Abstract
The Type III Secretion System (T3SS) is essential for the infectivity of many pathogenic Gram-negative bacteria. The T3SS contains proteins that form a channel in the inner and outer bacterial membranes, as well as an extracellular needle that is used for transporting and injecting effector proteins into a host cell. The homology between the T3SS and the bacterial flagellar system has been firmly established, based upon both sequence similarities between respective proteins in the two systems and the structural homology of the higher-order assemblies. It has previously been shown that the Shigella flexneri needle has a helical symmetry of ~ 5.6 subunits per turn, which is quite similar to that of the most intensively studied flagellar filament, from Salmonella typhimurium, which has ~ 5.5 subunits per turn. We now show that the S. typhimurium needle, expected by homology arguments to be more similar to the S. typhimurium flagellar filament than is the needle from Shigella, actually has ~ 6.3 subunits per turn. It is not currently understood how host cell contact, made at the tip of the needle, is communicated to the secretory system at the base. In contrast to the S. typhimurium flagellar filament, which shows a nearly crystalline order, the S. typhimurium needle has a highly variable symmetry, which could be used to transmit information about host cell contact.
Introduction
Many pathogenic Gram-negative bacteria contain T3SSs that are essential for infectivity1. These T3SSs serve as both a secretion and injection system for bacterial effector proteins that need to be passed through the inner and outer bacterial membranes and into a host cell. The strong morphological and sequence similarities between the T3SS and the bacterial flagellar system gave rise to the suggestion that these systems are homologs, having common origins2, 3. Consistent with this, it was shown4 that the needle of the S. flexneri T3SS has approximately the same helical symmetry (5.6 subunits per turn of a 24 Å pitch helix) as the best characterized flagellar filament5, from S. typhimurium (~5.5 subunits per turn of a 26 Å pitch helix). Since the contact between the bacterium and a host cell has been shown to switch the T3SS to a secretory state, attempts have been made to find changes in helical structure of the S. flexneri needle with mutations in the needle protein MxiH which lock the T3SS into a constitutive “on” state6. Given the homology between the T3SS and the flagellar system, it was thought that a switching of state of the needle might occur in a similar manner to the switching of the flagellar filament from a left-handed to a right-handed state of the protofilaments7. These studies6 failed to find any changes in the average helical parameters, leading to the suggestion that such changes in helical symmetry might be too small to be detected by the analysis methods used, or that signal transduction in this system occurs through some novel mechanism that has not yet been characterized.
Results
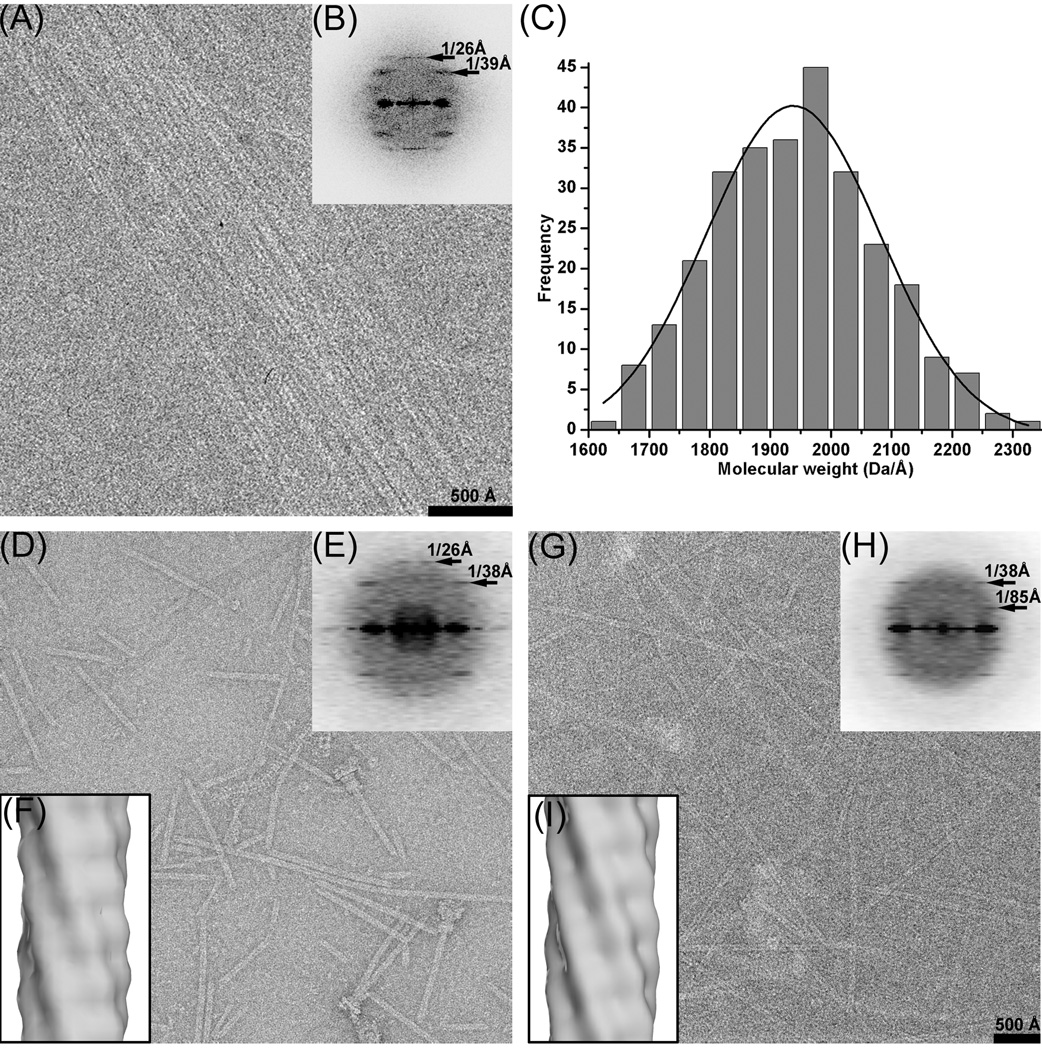
The S. typhimurium needles have been imaged by both negative stain (Fig. 1d) and cryo-EM (Fig. 1a,g). Fields of needles imaged by cryo-EM (Fig. 1a) were used to generate an averaged power spectrum (Fig. 1b) that is quite similar to a fiber diffraction pattern as it shows no sampling due to an ordered arrangement of filaments. This power spectrum shows only three layer lines: one at ~ 1/(26 Å) arising from a one-start helix, a more diffuse one at ~ 1/(38 Å), and a very diffuse one at ~ 1/(85 Å). The broadening of these other layer lines is a characteristic feature of a variability in twist of a helical structure8, where helical symmetry is not exactly conserved between every adjacent subunit. As a result, deviations in twist accumulate along a filament, and long range order does not exist. Indexing these two more diffuse layer lines was problematic, as multiple solutions are possible that are consistent with the diameter of these particles. The layer lines at 1/(38 Å) and 1/(85 Å) could have any Bessel order less than eight. The power spectrum at this resolution simply does not uniquely specify the helical symmetry, due to the fact that for many structures at limited resolution there are intrinsic ambiguities in determining helical symmetry9. Similarly, multiple solutions can be obtained with the Iterative Helical Real Space Reconstruction (IHRSR) method10. For example, a scheme with 1/(38 Å) being n=−3, and 1/(85 Å) being n=4 (3.7 units/turn), and a scheme with 1/(38 Å) being n=7, and 1/(85 Å) being n=−6 (6.3 units/turn), both led to stable solutions with reconstructions generating power spectra indistinguishable from the power spectra generated from the images. We therefore used Scanning Transmission Electron Microscopy (STEM) to get an accurate estimate of the mass per unit length in these needles11. The STEM measurements indicated that the average mass/length was ~ 2.0 kDa/Å (Fig. 1c). Since the component protein of the S. typhimurium needle, PrgI, contains 80 residues and is 8.4 kDa, this would suggest that the axial rise per subunit is on average ~ 4.2 Å, and that there are on average ~ 6.2 subunits per turn of the 26 Å pitch one-start helix.
Fig. 1.
(a) Cryo-EM image of bundles of needles. (b) averaged power spectrum generated from 14 non-overlapping boxes (512×512 px, 5.9 Å/px) taken from bundles such as in (a). (c) STEM mass histogram, with a gaussian curve fitted. (d) negatively stained needles. (e) averaged negative stain power spectrum from 259 non-overlapping boxes, each 480 px long (1.33 Å/px). (f) overall IHRSR three-dimensional negative stain reconstruction derived from 5,420 segments (120 px long, 5.32 Å/px), filtered to 24 Å resolution. This reconstruction converged to a helical symmetry of 57° rotation and an axial rise of 4.2 Å, which corresponds to 6.3 subunits per turn of a 26.5 Å pitch helix. (g) Cryo-EM of isolated filaments. (h) averaged cryo-EM power spectrum from 850 non-overlapping boxes, each 480 px long (1.33 Å/px). (i) overall IHRSR cryo-EM three-dimensional reconstruction derived from 22,834 segments at (120 px long, 2.66 Å/px), filtered to 24 Å resolution. This converged to the same symmetry as the negative stain reconstruction (57°/4.2 Å).
This knowledge of mass per length was then used to generate overall reconstructions of the needles from both negative stain (Fig. 1f) and cryo-EM (Fig. 1i). The failure of the IHRSR method to converge to the same solution from different starting symmetries, as well as the broad layer lines seen in the averaged power spectrum from bundles (Fig. 1b), strongly suggested that these samples were heterogeneous with respect to the helical parameters. Power spectra computed from subsets of the filaments after sorting (Supplementary Movie) show the very large variability in twist, with a range that extends beyond 6.2 to 6.5 subunits/turn. The Supplementary Movie displays a fixed n=7 layer line at ~ 1/(38 Å), with a very variable n=−6 layer line.
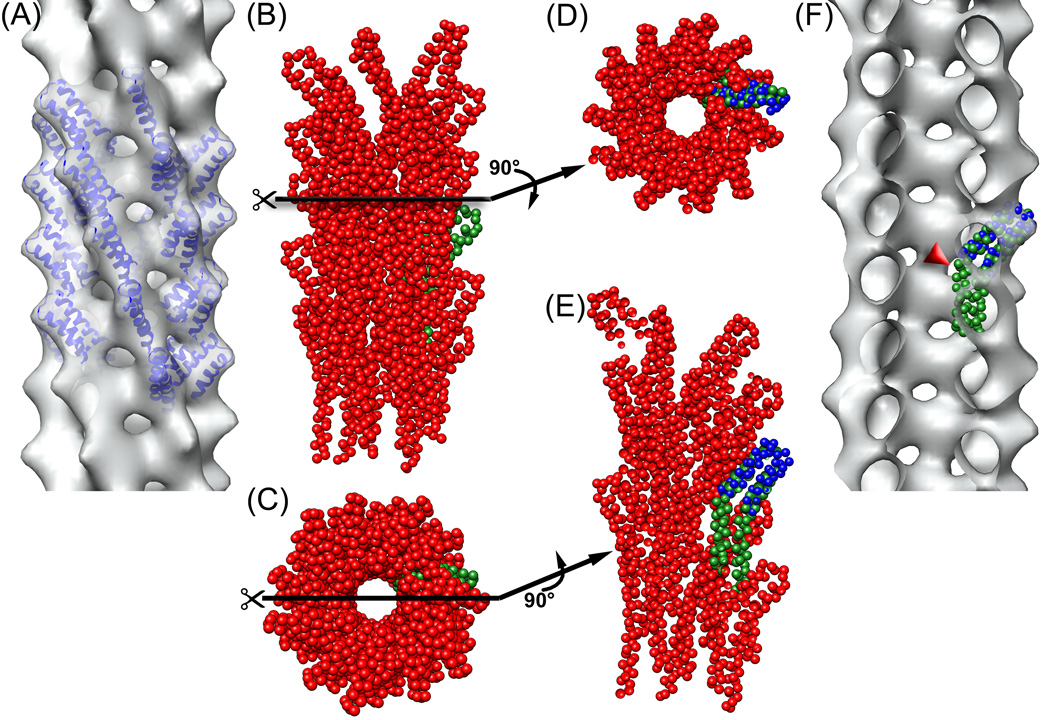
The best reconstruction (Fig. 2a) was obtained from the central bin (6.3 subunits/turn) of the cryo-EM filaments, using 9,366 segments from the total of 22,834 segments. The most conservative estimate12 of the resolution of this reconstruction is 19.5 Å, using a Fourier Shell Correlation of 0.5, and two completely independent reconstructions generated from different starting symmetries and using completely different images. This actually provides an estimate of the resolution for a reconstruction containing half of the total images, and must therefore be an underestimate of the resolution of the full data set. Filtering an atomic model of the two-helix bundle within the PrgI subunit13 (residues 18–60, Fig. 2a) to 18 Å resolution provides a good match to the reconstruction and suggests that this is a better estimate of the resolution. This resolution is insufficient to determine the absolute hand of the reconstruction, as the two enantiomorphic volumes that can be produced are fit equally well by the PrgI two helix bundle that projects out. We have therefore adopted the same hand (a right-handed one start helix) assumed for the S. flexneri needle4, 14, although the hand was not determined for that needle and assumed to be the same as that of the S. typhimurium flagellar hook and filament.
Fig. 2.
(a) reconstruction of the frozen-hydrated filaments filtered to 18 Å resolution, using 9,366 segments from the total 22,834 segments. The sorting was based on the variable twist and axial rise while keeping the n=+7 layer line fixed at 1/(38 Å). This class from the center of the distribution yielded a symmetry of 57°/4.2 Å). Residues 18 to 60 (blue ribbons) from the solution structure13 of the S. typhymurium needle subunit (pdb entry 2JOW) were manually docked into the reconstruction. (b) side view of the atomic model14 of S. flexneri T3SS needle (pdb entry 2V6L), with one of the protomers shown in green, and (c) top view of this model. (d) the atomic model of the S. flexneri T3SS needle in (b) cut along the plane perpendicular to the helical axis (black line). One of the protomers (green) was aligned with residues 18–60 of the S. typhymurium needle subunit (blue), positioned as fit to the reconstruction shown in (a). Despite the difference in symmetry, the paired helices of the needle protomers from both bacteria are located at the same radius and have the same orientation. (e) the atomic model of S. flexneri T3SS needle in (c) cut along the plane parallel to the helical axis (black line). It can be seen in this view, as well, that the paired helices in the models for both bacteria are located at the same radius and have the same orientation. (f) The reconstruction in (a) cut along the plane parallel to the helical axis. When the subunit from the atomic model14 of the S. flexneri needle (green) is docked into the reconstruction of the Salmonella needle, the N-terminal part of it sticks out from the volume (red arrowhead).
The outer diameter of the reconstructed volume is ~ 85 Å, and the central lumen has a diameter of ~ 25 Å. Docking the two helix bundle (residues 18–60) from the center of the PrgI sequence into the reconstructed volume (Fig. 2a) shows a remarkably good agreement (Fig. 2d,e) with the corresponding portion of the atomic model14 proposed for the S. flexneri needle. Unfortunately, the limited resolution of our present reconstruction, combined with the structural plasticity of the PrgI subunit outside of the fixed two helix bundle13, prevent us from generating a pseudo-atomic model for the remainder of the PrgI subunit that must be surrounding the lumen. It is quite likely that the residues on the N-terminal side of the two helix bundle are involved in crucial subunit-subunit interactions, as deletion of the first five residues renders the PrgI subunit incapable of polymerization13, 15,
Discussion
An atomic model of the S. typhimurium flagellar filament5 shows that the lumen in that hollow assembly is surrounded by coiled-coils (the D0 domain) formed by the N- and C-termini of the component subunit. These coiled-coils generate 11 nearly vertical protofilaments. The near atomic resolution for this system that was achieved by electron microscopy was possible due to the almost crystalline order of these filaments. In contrast, we have observed that the needle from these same bacteria is quite disordered and structurally heterogeneous. The average symmetry that we observe, ~ 6.3 subunits per turn of the 26 Å pitch helix, would generate 11-start helices with a very short pitch (~ 60 Å) Although we lack the resolution to directly observe the packing of subunits surrounding the lumen, the symmetry that exists is incompatible with long-pitch 11-start helices. It has recently been shown that although the S. typhimurium flagellar hook protein forms α-helical coiled-coils, the packing of these coiled-coils surrounding the lumen of the hook is very different from the packing in the flagellar filament, even though the helical symmetries of both are nearly identical16. We have previously shown17 that the flagellin subunits of S. typhimurium and Campylobacter jejuni, while highly conserved at the sequence level in the D0 and D1 coiled-coil domains responsible for filament polymerization in S. typhimurium, assemble into flagellar filaments with very different packing and helical symmetry. Thus, the ability of the extracellular flagellar and needle proteins to adopt different packing schemes around the lumens likely reflects the diversity in physical properties of these different polymers. While the packing of subunits surrounding the lumen of the needle remains unknown, there is a remarkably good match between the two-helix bundle that we can fit into our reconstruction and a model that has been previously proposed for the S. flexneri needle14.
How information about host cell contact is transmitted to turn on the secretory machinery located at the base of the T3SS remains a mystery. The likely role of the needle in transmitting such information was shown by the fact that mutants of the needle protein could be found in Shigella that induce a constitutively “on” secretion18. However, the needles formed from the mutant proteins unexpectedly showed no overall changes in the average helical symmetry6. The structural heterogeneity that we observe for the S. typhimurium needle raises new possibilities for how the needle might transmit information about host cell contact. The large variability in twist present in the S. typhimurium needles means that there must be a correspondingly large plasticity in how subunits are locally packed around the lumen. Such a plasticity could be used to transmit information along the needle. A testable prediction of this hypothesis, beyond the scope of the present paper, is that agents (such as crosslinks) that might lock the needle into a fixed symmetry should abrogate host cell sensing.
Materials and Methods
Needle complex preparation
To facilitate formation of elongated needle structures, a pWSK29 based plasmid was constructed, containing prgI including a 900 bp upstream region. The plasmid was transformed into a S. typhimurium strain (SB905, described elsewhere) carrying a plasmid containing an inducible hilA-gene19. The strain was grown under conditions promoting formation of the T3SS and induction of the regulatory protein HilA over night (LB, 300 mM NaCl, suitable antibiotics, 0,02% Arabinose, 37°C, 180 rpm).
Needle complexes were purified as described previously20 with minor modifications. In brief, cells from a 12 liter culture volume were harvested, resuspended in 300 ml of 500 mM sucrose in 150 mM Tris buffer at pH 8.0, and incubated after addition of 7.5 ml 500 mM EDTA solution and 15 ml lysozyme solution (30 mg/ml, 98381 IU/mg, Sigma-Aldrich) on ice for 60 minutes. Protease inhibitor was added (3 tablets Complete® Protease Inhibitor, Roche) and the temperature raised to 37° C for 30 minutes. Successively, 15 ml 10% Lauryldimethylamineoxide (LDAO), 33 ml of 5 M NaCl and 9 ml 1 M MgCl2 solution were added, before incubating the sample on ice for 60 minutes.
Aliquots (28 ml) were transferred into 36 ml ultracentrifugation tubes (Sorvall centrifuges. US) and a 3 ml layer of 36% w/v CsCl solution (36% w/v CsCl in 10 mM Tris, 500 mM NaCl, 5 mM EDTA, 0.5% LDAO, pH 8) was created below each sample. Solubilized needle complexes were centrifuged into the lower fractions of the CsCl-cushion (Sorvall Discovery 90SE, rotor AH-629, 28000 rpm, 5°C, 3 hours) to avoid pelleting directly onto the tube wall and damaging the filaments. Fractions containing needle filaments were pooled, and the density adjusted to equal a CsCl concentration of 35–36% w/v. The sample was split into 6 thinwall tubes (4.4 ml PA, Thermo Scientific, US) and centrifuged over night (Sorvall 90SE, rotor TH-660, 40,000rpm, 12h, 5° C). The needle complexes formed a visible band in the upper third part of the tube. This fraction was collected and pelleted (Sorvall RC M150 GX, rotor S100-AT4, 90 kprm, 30min, 5° C). The pellet was resuspended in 10 mM Tris-buffer containing 500 mM NaCl, 5 mM EDTA and 0,1% LDAO, pH 8.
Electron microscopy and image processing
Purified needle complex sample was applied to glow-discharged carbon-coated 400 mesh hexagonal Cu/Pd-grids. For negative stain images 5 µl of sample were put on the grid and incubated for 40 seconds. The grid was washed and subsequently stained with 2% PTA at neutral pH for 25 seconds. For electron cryo-microscopy 5 µl sample were applied to the glow-discharged grids, the sample was allowed to settle for 40 seconds and excess liquid was removed by blotting with double layered filter paper (Whatman 4) before vitrifying the sample by plunge freezing in liquid ethane. Low-dose data were collected with a FEI Tecnai Polara at 300 kV using a Gatan Ultrascan 4000 UHS CCD camera (16 mega-pixel, 4k X 4k, 15 micron pixel size). Images were acquired at 112,968-fold magnification (1.33 Å/pixel at the level of the specimen) with underfocus values ranging from 1.2–3.0 µm. Images were subsequently decimated to 2.66 Å, and boxes of 120 px length (320 Å) were used for IHRSR.
STEM
STEM experiments were performed at the Brookaven National Laboratory using tobacco mosaic virus as an internal standard for mass/length measurements. The preparation of samples and analysis of images were as described11.
Supplementary Material
Supplementary Movie) Segments of negatively stained needles were sorted using models having a 7-start helix with a fixed pitch of 271 Å (giving rise to a fixed layer line at ~ 1/39 Å) but with variable numbers of subunits along the 1-start helix. The averaged power spectra from five classes are shown: 6.40 units/turn (n=1,949), 6.35 units/turn (n=2,527), 6.30 units/turn (n=3,603), 6.25 units/turn (n=2,667) and 6.20 units/turn (n=2,517). The validity of this sorting was confirmed using the IHRSR algorithm for each class, and it was found that the segments in each class converged to a different symmetry, but one that was very close to the parameters of the models used for sorting. That is, segments selected using a model-based sorting as having a symmetry of ~ 6.2 units/turn converged to a symmetry of ~ 6.2 units/turn in the IHRSR method.
Acknowledgments
We thank Joe Wall and Martha Simon for the use of the STEM facility at Brookhaven National Laboratory. This work was supported by NIH EB001567 (to E.H.E.) and the Zentrum für Innovation und Technology through the Center of Molecular and Cellular Nanostructure (CMCN) Vienna (to T.C.M.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 2.Desvaux M, Hebraud M, Henderson IR, Pallen MJ. Type III secretion: what's in a name? Trends Microbiol. 2006;14:157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities 1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 5.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 6.Cordes FS, et al. Helical packing of needles from functionally altered Shigella type III secretion systems. J. Mol. Biol. 2005;354:206–211. doi: 10.1016/j.jmb.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita I, et al. Structure and switching of bacterial flagellar filaments studied by X-ray fiber diffraction. Nat. Struct. Biol. 1998;5:125–132. doi: 10.1038/nsb0298-125. [DOI] [PubMed] [Google Scholar]
- 8.Egelman EH, DeRosier DJ. The Fourier transform of actin and other helical systems with cumulative random angular disorder. Acta Cryst. 1982;A38:796–799. [Google Scholar]
- 9.Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J. Struct. Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 11.Wall JS, Hainfeld JF. Mass Mapping with the Scanning Transmission Electron Microscope. Annu. Rev. Biophys. Biophys. Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 12.Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 2007;371:1304–1314. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deane JE, et al. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darboe N, Kenjale R, Picking WL, Picking WD, Middaugh CR. Physical characterization of MxiH and PrgI, the needle component of the type III secretion apparatus from Shigella and Salmonella. Protein Sci. 2006;15:543–552. doi: 10.1110/ps.051733506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii T, Kato T, Namba K. Specific arrangement of alpha-helical coiled-coils in the core domain of the bacterial flagellar hook for the universal joint function. Structure. 2009 doi: 10.1016/j.str.2009.08.017. in press. [DOI] [PubMed] [Google Scholar]
- 17.Galkin VE, et al. Divergence of quaternary structures among bacterial flagellar filaments. Science. 2008;320:382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- 18.Kenjale R, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 19.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie) Segments of negatively stained needles were sorted using models having a 7-start helix with a fixed pitch of 271 Å (giving rise to a fixed layer line at ~ 1/39 Å) but with variable numbers of subunits along the 1-start helix. The averaged power spectra from five classes are shown: 6.40 units/turn (n=1,949), 6.35 units/turn (n=2,527), 6.30 units/turn (n=3,603), 6.25 units/turn (n=2,667) and 6.20 units/turn (n=2,517). The validity of this sorting was confirmed using the IHRSR algorithm for each class, and it was found that the segments in each class converged to a different symmetry, but one that was very close to the parameters of the models used for sorting. That is, segments selected using a model-based sorting as having a symmetry of ~ 6.2 units/turn converged to a symmetry of ~ 6.2 units/turn in the IHRSR method.