Abstract
Characterization of immune receptors found in phylogenetically disparate species at the genetic, structural and functional levels has provided unique insight into the evolutionary acquisition of immune function. The roles of variable- and intermediate-type immunoglobulin (Ig) domains in direct recognition of ligands and other functions are far wider than previously anticipated. Common mechanisms of multigene family diversification and expansion as well as unique adaptations that relate to function continue to provide unique insight into the numerous patterns, processes and complex interactions that regulate the host response to infectious challenge.
Keywords: immunoglobulin domain, functional diversity, evolutionary variation, chimeric function, adaptive and innate immunity
1. Introduction
Phylogenetic studies of the origins of immune diversity have been extremely productive in elucidating the likely evolutionary mechanisms that led to the establishment of complexity in both immune receptor structure and interactions of immune cells in modern metazoan forms. Striking variation in receptor structure and mechanisms of diversification has been noted for some mediators of immune function, whereas other integral features exhibit remarkably stable histories. Certain immune receptors, e.g. the immunoglobulins (Igs) and T cell antigen receptors (TCRs) that are encoded in complex multigene families, achieve levels of variation beyond simple genomic polymorphism through somatic modification of their corresponding genes in individual lymphoid cells. Variation in immune receptors, as well as immunity as an integrated system, is exceptional in its unique and diverse set of adaptations throughout phylogeny [1].
As a major class of protein structure, the Ig-type molecules exhibit an exceptionally broad range of immunological function that extends to include roles as adaptive immune receptors, innate immune molecules and accessory molecules. The defining structural characteristic of this protein family is the Ig domain, a compact, globular structure of approximately 110 amino acids, which typically contains an intrachain disulfide bond. Three major subclasses of Ig domains are recognized: variable (V), intermediate (I) and constant (C1 or C2), based on common structural features [2]. It is not uncommon for multiple domain types to be present in the same receptor molecule. Primary sequence criteria have been defined that correlate with solved structures, making subclassification of Ig domains relatively reliable.
Ig domains not only mediate a broad range of immunological functions but some, in addition to antibodies and TCRs, exhibit different forms of somatic variation, e.g. fibrinogen-related proteins (FREPs), which encode Ig domains in molluscs and are proposed to mediate immune functions [3]. VCBPs (variable region-containing chitin-binding proteins), discussed below, exhibit extraordinarily high degrees of germline polymorphism [4,5,6]. Dscam (Down syndrome cellular adhesion molecule) in insects is encoded in a multiexonic gene that undergoes complex differential processing. Although the primary role of the various Dscam isoforms appears to be in neuronal patterning, a subset of these Ig C2-type molecules in Drosophila also function in bacterial immunity [7]. In addition to these immune-type molecules, exceptionally high levels of variation are seen in Ig domain-encoding genes that are associated with recognition of self/nonself, such as seen in allorecognition mediated by protochordates [8,9] and cnidarians [10,11]. Collectively, findings of this latter type are extending the traditional roles of Ig domains in immune functions.
As alluded to above, the V domains of Ig and TCRs, and more specifically, their somatically diversified forms, are the central structural features of both humoral and cellular adaptive immune responses. During the maturation of both T and B lymphocytes, rearrangement of germline elements takes place through a DNA sequence-dependent mechanism that is essentially restricted to these cell lineages. Whereas some of the diversity within the V domains of Ig and TCR is formed from the combinatorial rearrangement and joining of the multiple V, D (diversity) and J (joining) genetic elements encoded in the germline, most diversity in both Ig and TCR is achieved through non-templated deletion and addition of nucleotides at the junctions of the rearranged segmental elements. If this process of somatic variation does not introduce a stop codon or frameshift mutation and assuming that the length and composition of the polypeptide folds appropriately, the product of the rearranged genes is expressed as a unique receptor on the surface of a single cell. If a cognate determinant interacts with that ligand, the receptor-bearing cell will undergo clonal expansion. Furthermore, in the case of B cells, the productive rearrangement of the Ig locus, which occurs on only one chromosome, becomes a target for somatic mutation, giving rise to mutants, of which some exhibit increased affinities for the ligand that in turn are expressed and selected. Successive rounds of mutation can significantly change the character of the initial antibody that serves as a receptor. In certain circumstances, further changes in the structure can occur through a secondary process of rearrangement, termed receptor editing. Ultimately, the B cell clone differentiates to a plasma cell at which point the mRNA encoding the “selected” V region-containing Ig converts from a membrane-bound to a secreted form through differential RNA processing. The rearrangement process is much the same for V regions of the TCRs (which in the case of TCRα/β recognize peptides presented by MHC I or MHC II); however, TCRs do not exhibit secondary changes as seen in Ig and are not secreted. This elaborate mechanism of somatic variation and clonal selection arose through relatively few innovations that likely involved the functional redirection of a forerunner of contemporary jawed vertebrate RAG [12], the paralogous divergence of TdT within the Polμ family of polymerases and the capturing of essential elements of the physiologically ubiquitous DNA repair system. Activation-induced cytidine deaminase, which derives from the APOBEC family, effects several different processes associated with B cell receptor function, i.e., somatic mutation, class switch recombination and gene conversion. The fluidity and adaptability of the immune system and its diverse Ig domain-containing receptors is reflected in the unusually large number of species-specific differences in both the genomic organization of the rearranging clusters as well as in the mechanistic details of the recombination process seen in both Ig and TCR gene loci. The emerging picture from studies of genes that resemble those encoding immune-type receptors in several different chordate model systems presents a unique and essentially unanticipated view of immune receptors at the genome level that are now being addressed at the structural and functional levels.
2. The monomorphic to modest size family of V-type receptors
The Ig domain’s capacity to serve as an unusually plastic scaffold is well documented; of the three major subclasses of domains, the V-type appears to have taken on the greatest number of forms. The evolutionary origins of the V-type domain, its patterns of divergence and both its conventional and alternative functions have been the focus of the primary efforts in our laboratory. In addition to their extraordinarily well characterized roles in adaptive immunity mediated by T cells and B cells in jawed vertebrates, V regions also constitute an integral feature of several families of innate immune receptors in mammalian species, e.g. NKp30, NKp44, triggering receptor expressed on myeloid cells (TREM), TREM-like transcript (TLT), T-cell Ig mucin (TIM) and CD300 (see below). Superantigen can bind directly to the V region, outside of the primary antigen (peptide) binding site in TCR Vβ. Superantigens, which also can bind to antigen presenting cells, do not need to undergo intracellular processing (peptide formation) and their binding to nonpolymorphic regions of MHC II is independent of polymorphism. V domains also are integral features of accessory molecules that factor in adaptive immunity, e.g., CD8.
Besides the V-type, other Ig domains also are involved in immunological functions. Mammalian Ig superfamily (IgSF) molecules have been annotated most comprehensively, of which >850 proteins are predicted in the human genome [13]. Several genes within this group are encoded in the extended human leukocyte receptor complex (LRC) on chromosome 19q13 and include: killer-cell Ig-like receptors (KIRs), leukocyte Ig-like receptors (LILRs, also known as LIRs/ILTs/mouse PIRs), LAIR-1 and LAIR-2, Siglecs and CEACAMs. The KIRs, which function as NK cell receptors, are the best characterized at the functional level; both KIRs and LILRs recognize MHC I and MHC I-like molecules. LILR signaling is complex and its physiological consequences thus far are not well understood. Ligation of different LILRs on monocytes is associated with markedly different functional outcomes. The range of ligand recognition by these IgSF receptors extends to include: 1) LAIR-1 and LAIR-2, which bind collagen [14,15]; 2) CEACAMs, which recognize bacterial and viral pathogens and also mediate intercellular adhesion through both homophilic and heterophilic interactions [16]; 3) Siglecs, which bind sialic acid and regulate responses of lymphoid and myeloid cells [17], and 4) SIRPs, which recognize CD47, SP-A and SP-D [18]. TREM-1 is an inflammatory amplifier that is involved in septic shock. TREM-2 may recognize both endogenous and exogenous microbial ligands [19]. TLT-1 and TLT-2 represent an additional class of IgSF molecules and TLT-2 appears to recognize B7-H3, although the specifics of this interaction are a matter of some controversy [20,21]. The FcR and FcRL molecules represent yet another extremely widespread group of IgSF molecules in vertebrates; the functions of FcRLs are only beginning to be elucidated [22,23].
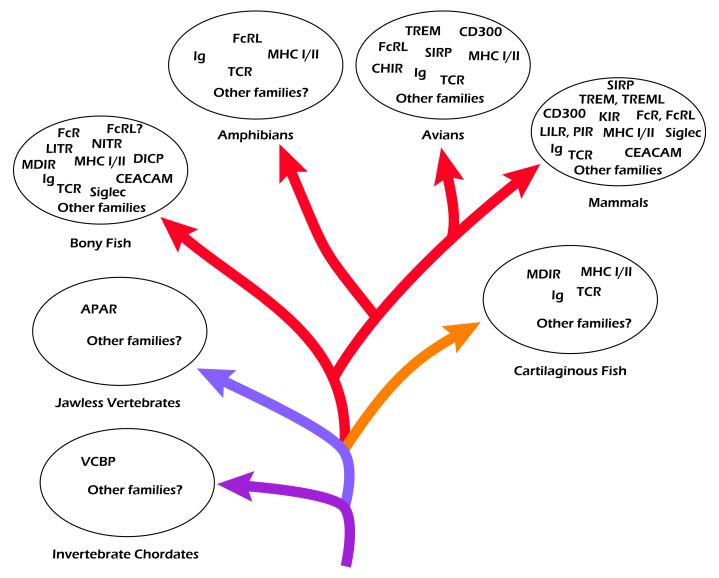
Despite their remarkable complexity, one consistent trend among IgSF receptors is that a large number of them are paired with related IgSF receptors that mediate antagonistic signaling. Many of these receptors appear to function in concert with cytokines, chemokines and other signals in the context of immunological synapses, at which their effects are concentrated and integration of signals is facilitated. Historically, studies of diverse IgSF molecules are approached one gene family or one receptor type at a time; however, it seems most likely from our current knowledge that all of these receptors function in an integrated system-wide manner that includes cooperative interactions as well as functional redundancy. Elucidation of these interactions, many of which are possibly very subtle, will be essential for advancing our understanding of the complex interactions that define responses by immune cells. Whereas the human IgSF dataset continues to represent the most extensive yet acquired, similar data are also beginning to emerge from studies in other vertebrate species (Fig. 1).
Fig. 1.
Phylogenetic distribution of representative immune-type molecules containing immunoglobulin superfamily (IgSF) domains in invertebrate (protochordate) and vertebrate (chordate) lineages. Arrows depict diversification of species over evolutionary time and are color-coded to illustrate the presence (or absence) of Ig and T cell antigen receptor (TCR) genes: red, combinatorial Ig and TCR; orange, combinatorial Ig and TCR plus noncombinatorial Ig; lavender, no definitive Ig or TCR but adaptive immune responses are mediated by variable lymphocyte receptors (VLRs); purple, no definitive Ig, TCR or VLR; variable region-containing chitin-binding protein (VCBP) is a putative antigen binding receptor. Select additional IgSF receptors and receptor families that are present in individual groups also are shown. Diversity of IgSF families among different chordate species is evident.
3. Novel immune-type receptors in bony fish
Given the primary role of the V region in forming specific antigen binding receptors, our initial focus was centered on potential alternative roles for this domain type. Several years ago we began to examine whether or not Ig-type V regions were employed by other systems of recognition in a manner that is equivalent or related to their role in antigen-binding by Ig and TCR molecules [24]. These investigations resulted in the discovery of the novel immune-type receptors (NITRs), an unexpectedly large family of Ig-V-domain-containing receptors in bony fish [24,25,26,27]. The vast majority of NITRs are chimeras of Ig-like and TCR-like extracellular V regions combined with transmembrane/cytoplasmic regions, reminiscent of paired regulatory receptors such as: KIRs, LILRs, FcRs and other IgSF molecules. NITRs are the only known multigene family of V-domain-containing immunoreceptors that approaches both the expanse and diversity of the segmental V elements found in the rearranging Ig and TCR genomic regions in jawed vertebrates; however, the cytoplasmic signaling domains and likely physiological functions of NITRs are very distinct from Ig and TCR in bony fish [28].
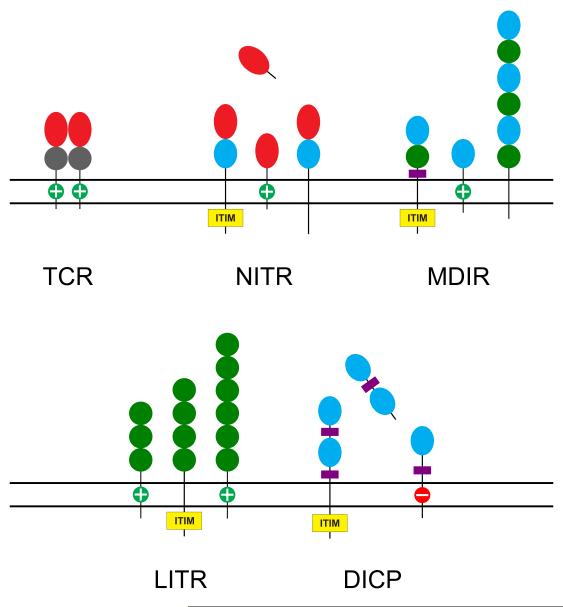
Twelve NITR gene families, defined as consisting of members that exhibit 70% or more identity at the predicted amino-acid-sequence level, initially were identified in zebrafish [27]; eight additional families were identified subsequently [29]. The 12 families contain a total of 36 different NITR genes, including both activating and inhibitory forms, and reside in a locus of ~350 kb on zebrafish chromosome 7. NITRs are composed of either one (V) or two (V plus I) ectodomains and most family members appear to be activating or inhibitory; however, several lack transmembrane domains (Fig. 2). Screening of both single-animal (Tübingen) BAC and multi-animal PAC libraries resulted in the identification of 138 different NITR alleles, implying high levels of polymorphism in the zebrafish population [30]. At that point, the only significant species to which the zebrafish NITRs could be compared in terms of sequence complexity and orthologous relationships was the pufferfish, in which NITR genes were described initially [25]. In subsequent studies, two additional NITR gene families were identified on zebrafish chromosome 14 [31]. Phylogenetic analyses showed that these genes are related more closely to each other than to the previously described 12 families of V regions. The relatively modest NITR locus on chromosome 14 consists of Nitr13, Nitr14a and Nitr14b. The chromosomal structure in the vicinity of Nitr13 exhibits features that suggest that this gene originated via retrotransposition.
Fig. 2.
Graphical representation of the general forms of novel immune-type receptor (NITR), modular domain immune-type receptor (MDIR), leukocyte immune-type receptor (LITR), diversified intermediate domain containing protein (DICP) and variable region-containing chitin-binding protein (VCBP) molecules in comparison to the mammalian TCR. LITRs have been annotated in multiple species of bony fish. DICPs also appear to be widespread in teleost fish (unpublished observations). Ig variable (V) domains are indicated by red ovals, intermediate (I) domains by cyan ovals, constant type 1 (C1) domains by gray circles and C2 domains by green circles. “+” indicates a positively charged transmembrane region predicted to interact with an activating transmembrane adaptor molecule. ITIM, immunoreceptor tyrosine-based inhibitory motif.
Although precise annotation of NITR gene families across all teleost lineages currently is not possible in the absence of genome resolution for most of these species, it appears that the patterns of diversification of NITRs among different teleost fishes are very distinct. The Japanese medaka fish (Oryzias latipes) is emerging as a developmental and genetic model system, sharing many of the advantages of zebrafish. Annotation of the genome of medaka led to the identification of 24 families of NITRs which were identified in clusters of genes on chromosomes 10, 18 and 21; as in the cases of pufferfish and zebrafish, extensive intrafamilial diversification is evident [32]. However, only minimal similarities in sequence are seen between the V regions of zebrafish, pufferfish and medaka NITRs. For instance, NITR V domain families in pufferfish are no more related to those seen in medaka than the different zebrafish NITR V domain families are related to each other. This pattern of sequence difference is consistent with a process of recent birth and death of genes. Thus, despite the separate divergence of NITRs from that of Ig and TCR, all of these diversified V-domain-containing receptors appear to be undergoing parallel types of evolution. Similar patterns of variation in Ig domains are seen in the non-rearranging primate KIR receptors in which rapid nucleotide polymorphism and haplotype variation, in the form of copy number variation, have been documented [33,34,35].
Examination of the structures of predicted NITR proteins suggests that the majority of NITRs function either through immunoreceptor tyrosine-based inhibition motif (ITIM)-based inhibitory signaling or through interaction with transmembrane adaptor molecules containing immunoreceptor tyrosine-based activation motifs (ITAMs) [28]. Genome annotation and cDNA studies in the zebrafish have identified six different predicted adaptor molecules that appear to have mammalian orthologs, namely Dap10, Dap12, CD3ζ, CD3ζ-like, FcRγ and FcRγ-like [36]. Cytotoxicity can be induced in human NK cells through transfection with the zebrafish activating receptor NITR9 [37]. Of the several isoforms of Nitr9 that have been identified, the Nitr9L (long) isoform has been shown to partner preferentially with the zebrafish ortholog of mammalian Dap12. Cross-linking of the zebrafish Nitr9L-Dap12 complex results in activation of the a pathway involving phosphytidylinositol 3-kinase, AKT and the extracellular signal-regulated kinase (ERK) [37]. Together, these data strongly suggest that NITRs share similar Dap12-based signaling pathways with those seen in mammalian FcRs and KIRs, underscoring the preservation of conserved primary signaling pathways during immune system evolution.
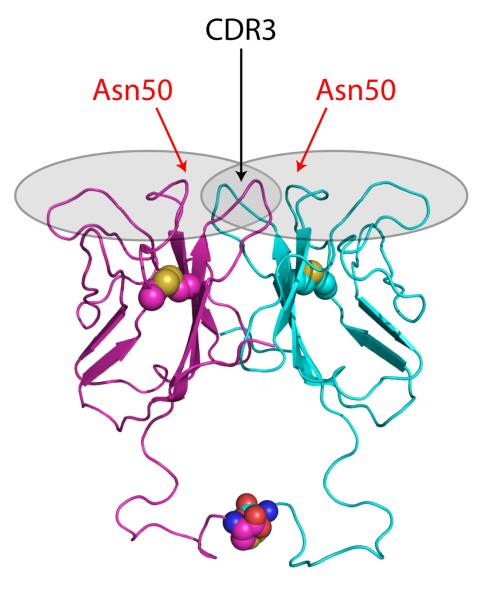
NITRs originally were hypothesized to be involved in allogeneic recognition based on the resemblance of the positively-charged transmembrane domains of some NITRs and the ITIM-containing cytoplasmic regions of others to similar features seen in the mammalian KIRs (among other IgSF molecules); the diversity of NITR V domains is reminiscent of the diversity of the C2 domains of KIR molecules and supports this possibility [25]. Multiple NITR transcripts were shown to be expressed by a channel catfish (Ictalurus punctatus) NK-like cell line that exhibits allogeneic cytotoxicity [26,38]. Cell-based assays in which different chimeric catfish NITR/mouse CD3ζ fusion proteins were used to induce GFP fluorescence in a reporter cell line after exposure to candidate target cells indicated that NITR11 interacts specifically with an allogeneic (MHC I- and MHC II-disparate) target B cell line. Subsequently, soluble chimeric catfish NITR11-human IgG1 Fc (NITR11-Fcγ) fusion proteins were shown to bind to the same B cell allogeneic target cell line. Through a series of site-directed mutagenesis experiments, in which both loss and gain of function NITR mutants were derived, the allogeneic interaction of catfish NITR11 was shown to be dependent on a single amino acid (Asn50) within the V domain [39]; mutation of this amino acid (Asn50→Asp50) completely abrogated allogeneic binding. Catfish NITR10, which did not exhibit allogeneic specificity in these assays but is 93% identical at the amino acid level to the corresponding domain of catfish NITR11, also was used as a substrate for mutagenesis studies. Mutation of Asp50→Asn50 in NITR10 resulted in a gain of allogeneic cell binding function similar to NITR11. Based on these observations, NITR10, NITR11, and their respective gain and loss of function single-site mutants were crystallized in both native and selenomethionyl forms. The structures of these eight NITR proteins, as well as a native, dimeric NITR11 molecule containing an interchain disulfide bond, were solved [39,40]. Collectively, these data demonstrate that Asn50 of NITR11, which is located in a region corresponding to CDR1 in Ig and TCR, is critical for allogeneic binding specificity that likely is dependent on charge distributions at the binding surface. Although strong similarities in tertiary and quaternary structure were noted between the adaptive antigen-binding receptors (Ig and TCR) and NITRs, the crucial amino acid at position 50 in NITR11 resides in the CDR1-equivalent region (Fig. 3). By contrast, CDR3 is derived somatically and is the most highly variable region of the adaptive receptors and is involved directly in ligand binding in both Ig and TCR.
Fig. 3.
Solved structure of a dimer of a novel immune-type receptor (NITR). The position of Asn50, critical for interaction with ligand(s) in catfish NITR11, is indicated. Gray shading defines the conventional antigen binding receptor complementarity determining region (CDR)-corresponding regions; the position of CDR3 is specified further. Disulfide-linked cysteine residues are depicted by color-coded spheres (carbon, magenta and cyan; sulfur, yellow; nitrogen, blue; oxygen, red).
Binding of catfish NITRs to determinants on allogeneic cells may account for cytotoxic reactivity and thereby resemble the functions of several families of mammalian NK cell receptors, such as KIR and Ly49, and suggests a previously unappreciated relationship at the receptor level between innate and adaptive immune recognition. As alluded to above, V regions are also found in mammalian NK-type receptors such as NKp30 and NKp44; however, these receptors are encoded by single-copy, minimally polymorphic genes. In contrast, the diversified V domains of NITRs appear to have been derived from the Ig and TCR loci themselves but, during the considerable time after that separation, have diverged in both structure and function [39]. Although still speculative, it appears that at least one genetic recombination event between a V element of a rearranging antigen-binding receptor and a member of another gene family involved in activating/inhibitory signaling may have occurred in the ancestors of NITR genes.
Although the V regions of NITRs do not undergo rearrangement and represent a functional departure from the rearranging antigen-binding receptors, their retention of the core V-frame residues and intimate participation of one CDR in creating a ligand binding specificity preserve two central characters of V regions associated with the rearranging antigen-binding receptors. Their function (from one well characterized example) appears to be in line with that exhibited by mammalian NK cells, i.e., recognition of an allogeneic determinant. Although, the molecular character of the determinant that is recognized is not understood, the NITRs seem exceptionally complex and there is a strong possibility that certain members of the NITR gene family may have taken on very different roles in terms of the range of ligands they may recognize. Not only are the families large but their interfamily and interspecies relatedness implies very rapid evolution, a phenomenon that also is attributed to conserved rearranging antigen-binding receptors, NK receptors and certain MHC genes.
4. Modular domain immune-type receptors
NITR-V-region consensus sequences were used to design primers for analysis of cDNA recovered from the clearnose skate Raja eglanteria (a cartilaginous fish) using the “Amptrap” technique, a RACE-PCR cloning method that we had previously developed to isolate cDNAs encoding secreted or membrane proteins containing single, very short, conserved amino-acid-motifs [4]. Like bony fishes, the cartilaginous fishes exhibit complex humoral and cellular immune responses and possess T cells and B cells; these species also possess lymphoid organs with clear mammalian orthologs, such as spleen and thymus, as well as organs that are unique to these groups, such as the Leydig and epigonal organs and rectal gland [41]. Our initial efforts were directed towards identifying NITR sequence orthologs in cartilaginous fish species; however, such sequences were not obtained, raising the possibility that the distribution of NITRs is limited to teleost fish. However, these experiments identified a complex IgSF receptor family encoding modular domain immune-type receptors (MDIRs) [42], composed of I-type and C2-type IgSF domains. The basis for the identification of MDIRs using a V region-direct approach involves the sharing of sequence motifs (but not length [spacing] relationships) between V and other Ig domain types. The current set of predicted MDIRs encodes between one and six individual extracellular IgSF domains; like NITRs, the MDIR family exists in both activating and inhibitory forms (Fig. 2). In marked contrast to NITRs, orthologs of MDIRs have been identified in a number of phylogenetically disparate vertebrate groups, including teleost fish, amphibians, birds and mammals. In terms of predicted peptide structure, MDIRs in cartilaginous fish are most similar to mammalian CD300 genes, which encode a family of Ig-V-domain-containing receptors that are predicted to modulate the activation of immune cells [43,44]. MDIRs also exhibit relatedness to the extracellular domains of the TREM, TLT, TIM and polymeric Ig receptor (pIgR) families, implying that MDIR, CD300, TREM, TLT, TIM and pIgR genes all diverged from a common, very ancient ancestor and subsequently dispersed in both structure and function [42]. MDIR sequence orthologs are not yet apparent in any species outside of the jawed vertebrates (including in the in-process genome sequence of the sea lamprey, a jawless vertebrate). It is perhaps noteworthy that the jawless vertebrates also do not posses unequivocal orthologs of Ig, TCR and major histocompatibility complex (MHC) class I and II genes.
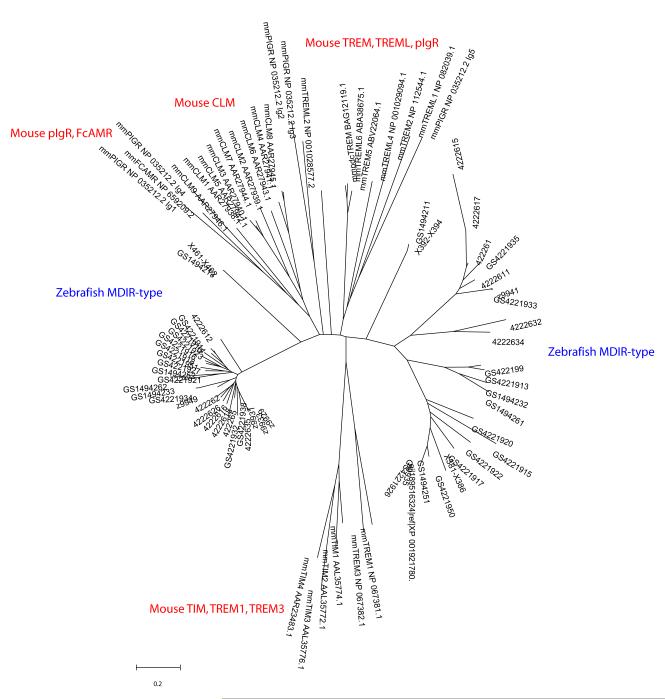
Using both skate MDIR and mouse CD300 domains as BLAST queries, a large set of MDIR-type genes can be identified in the nearly complete zebrafish genome sequence. Similar to the findings in skate, zebrafish MDIR-type genes appear to be dramatically expanded in number relative to MDIR-type genes found in mammals. The genes reside in two major complexes, each consisting of multiple loci, as well as in several other genomic regions. A preliminary examination of these loci revealed at least 20 MDIR-type domains of two types as linked pairs.
From the above data it appears that MDIRs and their mammalian relatives comprise a very large gene family that is found throughout the jawed vertebrates and has diversified considerably during the evolution of extant vertebrate forms (Fig. 4). The pronounced divergence of MDIR ectodomains, even in relatively closely related species (e.g., human and mouse), suggests that MDIR specificities may be redirected in species-specific ways that are under rapid selection. Recent data indicate that skate and mouse MDIR-related genes may share similar ligand-binding functions (Cannon, unpublished observations).
Fig. 4.
Diversity of MDIR-related domains encoded in the zebrafish genome. Neighbor-joining tree analyses depict phylogenetic relationships among (predicted) Ig I-type domains in MDIR-related sequences identified in zebrafish. Ensemble version 8 as well as additional alleles identified by our laboratory (not described previously) are included in the set of zebrafish sequences. MDIR domain sequences are compared to known reference mouse MDIR-related domains. Trees are unrooted. Certain groups of domains are specified.
In addition to MDIRs and NITRs, zebrafish encode representatives of leukocyte immune-type receptor genes described originally in catfish, as well as an additional large set of receptors that we provisionally have designated diverse I domain-containing proteins (DICPs) (Fig. 2). Like NITRs, DICPs appear to be a bony fish-specific innovation; their function is unknown. The presence of several large families of IgSF receptors in all vertebrates examined thus far at the whole-genome level argues for a profound role for these families in initiation, elaboration or regulation of immune responses.
5. Variable region-containing chitin-binding proteins in amphioxus
Like all species outside of the jawed vertebrates, the protochordate amphioxus (Branchiostoma floridae, a representative species of cephalochordate) as well as other invertebrates lack definitive sequence orthologs of Ig and TCR. The Amptrap technique was applied originally in efforts to recover IgSF-encoding cDNAs and resulted in the identification of the VCBP genes, which encode soluble proteins possessing tandem V-type domains fused to a C-terminal chitin-binding domain (Fig. 2). VCBPs exhibit very high levels of localized sequence polymorphism that is focused at the N-termini of the V domains [4,5]. Since their original description, resolution of the amphioxus genome has allowed the enumeration and annotation of the VCBP loci, which are comprised of one main complex encoding four of the five VCBP subfamilies; a fifth family (VCBP3) maps to a separate chromosomal region [6].
Extensive sequence surveys of VCBP genes from wild amphioxus populations demonstrate a high level of haplotypic variation and the apparent maintenance of an exceptionally large pool of allelic variants. Nearly every individual amphioxus encodes a distinct repertoire of VCBPs, despite the presence of only a limited number of VCBP genes in a given individual [5]. Localized hypervariation in the V as well as C-terminal chitin-binding domains of VCBPs represent two structural features of potential immunological significance. VCBP molecules presumably are bifunctional at a biochemical level and could represent an important transitional form of molecule in the evolution of V region-mediated immunity, bridging critical features of conventional innate and adaptive immune function [45].
The two major sites of hypervariation in the V domains of VCBPs are displaced from the sequence regions that form the antigen combining site of Ig and TCR molecules. To examine their structural relationships, two variants of a VCBP3 molecule were expressed in bacteria, refolded, crystallized and used to solve the structure of the V domains (V1•V2) (Fig. 5) [46,47]. The patterns of polypeptide folding and secondary structure of each V domain are essentially superimposable with those of Ig and TCR V domains. A convincing rationale for the observed genetic hypervariation is provided by contiguous sequence variation along the back sheet sequence of V1•V2. However, the hypothetical VCBP counterparts of the CDRs, which by analogy to Ig and TCR V-domain heterodimers would be expected to reside at the sites of highest sequence variation and presumed ligand binding, are opposed in orientation by 180° relative to that seen in conventional antigen-binding receptors. These observations suggest strongly that VCBPs bind to their hypothesized ligands through a distinct structural mechanism [47]. The pattern of utilization of different V folds to create the potential recognition site in VCBPs is markedly different from that seen in Ig and TCR as well as in an NITR [38]. The in vivo ligand specificities and functions of VCBPs currently are under investigation.
Fig. 5.
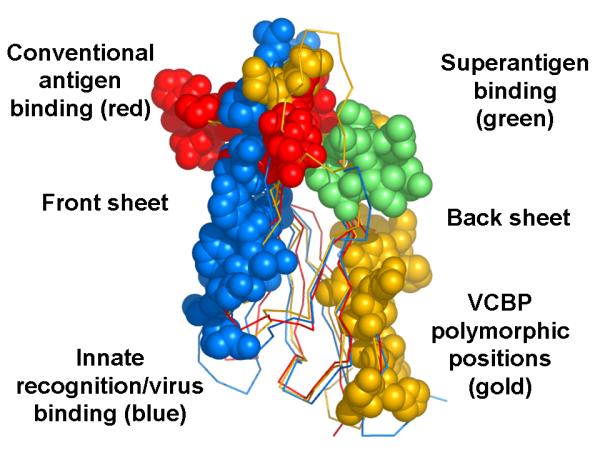
Functionally defined regions in the V-type fold of VCBP3. The diversity of recognition-interaction functions of the V domain and the diversity in defined and predicted (in this case VCBPs) binding sites are emphasized.
The domain configuration of VCBPs may represent a common feature of immune-type receptors in invertebrates, i.e. the juxtaposition of Ig domains with a C-terminal domain of another general class, as such an arrangement also has been reported in the FREPs of the snail Biomphalaria glabrata. FREPs, which interact with parasite-derived molecules, contain N-terminal IgSF domains and C-terminal fibrinogen-like domains [48,49]. In addition, predicted gene products that encode Ig variable domains fused to chitin-binding domains have been identified in the genome sequences of representative Annelid, Mollusc and Cnidarian species. Although these relationships have not been confirmed yet at the cDNA level, they argue for a widespread utility of this particular domain combination (Haire and Dishaw, unpublished).
6. Concluding remarks
Immune receptors are ubiquitous in all metazoans and occur in a remarkable array of different molecular forms that serve to combat pathogenic threats. Even when viewed only from the standpoint of the IgSF as a general structural class, this diversity of form and specialization of direct function, such as nonself ligand binding or interaction with endogenous molecules, is remarkable. Among the various subtypes of IgSF domains, the V domain remains the defining structural feature of the antigen binding receptors that mediate adaptive immune recognition in jawed vertebrates.
Ig- and TCR-mediated adaptive immunity, which derived early in the evolution of the jawed vertebrate form, likely conferred a tremendous selective advantage. For this reason, the lineage of IgSF receptors that includes Ig and TCR likely became fixed in structure and function, ultimately diversifying into the highly complex multigene families found in modern organisms. Likewise, it appears that molecules such as NITRs were recruited to separate functions in the teleost lineage, whereas MDIR and MDIR-like genes became associated with still another range of function that may be fundamental to immunity in all jawed vertebrates. These differences in structure and function are of significant interest as they underscore the remarkable plasticity of the Ig V domain (and in the case of MDIRs, I domain) and accentuate its role in shaping modern immune systems as they have evolved to respond rapidly to pathogenic organisms that are under continuous selection for variation of their molecular signatures. According to a model of this type, the long-sought definitive assignment of sequence orthologs of Ig and TCR in species outside jawed vertebrates may prove an intractable issue and, at best, could be incidental based on stochastic similarity of exceptionally derived and heavily diversified sequences. Rather, elucidation of the specific, interrelated functions of all of the modern sets of IgSF (and other) receptors that simultaneously signal during an immune response has begun to appear much more relevant to our complete understanding of the immune response and is the current focus of our present efforts.
ACKNOWLEDGEMENTS
We wish to thank Barbara Pryor for editorial assistance. This research was supported by grants R01 AI23338 and R01 AI57559 to GWL from the National Institutes of Health and a grant to JPC from the All Children’s Hospital Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Litman GW, Cooper MD. Commentary: Why study the evolution of immunity? Nat Immunol. 2007;8:547–548. doi: 10.1038/ni0607-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- [3].Zhang S-M, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- [4].Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. 2002;3:1200–1207. doi: 10.1038/ni849. [DOI] [PubMed] [Google Scholar]
- [5].Cannon JP, Haire RN, Schnitker N, Mueller MG, Litman GW. Individual protochordates possess unique immune-type receptor repertoires. Curr Biol. 2004;14:R465–R466. doi: 10.1016/j.cub.2004.06.009. [DOI] [PubMed] [Google Scholar]
- [6].Dishaw LJ, Mueller MG, Gwatney N, Cannon JP, Haire RN, Litman RT, et al. Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genomics. 2008;9:78. doi: 10.1186/1471-2156-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- [8].De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, et al. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nyholm SV, Passegue E, Ludington WB, Voskoboynik A, Mitchel K, Weissman IL, et al. fester, A candidate allorecognition receptor from a primitive chordate. Immunity. 2006;25:163–173. doi: 10.1016/j.immuni.2006.04.011. [DOI] [PubMed] [Google Scholar]
- [10].Cadavid LF, Powell AE, Nicotra ML, Moreno M, Buss LW. An invertebrate histocompatibility complex. Genetics. 2004;167:357–365. doi: 10.1534/genetics.167.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nicotra ML, Powell AE, Rosengarten RD, Moreno M, Grimwood J, Lakkis FG, et al. A hypervariable invertebrate allodeterminant. Curr Biol. 2009;19:583–589. doi: 10.1016/j.cub.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci USA. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- [14].Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) J Leukoc Biol. 2008;83:799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- [16].Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- [17].Crocker PR. Siglecs in innate immunity. Curr Opin Pharmacol. 2005;5:431–437. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- [18].Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- [19].Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- [20].Hashiguchi M, Kobori H, Ritprajak P, Kamimura Y, Kozono H, Azuma M. Triggering receptor expressed on myeloid cell-like transcript 2 (TLT-2) is a counter-receptor for B7-H3 and enhances T cell responses. Proc Natl Acad Sci USA. 2008;105:10495–10500. doi: 10.1073/pnas.0802423105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Leitner J, Klauser C, Pickl WF, Stockl J, Majdic O, Bardet AF, et al. B7-H3 is a potent inhibitor of human T-cell activation: No evidence for B7-H3 and TREML2 interaction. Eur J Immunol. 2009;39:1754–1764. doi: 10.1002/eji.200839028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nimmerjahn F, Ravetch JV. Fc-receptors as regulators of immunity. Adv Immunol. 2007;96:179–204. doi: 10.1016/S0065-2776(07)96005-8. [DOI] [PubMed] [Google Scholar]
- [23].Davis RS. Fc receptor-like molecules. Ann Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- [24].Rast JP, Haire RN, Litman RT, Pross S, Litman GW. Identification and characterization of T-cell antigen receptor related genes in phylogenetically diverse vertebrate species. Immunogenet. 1995;42:204–212. doi: 10.1007/BF00191226. [DOI] [PubMed] [Google Scholar]
- [25].Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, et al. A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci USA. 1999;96:15080–15085. doi: 10.1073/pnas.96.26.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hawke NA, Yoder JA, Haire RN, Mueller MG, Litman RT, Miracle AL, et al. Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci USA. 2001;98:13832–13837. doi: 10.1073/pnas.231418598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, et al. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian lymphocyte receptor cluster. Proc Natl Acad Sci USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoder JA. Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol. 2009;33:135–144. doi: 10.1016/j.dci.2008.09.004. [DOI] [PubMed] [Google Scholar]
- [29].Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, et al. Resolution of the NITR gene cluster in zebrafish. Proc Natl Acad Sci USA. 2004;101:15706–15711. doi: 10.1073/pnas.0405242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Traver T, Herbomel P, Patton EE, Murphy R, Yoder JA, Litman GW, et al. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- [31].Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW. Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenet. 2008;60:257–265. doi: 10.1007/s00251-008-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Desai S, Heffelfinger AK, Orcutt TM, Litman GW, Yoder JA. The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. Evol Biol. 2008;8:177–188. doi: 10.1186/1471-2148-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ota T, Sitnikova T, Nei M. Evolution of vertebrate immunoglobulin variable gene segments. Curr Top Microbiol Immunol. 2000;248:221–245. doi: 10.1007/978-3-642-59674-2_10. [DOI] [PubMed] [Google Scholar]
- [34].Martinez-Borra J, Khakoo SI. Speed and selection in the evolution of killer-cell immunoglobulin-like receptors. Int J Immunogenet. 2008;35:89–96. doi: 10.1111/j.1744-313X.2008.00756.x. [DOI] [PubMed] [Google Scholar]
- [35].Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- [36].Yoder JA, Orcutt TM, Traver D, Litman GW. Structural characteristics of zebrafish orthologs of adaptor molecules that associate with transmembrane immune receptors. Gene. 2007;401:154–164. doi: 10.1016/j.gene.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wei S, Zhou J, Chen X, Shah RN, Liu J, Orcutt TM, et al. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenet. 2007;59:813–821. doi: 10.1007/s00251-007-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shen L, Stuge TB, Bengten E, Wilson M, Chinchar VG, Naftel JP, et al. Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus) Dev Comp Immunol. 2004;28:139–152. doi: 10.1016/s0145-305x(03)00119-8. [DOI] [PubMed] [Google Scholar]
- [39].Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Prada JA Hernandez, et al. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity. 2008;29:228–237. doi: 10.1016/j.immuni.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ostrov DA, Prada JA Hernandez, Haire RN, Cannon JP, Magis AT, Bailey K, et al. Crystallization and X-ray diffraction analysis of a novel immune-type receptor from Ictalurus punctatus and phasing by selenium anomalous dispersion methods. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:1035–1037. doi: 10.1107/S1744309107054231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Miracle AL, Anderson MK, Litman RT, Walsh CJ, Luer CA, Rothenberg EV, et al. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol. 2001;13:567–580. doi: 10.1093/intimm/13.4.567. [DOI] [PubMed] [Google Scholar]
- [42].Cannon JP, Haire RN, Mueller MG, Litman RT, Eason DD, Tinnemore D, et al. Ancient divergence of a complex family of immune-type receptor genes. Immunogenet. 2006;58:362–373. doi: 10.1007/s00251-006-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yotsumoto K, Okoshi Y, Shibuya K, Yamazaki S, Tahara-Hanaoka S, Honda S, et al. Paired activating and inhibitory immunoglobulin-like receptors, MAIR-I and MAIR-II, regulate mast cell and macrophage activation. J Exp Med. 2003;198:223–233. doi: 10.1084/jem.20021825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chung DH, Humphrey MB, Nakamura MC, Ginzinger DG, Seaman WE, Daws MR. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclase formation. J Immunol. 2003;171:6541–6548. doi: 10.4049/jimmunol.171.12.6541. [DOI] [PubMed] [Google Scholar]
- [45].Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: new perspectives. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Prada JA Hernandez, Haire RN, Cannon JP, Litman GW, Ostrov DA. Crystallization and preliminary x-ray analysis of VCBP3 from Branchiostoma floridae. Acta Crystallogr D Biol Crystallogr. 2004;60:2022–2024. doi: 10.1107/S0907444904020827. [DOI] [PubMed] [Google Scholar]
- [47].Prada JA Hernandez, Haire RN, Allaire M, Jakoncic J, Stojanovic N, Cannon JP, et al. Ancient evolutionary origin of diversified variable regions revealed by crystal structures of an immune-type receptor in amphioxus. Nat Immunol. 2006;7:875–882. doi: 10.1038/ni1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Leonard PM, Adema CM, Zhang S-M, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalaria glabrata. Gene. 2001;269:155–165. doi: 10.1016/s0378-1119(01)00444-9. [DOI] [PubMed] [Google Scholar]