049/jimmunol.178.3.1938. [DOI] [PubMed] [Google Scholar]
- 23.Deng GM, Tsokos GC. Cholera toxin B accelerates disease progression in lupus-prone mice by promoting lipid raft aggregation. J Immunol. 2008;181:4019–4026. doi: 10.4049/jimmunol.181.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyttaris VC, et al. Increased levels of NF-ATc2 differentially regulate CD154 and IL-2 genes in T cells from patients with systemic lupus erythematosus. J Immunol. 2007;178:1960–1966. doi: 10.4049/jimmunol.178.3.1960. [DOI] [PubMed] [Google Scholar]
- 25.Tenbrock K, et al. The cyclic adenosine 5′-monophosphate response element modulator suppresses IL-2 production in stimulated T cells by a chromatin-dependent mechanism. J Immunol. 2003;170:2971–2976. doi: 10.4049/jimmunol.170.6.2971. [DOI] [PubMed] [Google Scholar]
- 26.Juang YT, et al. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J Clin Invest. 2005;115:996–1005. doi: 10.1172/JCI200522854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsiari CG, et al. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest. 2005;115:3193–3204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juang YT, et al. Defective production of functional 98-kDa form of Elf-1 is responsible for the decreased expression of TCR zeta-chain in patients with systemic lupus erythematosus. J Immunol. 2002;169:6048–6055. doi: 10.4049/jimmunol.169.10.6048. [DOI] [PubMed] [Google Scholar]
- 29.Juang YT, et al. PP2A dephosphorylates Elf-1 and determines the expression of CD3zeta and FcRgamma in human systemic lupus erythematosus T cells. J Immunol. 2008;181:3658–3664. doi: 10.4049/jimmunol.181.5.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korn T, et al. IL-17 and Th17 Cells. Annu. Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 31.Nalbandian A, et al. Interleukin-17 and systemic lupus erythematosus: current concepts. Clin Exp Immunol. 2009;157:209–215. doi: 10.1111/j.1365-2249.2009.03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doreau A, et al. Interleukin 17 acts in synergy with B cell-activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nat Immunol. 2009;10:778–785. doi: 10.1038/ni.1741. [DOI] [PubMed] [Google Scholar]
- 33.Crispin JC, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol. 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, et al. Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1472–1483. doi: 10.1002/art.24499. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. The role of IL-23/IL-17 axis in lupus nephritis. J Immunol. 2009;183:3160–3169. doi: 10.4049/jimmunol.0900385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espinosa A, et al. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J Exp Med. 2009;206:1661–1671. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fanzo JC, et al. Loss of IRF-4-binding protein leads to the spontaneous development of systemic autoimmunity. J Clin Invest. 2006;116:703–714. doi: 10.1172/JCI24096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q, et al. IRF-4-binding protein inhibits interleukin-17 and interleukin-21 production by controlling the activity of IRF-4 transcription factor. Immunity. 2008;29:899–911. doi: 10.1016/j.immuni.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linker-Israeli M, et al. Elevated levels of endogenous IL-6 in systemic lupus erythematosus. A putative role in pathogenesis. J Immunol. 1991;147:117–123. [PubMed] [Google Scholar]
- 40.Harada T, et al. Increased expression of STAT3 in SLE T cells contributes to enhanced chemokine-mediated cell migration. Autoimmunity. 2007;40:1–8. doi: 10.1080/08916930601095148. [DOI] [PubMed] [Google Scholar]
- 41.Wong CK, et al. Elevated Production of B Cell Chemokine CXCL13 is Correlated with Systemic Lupus Erythematosus Disease Activity. J Clin Immunol. 2009 doi: 10.1007/s10875-009-9325-5. [DOI] [PubMed] [Google Scholar]
- 42.Odegard JM, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bubier JA, et al. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1518–1523. doi: 10.1073/pnas.0807309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herber D, et al. IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J Immunol. 2007;178:3822–3830. doi: 10.4049/jimmunol.178.6.3822. [DOI] [PubMed] [Google Scholar]
- 45.Crispin JC, et al. Immunoregulatory T cells in autoimmunity. Autoimmun. Rev. 2004;3:45–51. doi: 10.1016/S1568-9972(03)00086-7. [DOI] [PubMed] [Google Scholar]
- 46.Dai Z, et al. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J Exp Med. 2009;206:793–805. doi: 10.1084/jem.20081648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, et al. Regulatory T cell (Treg) subsets return in patients with refractory lupus following stem cell transplantation, and TGF-beta-producing CD8+ Treg cells are associated with immunological remission of lupus. J Immunol. 2009;183:6346–6358. doi: 10.4049/jimmunol.0901773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stohl W. Impaired polyclonal T cell cytolytic activity. A possible risk factor for systemic lupus erythematosus. Arthritis Rheum. 1995;38:506–516. doi: 10.1002/art.1780380408. [DOI] [PubMed] [Google Scholar]
- 49.Puliaeva I, et al. Therapeutic potential of CD8+ cytotoxic T lymphocytes in SLE. Autoimmun. Rev. 2009;8:219–223. doi: 10.1016/j.autrev.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng SL, et al. Perforin protects against autoimmunity in lupus-prone mice. J Immunol. 1998;160:652–660. [PubMed] [Google Scholar]
- 51.Crispin JC, Tsokos GC. Human TCR-αβ+ CD4- CD8- T cells can derive from CD8+ T cells and display an inflammatory effector phenotype. J Immunol. 2009;183:4675–4681. doi: 10.4049/jimmunol.0901533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estess P, et al. Functional activation of lymphocyte CD44 in peripheral blood is a marker of autoimmune disease activity. J Clin Invest. 1998;102:1173–1182. doi: 10.1172/JCI4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crispin JC, et al. Expression of CD44v3 and CD44v6 isoforms is increased on T cells from patients with systemic lupus erythematosus and correlates with disease activity. Arthritis Rheum. 2009 doi: 10.1002/art.27385. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen RA, et al. T cells and in situ cryoglobulin deposition in the pathogenesis of lupus nephritis. Clin Immunol. 2008;128:1–7. doi: 10.1016/j.clim.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yurasov S, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med. 2005;201:703–711. doi: 10.1084/jem.20042251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gottlieb AB, et al. Immune function in systemic lupus erythematosus. Impairment of in vitro T-cell proliferation and in vivo antibody response to exogenous antigen. J Clin Invest. 1979;63:885–892. doi: 10.1172/JCI109388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odendahl M, et al. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. J Immunol. 2000;165:5970–5979. doi: 10.4049/jimmunol.165.10.5970. [DOI] [PubMed] [Google Scholar]
- 58.Jacobi AM, et al. Correlation between circulating CD27high plasma cells and disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2003;48:1332–1342. doi: 10.1002/art.10949. [DOI] [PubMed] [Google Scholar]
- 59.Liossis SN, et al. B cells from patients with systemic lupus erythematosus display abnormal antigen receptor-mediated early signal transduction events. J Clin Invest. 1996;98:2549–2557. doi: 10.1172/JCI119073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su K, et al. Expression profile of FcgammaRIIb on leukocytes and its dysregulation in systemic lupus erythematosus. J Immunol. 2007;178:3272–3280. doi: 10.4049/jimmunol.178.5.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackay M, et al. Selective dysregulation of the FcgammaIIB receptor on memory B cells in SLE. J Exp Med. 2006;203:2157–2164. doi: 10.1084/jem.20051503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boackle SA, et al. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity. 2001;15:775–785. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- 63.Blank MC, et al. Decreased transcription of the human FCGR2B gene mediated by the -343 G/C promoter polymorphism and association with systemic lupus erythematosus. Hum. Genet. 2005;117:220–227. doi: 10.1007/s00439-005-1302-3. [DOI] [PubMed] [Google Scholar]
- 64.Liossis SN, et al. B-cell kinase lyn deficiency in patients with systemic lupus erythematosus. J Investig. Med. 2001;49:157–165. doi: 10.2310/6650.2001.34042. [DOI] [PubMed] [Google Scholar]
- 65.Cornall RJ, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 66.Kumar KR, et al. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–1669. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- 67.Koller M, et al. Phenotypic and functional deficiencies of monocyte-derived dendritic cells in systemic lupus erythematosus (SLE) patients. Int. Immunol. 2004;16:1595–1604. doi: 10.1093/intimm/dxh160. [DOI] [PubMed] [Google Scholar]
- 68.Ding D, et al. Aberrant phenotype and function of myeloid dendritic cells in systemic lupus erythematosus. J Immunol. 2006;177:5878–5889. doi: 10.4049/jimmunol.177.9.5878. [DOI] [PubMed] [Google Scholar]
- 69.Blanco P, et al. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 70.Decker P, et al. Nucleosome, the main autoantigen in systemic lupus erythematosus, induces direct dendritic cell activation via a MyD88-independent pathway: consequences on inflammation. J Immunol. 2005;174:3326–3334. doi: 10.4049/jimmunol.174.6.3326. [DOI] [PubMed] [Google Scholar]
- 71.Means TK, et al. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinman RM, et al. Dendritic cell function in vivo during the steady state: a role in peripheral tolerance. Ann N. Y. Acad. Sci. 2003;987:15–25. doi: 10.1111/j.1749-6632.2003.tb06029.x. [DOI] [PubMed] [Google Scholar]
- 73.Stuart LM, et al. Inhibitory effects of apoptotic cell ingestion upon endotoxin-driven myeloid dendritic cell maturation. J Immunol. 2002;168:1627–1635. doi: 10.4049/jimmunol.168.4.1627. [DOI] [PubMed] [Google Scholar]
- 74.Kaplan MJ. Apoptosis in systemic lupus erythematosus. Clin Immunol. 2004;112:210–218. doi: 10.1016/j.clim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 75.Herrmann M, et al. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41:1241–1250. doi: 10.1002/1529-0131(199807)41:7<1241::AID-ART15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 76.Verbovetski I, et al. Opsonization of apoptotic cells by autologous iC3b facilitates clearance by immature dendritic cells, down-regulates DR and CD86, and up-regulates CC chemokine receptor 7. J Exp Med. 2002;196:1553–1561. doi: 10.1084/jem.20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krispin A, et al. Apoptotic cell thrombospondin-1 and heparin-binding domain lead to dendritic-cell phagocytic and tolerizing states. Blood. 2006;108:3580–3589. doi: 10.1182/blood-2006-03-013334. [DOI] [PubMed] [Google Scholar]
- 78.Tian J, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 79.Urbonaviciute V, et al. Factors masking HMGB1 in human serum and plasma. J Leukoc. Biol. 2007;81:67–74. doi: 10.1189/jlb.0306196. [DOI] [PubMed] [Google Scholar]
- 80.Urbonaviciute V, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J Exp Med. 2008;205:3007–3018. doi: 10.1084/jem.20081165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patole PS, et al. Viral double-stranded RNA aggravates lupus nephritis through Toll-like receptor 3 on glomerular mesangial cells and antigen-presenting cells. J Am Soc Nephrol. 2005;16:1326–1338. doi: 10.1681/ASN.2004100820. [DOI] [PubMed] [Google Scholar]
- 82.Rahman AH, Eisenberg RA. The role of toll-like receptors in systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:131–143. doi: 10.1007/s00281-006-0034-3. [DOI] [PubMed] [Google Scholar]
- 83.Boule MW, et al. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–1640. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savarese E, et al. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood. 2006;107:3229–3234. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- 85.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 86.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 87.Schroder K, et al. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr. Biol. 2009;19:R262–R265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 88.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 89.Choubey D, Panchanathan R. Interferon-inducible Ifi200-family genes in systemic lupus erythematosus. Immunol Lett. 2008;119:32–41. doi: 10.1016/j.imlet.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mondini M, et al. Role of the interferon-inducible gene IFI16 in the etiopathogenesis of systemic autoimmune disorders. Ann N. Y. Acad. Sci. 2007;1110:47–56. doi: 10.1196/annals.1423.006. [DOI] [PubMed] [Google Scholar]
- 91.Botto M, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 92.Mevorach D, et al. Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med. 1998;188:2313–2320. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reis ES, et al. Complement components, regulators and receptors are produced by human monocyte-derived dendritic cells. Immunobiology. 2007;212:151–157. doi: 10.1016/j.imbio.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 94.Castellano G, et al. Maturation of dendritic cells abrogates C1q production in vivo and in vitro. Blood. 2004;103:3813–3820. doi: 10.1182/blood-2003-09-3046. [DOI] [PubMed] [Google Scholar]
- 95.Manderson AP, et al. The role of complement in the development of systemic lupus erythematosus. Annu. Rev Immunol. 2004;22:431–456. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- 96.Castellano G, et al. Immune modulation of human dendritic cells by complement. Eur. J Immunol. 2007;37:2803–2811. doi: 10.1002/eji.200636845. [DOI] [PubMed] [Google Scholar]
- 97.Csomor E, et al. Complement protein C1q induces maturation of human dendritic cells. Mol. Immunol. 2007;44:3389–3397. doi: 10.1016/j.molimm.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 98.Feng X, et al. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- 99.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 100.Farkas L, et al. Plasmacytoid dendritic cells (natural interferon- alpha/beta-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol. 2001;159:237–243. doi: 10.1016/s0002-9440(10)61689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahman A, Isenberg DA. Systemic lupus erythematosus. N. Engl. J Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 102.Ramos-Casals M, et al. Acute viral infections in patients with systemic lupus erythematosus: description of 23 cases and review of the literature. Medicine (Baltimore) 2008;87:311–318. doi: 10.1097/MD.0b013e31818ec711. [DOI] [PubMed] [Google Scholar]
- 103.Aslanidis S, et al. Parvovirus B19 infection and systemic lupus erythematosus: Activation of an aberrant pathway? Eur. J Intern. Med. 2008;19:314–318. doi: 10.1016/j.ejim.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 104.McClain MT, et al. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11:85–89. doi: 10.1038/nm1167. [DOI] [PubMed] [Google Scholar]
- 105.James JA, et al. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100:3019–3026. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kang I, et al. Defective control of latent Epstein-Barr virus infection in systemic lupus erythematosus. J Immunol. 2004;172:1287–1294. doi: 10.4049/jimmunol.172.2.1287. [DOI] [PubMed] [Google Scholar]
- 107.Toussirot E, Roudier J. Epstein-Barr virus in autoimmune diseases. Best. Pract. Res. Clin Rheumatol. 2008;22:883–896. doi: 10.1016/j.berh.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 108.Cohen-Solal JF, et al. Hormonal regulation of B-cell function and systemic lupus erythematosus. Lupus. 2008;17:528–532. doi: 10.1177/0961203308089402. [DOI] [PubMed] [Google Scholar]
- 109.Smith-Bouvier DL, et al. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanchez-Guerrero J, et al. A trial of contraceptive methods in women with systemic lupus erythematosus. N. Engl. J Med. 2005;353:2539–2549. doi: 10.1056/NEJMoa050817. [DOI] [PubMed] [Google Scholar]
- 111.Doria A, et al. Steroid hormones and disease activity during pregnancy in systemic lupus erythematosus. Arthritis Rheum. 2002;47:202–209. doi: 10.1002/art.10248. [DOI] [PubMed] [Google Scholar]
- 112.Sequeira JF, et al. Systemic lupus erythematosus: sex hormones in male patients. Lupus. 1993;2:315–317. doi: 10.1177/096120339300200507. [DOI] [PubMed] [Google Scholar]
- 113.Chang DM, et al. Dehydroepiandrosterone treatment of women with mild-to-moderate systemic lupus erythematosus: a multicenter randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2002;46:2924–2927. doi: 10.1002/art.10615. [DOI] [PubMed] [Google Scholar]
- 114.Ballestar E, et al. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176:7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- 115.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity - DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41:278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Balada E, et al. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124:339–347. doi: 10.1111/j.1365-2567.2007.02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lei W, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand. J Rheumatol. 2009:1–6. doi: 10.1080/03009740902758875. [DOI] [PubMed] [Google Scholar]
- 118.Sunahori K, et al. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol. 2009;182:1500–1508. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gorelik G, et al. Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol. 2007;179:5553–5563. doi: 10.4049/jimmunol.179.8.5553. [DOI] [PubMed] [Google Scholar]
- 120.Garaud S, et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol. 2009;182:5623–5632. doi: 10.4049/jimmunol.0802412. [DOI] [PubMed] [Google Scholar]
- 121.Tenbrock K, et al. The transcriptional repressor cAMP response element modulator alpha interacts with histone deacetylase 1 to repress promoter activity. J Immunol. 2006;177:6159–6164. doi: 10.4049/jimmunol.177.9.6159. [DOI] [PubMed] [Google Scholar]
- 122.Martin M, et al. Protein phosphatase 2A controls the activity of histone deacetylase 7 during T cell apoptosis and angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4727–4732. doi: 10.1073/pnas.0708455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mishra N, et al. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2628–2633. doi: 10.1073/pnas.051507098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li X, et al. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes Immun. 2009;10:380–389. doi: 10.1038/gene.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Girardi G, et al. Heparin prevents antiphospholipid antibody-induced fetal loss by inhibiting complement activation. Nat Med. 2004;10:1222–1226. doi: 10.1038/nm1121. [DOI] [PubMed] [Google Scholar]
- 126.Kowal C, et al. Human lupus autoantibodies against NMDA receptors mediate cognitive impairment. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19854–19859. doi: 10.1073/pnas.0608397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fleming SD, et al. Accelerated ischemia/reperfusion-induced injury in autoimmunity-prone mice. J Immunol. 2004;173:4230–4235. doi: 10.4049/jimmunol.173.6.4230. [DOI] [PubMed] [Google Scholar]
- 128.Fleming SD, et al. Anti-phospholipid antibodies restore mesenteric ischemia/reperfusion-induced injury in complement receptor 2/complement receptor 1-deficient mice. J Immunol. 2004;173:7055–7061. doi: 10.4049/jimmunol.173.11.7055. [DOI] [PubMed] [Google Scholar]
- 129.Bagavant H, Fu SM. Pathogenesis of kidney disease in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2009;21:489–494. doi: 10.1097/BOR.0b013e32832efff1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu K, et al. Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans. J Clin Invest. 2009;119:911–923. doi: 10.1172/JCI36728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takada K, et al. Cyclophosphamide for the treatment of systemic lupus erythematosus. Lupus. 2001;10:154–161. doi: 10.1191/096120301671376017. [DOI] [PubMed] [Google Scholar]
- 132.Ippolito A, Petri M. An update on mortality in systemic lupus erythematosus. Clin Exp Rheumatol. 2008;26:S72–S79. [PubMed] [Google Scholar]
- 133.Moore PA, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 134.Navarra S, et al. Belimumab, a BLyS-Specific Inhibitor, Reduced Disease Activity, Flares and Prednisone Use in Patients with Active SLE: Efficacy and Safety Results From the Phase 3 BLISS-52 Study. Arthritis Rheum. 2009 [Google Scholar]
- 135.Kishimoto T, Hirano T. Molecular regulation of B lymphocyte response. Annu. Rev Immunol. 1988;6:485–512. doi: 10.1146/annurev.iy.06.040188.002413. [DOI] [PubMed] [Google Scholar]
- 136.Tsai CY, et al. Increased excretions of beta2-microglobulin, IL-6, and IL-8 and decreased excretion of Tamm-Horsfall glycoprotein in urine of patients with active lupus nephritis. Nephron. 2000;85:207–214. doi: 10.1159/000045663. [DOI] [PubMed] [Google Scholar]
- 137.Sfikakis PP, et al. Rituximab anti-B-cell therapy in systemic lupus erythematosus: pointing to the future. Curr. Opin. Rheumatol. 2005;17:550–557. doi: 10.1097/01.bor.0000172798.26249.fc. [DOI] [PubMed] [Google Scholar]
- 138.Alarcon-Segovia D, et al. LJP 394 for the prevention of renal flare in patients with systemic lupus erythematosus: results from a randomized, double-blind, placebo-controlled study. Arthritis Rheum. 2003;48:442–454. doi: 10.1002/art.10763. [DOI] [PubMed] [Google Scholar]
- 139.Sidiropoulos PI, Boumpas DT. Lessons learned from anti-CD40L treatment in systemic lupus erythematosus patients. Lupus. 2004;13:391–397. doi: 10.1191/0961203304lu1032oa. [DOI] [PubMed] [Google Scholar]