Abstract
Background
Over two hundred asthma candidate genes have been examined in human association studies or identified with knockout mouse approaches. However, many have not been systematically replicated in human populations, especially those containing a large number of tagging single nucleotide polymorphisms (SNPs).
Objective
We comprehensively evaluated the association of previously implicated asthma candidate genes with childhood asthma in a Mexico City population.
Methods
We identified, from the literature, candidate genes with at least one positive report of association with asthma phenotypes in humans or implicated in asthma pathogenesis by knockout mouse experiments. We performed a genome-wide association study in 492 asthmatic children aged 5 to 17 years and both parents using the Illumina HumanHap 550v3 BeadChip. Separate candidate gene analyses were performed for 2,933 autosomal SNPs in the 237 selected genes using the log-linear method with a log-additive risk model.
Results
Sixty-one of the 237 genes had at least one SNP with p < 0.05 for association with asthma. The nine most significant results were observed for rs2241715 in TGFB1 (p=3.3×10−5), rs13431828 and rs1041973 in IL1RL1 (p=2×10−4 and 3.5×10−4), five SNPs in DPP10 (p=1.6×10−4 to 4.5×10−4), and rs17599222 in CYFIP2 (p=4.1×10−4). False discovery rates were <0.1 for all 9 SNPs. Multimarker analysis identified TGFB1, IL1RL1, IL18R1, and DPP10 as the genes most significantly associated with asthma.
Conclusions
This comprehensive analysis of literature-based candidate genes suggests that SNPs in several candidate genes including TGFB1, IL1RL1, IL18R1 and DPP10 may contribute to childhood asthma susceptibility in a Mexican population.
Keywords: Allergy, asthma, genetic predisposition to disease, genome-wide association study (GWAS), single nucleotide polymorphism (SNP)
INTRODUCTION
Asthma is a complex disease caused by multiple genetic and environmental factors. Two traditional approaches for identification of asthma susceptibility genes are association studies of candidate genes and linkage studies followed by positional cloning. Candidate gene association studies focus on genes plausibly involved in disease pathogenesis or located in a region of linkage for the disease. The majority of proposed asthma susceptibility genes are biological candidate genes.
In recent years, it has become feasible to interrogate single nucleotide polymorphisms (SNPs) across the genome to identify novel disease susceptibility genes, an approach known as the genome-wide association study (GWAS). A novel asthma gene, ORM1-like 3 (ORMDL3) has been identified using the GWAS approach.1 Incorporating a priori knowledge of disease etiology into the statistical analysis and evaluating prioritized SNPs in predefined candidate genes separately can achieve more efficient use of the GWAS data.2
Over 200 asthma candidate genes have been proposed using human association, positional cloning, and knockout mouse approaches in the past decade.3, 4 However, many of them have not been systematically replicated in additional human populations, including genes with a large number of tagging SNPs, such as dipeptidyl-peptidase 10 (DPP10) and estrogen receptor 1 (ESR1). Replication of associations in different populations is crucial for identifying complex disease susceptibility genes.5 A total of 39 candidate genes from the literature were recently examined for association with childhood asthma using GWAS data in a non-Hispanic white North American population.6 In a GWAS of case-parent triads from Mexico City, we comprehensively evaluated associations of over 200 previously reported candidate genes with childhood asthma.
METHODS
Study design and subject enrollment
Using the case-parent triad design,7, 8 we recruited nuclear families consisting of asthmatic children and both their parents. The cases were children aged 4–17 years with asthma diagnosed by a pediatric allergist at the allergy referral clinic of a large public pediatric hospital in central Mexico City (Hospital Infantil de México, Federico Gómez). Children and parents provided blood samples as sources of DNA. A parent, nearly always the mother, completed a questionnaire on the child’s symptoms and risk factors for asthma including parental smoking and residential history.
The protocol was reviewed and approved by the Institute Review Boards of the Mexican National Institute of Public Health, the Hospital Infantil de México, Federico Gómez, and the U.S. National Institute of Environmental Health Sciences. Parents provided the written informed consent for the child’s participation. Children also gave their informed assent.
Clinical evaluation
Detailed protocols for clinical evaluation were described in the Online Repository. In brief, the diagnosis of asthma was based on clinical symptoms and response to treatment by pediatric allergists at a major referral hospital.9, 10 At a later date, for research purposes, pulmonary function was measured according to ATS specifications.11 Atopy was determined using skin prick tests to a battery of 24 environmental aeroallergens common in Mexico City. Children were considered atopic if the diameter of the skin wheal to at least one allergen exceeded 4 mm.
Candidate gene selection
We included all 118 human asthma candidate genes listed by Ober and Hoffjan in 2006.3 To update the previous review,3 we searched in PubMed for the period June 1, 2005 to July 31, 2008 for genes that had at least one positive association of SNPs with asthma phenotypes in humans. We used keywords “genetic polymorphism” together with “asthma” or “bronchial or airway” or “hyperreactivity or hyperresponsiveness or hypersensitivity”. We also identified genes directly related to asthma phenotypes using a knockout mouse approach. For the knockout mouse studies, we used the keywords “mouse or mice or murine” and “wildtype or knockout" and "disease models, animal" together with “bronchial or airway" and “asthma or inflammation or hyperresponsiveness”. The updated review indentified 156 genes not referenced by Ober and Hoffjan3 for a total of 274 genes.
Among the 274 genes, 19 were not represented on the Illumina HumanHap 550v3 BeadChip (Table E1 in the Online Repository). We also excluded 5 genes on X chromosome and 4 genes with more than 300 SNPs within the gene region (5 kb upstream of the 5’ end through 1 kb downstream of the 3’ end) on the Illumina 550v3 BeadChip, leaving 246 autosomal genes for analysis (Table E1 in the Online Repository). The total number of SNPs was 3,326. We selected candidate genes prior to analysis of genotyping data.
Genotyping and quality control
Genotyping was done using the Illumina HumanHap 550v3 BeadChip (Illumina, San Diego, California) at the University of Washington, Department of Genome Sciences. Standard quality control of GWAS genotyping data was conducted using PLINK12 or GLU,13 as described in the Online Repository.
For the candidate gene analysis, more stringent SNP exclusion thresholds were used: minor allele frequency (MAF) less than 3% and Hardy Weinberg equilibrium p-value less than 1 × 10−6. Of the 3,326 autosomal SNPs in 246 selected candidate genes, 2,933 SNPs in 237 genes (Table E1 in the Online Repository) were analyzed in 492 complete case-parent trios.
The SNP coverage for these 237 genes by the Illumina 550v3 BeadChip is listed in Table E5 in the Online Repository.14, 15
Statistical analysis
We used a log-linear likelihood approach to analyze associations between asthma and the 2,933 individual SNPs.7 Details regarding the log-linear method are described in the Online Repository. The log-linear method was implemented using the LEM computer program16 with a one degree-of-freedom log-additive risk model specified. P-values were generated to assess statistical significance, and the relative risk of carrying one copy of the risk allele was calculated to assess the direction and magnitude of association under the log-additive model.
To account for multiple comparisons, we calculated the false discovery rate (FDR) q value for each p value for all of the 2,933 SNPs analyzed using the method of Storey.17 The FDR is the expected proportion of false positives incurred when a particular test is called significant. However, these corrections will be conservative because the false discovery rate does not take into account the correlation between SNPs. We used the FDR threshold of 0.1 for declaring significance because Van den Oord and Sullivan18 showed that it achieved a good balance between avoiding false discoveries and detecting true effects.
There is a higher chance of observing SNPs with significant p values for genes with more SNPs. To address this issue, we used a multimarker approach, TRIad MultiMarker test (TRIMM), to test the association of asthma with sets of SNPs.19 This procedure achieves a natural correction for multiple comparisons by treating multiple SNPs as a set and using permutation procedure to evaluate the test significance. In our analysis, all SNPs in a gene (5 kb upstream of the 5’ end through 1kb downstream of the 3’ end) were defined as a set, and a p value was calculated for each gene. For the largest gene on our candidate gene list, DPP10 that spans 1.4 Mb on Chromosome 2, the SNPs were divided into 7 sets along the chromosome based on the linkage disequilibrium (LD) structure of the gene (Table E2 in the Online Repository). The p values were estimated for each DPP10 block and the whole gene. We implemented the TRIMM procedure in R (http://www.r-project.org). The R code is available at http://www.niehs.nih.gov/research/atniehs/labs/bb/staff/weinberg.
RESULTS
Detailed characteristics of the 492 asthmatic children are presented in Table I and described in the Online Repository. The mean age of cases was 9.0 years (range 5–17 years). Most had mild as opposed to moderate or severe asthma. Ninety-two percent of cases had at least one positive skin test.
Table I.
Demographic and clinical characteristics of the 492 asthmatic children.
| Clinical characteristics | % |
|---|---|
| Age [years (mean ± SD)] | 9.0 ± 2.4 |
| Sex (male) | 58.7 |
| Asthma severity (n = 469) a | |
| Mild | 72.3 |
| Moderate to severe | 27.7 |
| Asthma Medication in the past 12 mo (n = 486) a | 98.2 |
| FEV1 [percent predicted (mean ± SD)] (n = 371) a | 90.5 ± 16.8 |
| Skin test positivity (of 24 aeroallergens) (n = 445) a | |
| ≥ 1 allergen | 91.7 |
| ≥ 5 allergens | 51.5 |
| Parental smoking (n = 486) a | |
| Mother smoked during pregnancy | 4.8 |
| Current smoking parent | 52.1 |
Number in parenthesis indicates total with non-missing data for each characteristic.
Many of the 2,933 analyzed SNPs are in high LD with each other in our Mexican population. Using the LD based SNP pruning procedure implemented in PLINK (using parameters of window size = 50, number of SNPs to shift at each step = 5, variance inflation factor = 2), we calculated that 1,125 SNPs were in approximate linkage equilibrium (variance inflation factor < 2) with each other.
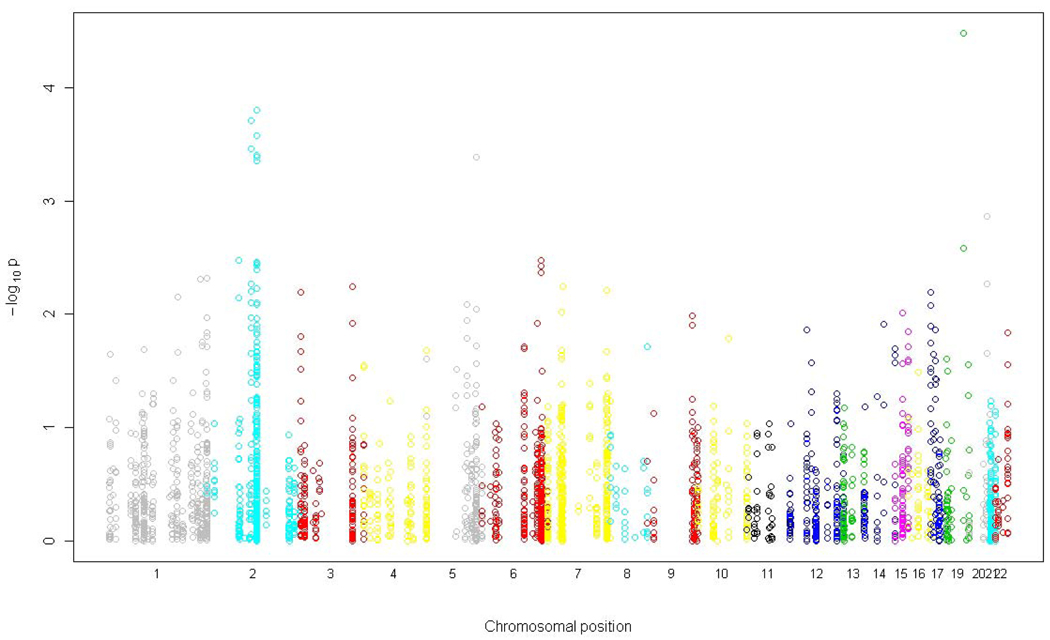
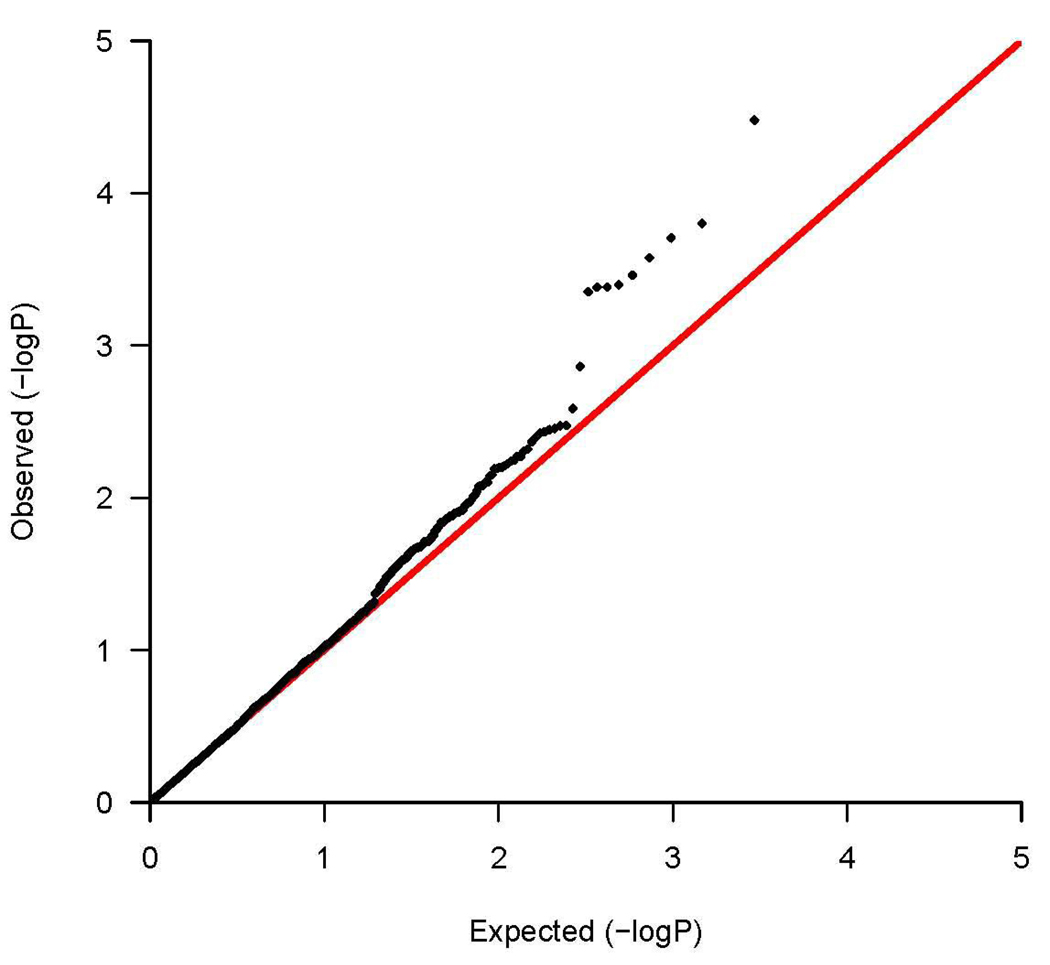
Figure 1a shows the chromosomal position of all candidate gene SNPs tested for association with asthma and their corresponding significance levels. Figure 1b shows the quantile-quantile plot of the p values indicating the number of observed significant associations exceeding the expected p values under the null hypothesis of no association. Among the 237 asthma candidate genes, 61 genes had at least one SNP with p < 0.05 for association with asthma (Table II for SNPs with p<0.01 and Table E3 in the Online Repository for SNPs with 0.01≤ p< 0.05). Using conservative Bonferroni correction for 1,125 independent tests (number of SNPs in approximate linkage equilibrium), only rs2241715 in transforming growth factor, beta 1 (TGFB1) met the significance level of 4.4×10−5. However, given that the genes were selected based on prior evidence, Bonferroni correction is overly conservative. Nine SNPs met the FDR q-value significance threshold of less than 0.1, including rs2241715 in TGFB1 on chromosome 19 (p=3.3×10−5, FDR q=0.059), rs13431828 and rs1041973 in interleukin 1 receptor-like 1 (IL1RL1) on chromosome 2 (p=2.0×10−4 for rs13431828 and 3.5×10−4 for rs1041973, FDR q=0.087 for both), rs980317, rs7421482, rs980316, rs949577, and rs12469474 in DPP10 on chromosome 2 (p=1.6×10−4 to 4.5×10−4, FDR q=0.087 for all), and rs17599222 in cytoplasmic FMR1 interacting protein 2 (CYFIP2) on chromosome 5 (p=4.1×10−4, FDR q=0.087).
Figure 1.
(a) A summary of associations between 2,933 autosomal SNPs in 237 candidate genes and childhood asthma in a Mexican population. The x axis indicates the genomic position of all SNPs divided by chromosome. The y axis shows the degree of association indicated as -log10P value. (b) Quantile –quantile plot. Distribution of the observed p values for the 2,933 SNPs compared to the p values expected under the null hypothesis of no association. Observed -log10P values are ranked in order on the y axis, and plotted against the corresponding expected -log10P values on the x axis. The red line indicates the null distribution.
Table II.
Genetic associations between SNPs in candidate genes and childhood asthma in a Mexican population (p < 0.01).
| Gene | Chr | SNP | SNP type | Minor allele |
MAF | RR a | Lower 95% CI |
Upper 95% CI |
P value |
|---|---|---|---|---|---|---|---|---|---|
| TGFB1 | 19 | rs2241715 | intron | G | 0.49 | 0.68 | 0.56 | 0.81 | 0.000033 |
| DPP10 | 2 | rs980317 | intron | C | 0.26 | 0.68 | 0.55 | 0.83 | 0.00016 |
| IL1RL1 | 2 | rs13431828 | UTR | T | 0.05 | 0.45 | 0.29 | 0.70 | 0.00020 |
| DPP10 | 2 | rs7421482 | intron | T | 0.24 | 0.68 | 0.55 | 0.84 | 0.00027 |
| IL1RL1 | 2 | rs1041973 | nonsynon | A | 0.10 | 0.58 | 0.43 | 0.78 | 0.00035 |
| DPP10 | 2 | rs980316 | intron | C | 0.33 | 0.71 | 0.59 | 0.86 | 0.00040 |
| CYFIP2 | 5 | rs17599222 | intron | G | 0.36 | 0.71 | 0.59 | 0.86 | 0.00041 |
| DPP10 | 2 | rs949577 | intron | C | 0.23 | 0.68 | 0.55 | 0.85 | 0.00041 |
| DPP10 | 2 | rs12469474 | intron | A | 0.24 | 0.69 | 0.56 | 0.85 | 0.00045 |
| MMP9 | 20 | rs4810482 | flanking | C | 0.20 | 1.44 | 1.15 | 1.79 | 0.0014 |
| TGFB1 | 19 | rs4803455 | intron | A | 0.30 | 0.74 | 0.61 | 0.90 | 0.0026 |
| ESR1 | 6 | rs9478265 | intron | A | 0.04 | 0.51 | 0.32 | 0.81 | 0.0034 |
| TACR1 | 2 | rs17010698 | intron | T | 0.20 | 0.72 | 0.57 | 0.90 | 0.0034 |
| DPP10 | 2 | rs1396932 | intron | A | 0.37 | 0.76 | 0.63 | 0.92 | 0.0035 |
| DPP10 | 2 | rs10496465 | intron | G | 0.07 | 1.71 | 1.18 | 2.47 | 0.0036 |
| DPP10 | 2 | rs2175176 | intron | G | 0.42 | 0.77 | 0.64 | 0.92 | 0.0037 |
| ESR1 | 6 | rs9371236 | intron | G | 0.04 | 0.49 | 0.30 | 0.81 | 0.0038 |
| DPP10 | 2 | rs4491738 | intron | C | 0.31 | 0.75 | 0.62 | 0.91 | 0.0040 |
| ESR1 | 6 | rs9340941 | intron | T | 0.04 | 0.50 | 0.31 | 0.82 | 0.0043 |
| KMO | 1 | rs12138459 | intron | A | 0.14 | 0.69 | 0.53 | 0.90 | 0.0048 |
| EPHX1 | 1 | rs2740170 | intron | T | 0.08 | 1.62 | 1.15 | 2.28 | 0.0049 |
| IL18R1 | 2 | rs3213733 | intron | T | 0.07 | 0.61 | 0.43 | 0.87 | 0.0054 |
| MMP9 | 20 | rs17576 | nonsynon | G | 0.18 | 1.37 | 1.10 | 1.72 | 0.0054 |
| CD86 | 3 | rs3792285 | intron | A | 0.04 | 1.86 | 1.19 | 2.92 | 0.0057 |
| AOAH | 7 | rs12540585 | intron | A | 0.17 | 0.72 | 0.57 | 0.91 | 0.0058 |
| DPP10 | 2 | rs983829 | intron | T | 0.31 | 0.76 | 0.63 | 0.93 | 0.0060 |
| TRB@ | 7 | rs17274 | intron | T | 0.17 | 0.72 | 0.56 | 0.91 | 0.0061 |
| IL18R1 | 2 | rs1420094 | flanking | A | 0.18 | 0.73 | 0.58 | 0.91 | 0.0063 |
| IL18R1 | 2 | rs2287033 | intron | G | 0.18 | 0.73 | 0.58 | 0.91 | 0.0063 |
| NOS2A | 17 | rs3794764 | intron | A | 0.33 | 1.30 | 1.08 | 1.57 | 0.0064 |
| IL5RA | 3 | rs9869655 | intron | A | 0.10 | 0.66 | 0.49 | 0.89 | 0.0065 |
| TNFSF4 | 1 | rs10489266 | flanking | C | 0.03 | 2.11 | 1.20 | 3.69 | 0.0070 |
| TACR1 | 2 | rs3755458 | intron | T | 0.18 | 0.73 | 0.58 | 0.92 | 0.0072 |
| DPP10 | 2 | rs6542256 | intron | C | 0.14 | 1.43 | 1.10 | 1.86 | 0.0079 |
| IL18R1 | 2 | rs4851004 | intron | T | 0.18 | 0.73 | 0.58 | 0.92 | 0.0079 |
| C5orf20 | 5 | rs13173226 | intron | C | 0.41 | 0.78 | 0.65 | 0.94 | 0.0083 |
| DPP10 | 2 | rs2420815 | intron | C | 0.22 | 1.34 | 1.08 | 1.67 | 0.0083 |
| NOS2A | 17 | rs2274894 | intron | T | 0.39 | 0.78 | 0.65 | 0.94 | 0.0084 |
| CYFIP2 | 5 | rs6555977 | intron | C | 0.21 | 0.74 | 0.59 | 0.93 | 0.0090 |
| AOAH | 7 | rs10499593 | intron | A | 0.19 | 1.36 | 1.08 | 1.73 | 0.0096 |
| SMAD3 | 15 | rs11637659 | intron | A | 0.20 | 0.74 | 0.59 | 0.93 | 0.0098 |
Abbreviations: Chr, chromosome; MAF, minor allele frequency; RR, relative risk; nonsynon, coding non-synonymous SNP; UTR, un-translated region.
Relative risk for carrying one copy of the minor allele compared to carrying no copies.
Phenotypic heterogeneity is a potential factor contributing to failure of replication among different studies. In addition to the primary analysis among the 492 trios, we repeated the log-linear analysis among 378 trios including children with non-missing skin test and questionnaire data who had positive skin tests and whose mothers did not smoke during pregnancy. The magnitude and direction of the association did not differ appreciably when we analyzed this smaller dataset (Table E4 in the Online Repository).
Results from the multimarker analysis, which corrects for the number of SNPs analyzed in a gene, were consistent with the single SNP findings (Table III and Table E5 in the Online Repository). The candidate genes that were most significantly associated with asthma were TGFB1 (global p=2.8×10−4) on chromosome 19q13, IL1RL1 (global p=2.2×10−4) and the adjacent interleukin 18 receptor 1 (IL18R1) (global p=9×10−3) on chromosome 2q12, and DPP10 (global p=7.8×10−4 for DPP10_block 3 and 0.05 for the whole gene) on chromosome 2q14.
Table III.
Multimarker analysis of associations between candidate genes and childhood asthma in a Mexican population.
| Gene | Number of SNPs a | P Value |
|---|---|---|
| IL1RL1 | 11 | 0.00022 |
| TGFB1 | 4 | 0.00028 |
| DPP10_Block3 b | 30 | 0.00078 |
| IL18R1 | 9 | 0.0090 |
| MMP9 | 4 | 0.012 |
| IL5RA | 26 | 0.025 |
| ZPBP2 | 3 | 0.031 |
| TNFSF4 | 6 | 0.032 |
| TLR6 | 2 | 0.034 |
| IL1R1 | 9 | 0.044 |
| NOS2A | 16 | 0.046 |
| CYFIP2 | 37 | 0.047 |
| EPHX1 | 7 | 0.050 |
| DPP10 b | 253 | 0.050 |
| PTGER4 | 2 | 0.050 |
All SNPs in a gene were treated as a set in the multimarker analysis using the TRIMM program.
P values were calculated for the whole DPP10 gene and 7 DPP10 LD blocks separately. See Table E2 in the Online Repository for the definition of the 7 sets of DPP10 SNPs.
IL1RL1 is adjacent to IL18R1, located 12 Mb upstream of DPP10 on chromosome 2. Figure E1 in the Online Repository shows the pairwise LD (r2) between IL1RL1, IL18R1, and DPP10 SNPs with p less than 0.05 for association with asthma. IL1RL1 and IL18R1 resided in a LD block. The two IL1RL1 SNPs, rs13431828 and rs1041973 that were significantly associated with asthma at FDR q-value less than 0.1 are in moderate LD (r2 = 0.46) with each other. These two SNPs are potentially functional. The SNP rs13431828 is located in the 5’ untranslated region (5’-UTR) of IL1RL1, and rs1041973 is a coding non-synonymous SNP (Glu/Ala) in exon 2. Three additional tightly linked coding non-synonymous IL1RL1 SNPs, rs10204137, rs10192157, and rs10206753 (r2 = 0.97 to 1) also showed moderate associations with asthma (p = 0.013 for all three SNPs). The 5 DPP10 SNPs rs980317, rs7421482, rs980316, rs949577, and rs12469474 that were significantly associated with asthma at FDR q-value less than 0.1 are in moderate to tight LD (r2 = 0.39 to 0.93) with each other and located within the LD block DPP10_block 3 (Figure E1 and Table E2 in the Online Repository).
DISCUSSION
We comprehensively evaluated the association of previously reported asthma genes with childhood asthma in Mexico City within the context of a genome-wide association genotyping platform. Candidate genes were identified from a systematic literature review completed before analysis of the genotyping data. Single SNP analyses showed that SNPs in TGFB1, DPP10, IL1RL1, and CYFIP2 were significantly associated with childhood asthma in a Mexican population after correction for multiple comparisons using a false discovery rate approach (FDR q-value < 0.1). Our multimarker analysis accounted for gene-wide multiple comparisons by generating a global p value for all SNPs in a region, and these results confirmed that several genes including TGFB1, DPP10, and IL1RL1 are related to childhood asthma susceptibility.
Compared to traditional candidate gene and linkage studies, the GWAS approach has the advantage of interrogating SNPs across the whole genome to identify novel disease susceptibility genes unrestrained by prior knowledge. However, questions regarding how to make optimal use of the GWAS data remain unanswered. Li et al2 have shown that pre-selecting SNPs from candidate genes and analyzing this prioritized subset of SNPs separately can improve the power of detecting a disease susceptibility locus in GWAS.
Many candidate genes have been studied for asthma.3, 4 A candidate gene association study usually examines only a relatively small number of SNPs in few selected genes. Many of the published asthma candidate genes, especially large genes with many tagging SNPs such as DPP10, have not been comprehensively evaluated in additional human populations. Thirty-nine candidate genes were recently evaluated for associations with childhood asthma using GWAS data from a non-Hispanic white North American population.6 We examined a much larger number of candidate genes in a population that has not been well-studied.
TGFB1 is a multi-functional cytokine that may influence asthma by modulating allergic airway inflammation and airway remodeling. TGFB1 is one of the most replicated asthma candidate genes, and SNPs in TGFB1 have been associated with asthma phenotypes in approximately 10 published studies.20 We previously reported that three of five genotyped TGFB1 SNPs, rs1800469 (C-509T, a promoter SNP), rs1982073 (T869C, a non-synonymous SNP), and rs7258445 (an intronic SNP) were associated with asthma in the Mexican population.21 In the present analysis, we examined three additional TGFB1 SNPs, rs2241715, rs4803455, and rs8110090. Figure E2 in the Online Repository shows the pairwise LD (r2) between the 8 TGFB1 SNPs that have been examined in our study population to date. The SNP rs2241715 that was significantly associated with asthma in the present analysis was in moderate to high LD (r2 = 0.5 to 0.95) with the three asthma-associated SNPs reported in our previous paper.21 Two asthma-associated SNPs, rs1800469 and rs1982073 are functional. Rs1800469, also referred to as C-509T, is located in the promoter region, and this SNP can influence TGFB1 function, promoter activity, and circulating TGFB1 levels.21 Rs1982073, also referred to as T869C, is a non-synonymous SNP, and the T to C substitution leads to an amino acid change from leucine to proline in the signal peptide resulting in increased secretion of TGFB1 in vitro and increased circulating TGFB1 concentration.21
IL1RL1 is adjacent to IL18R1 and located in an interleukin 1 (IL1) receptor gene cluster on chromosome 2q12.22 Gene products of IL1RL1 and IL18R1 both belong to the IL1 receptor family whose members mediate the signal transduction of IL1 cytokines during inflammation and host defense.23 IL1RL1 binds IL-33 and plays important roles in regulation of T helper type2 (TH2) cell-mediated allergic airway inflammation24, 25 and eosinophil-mediated inflammation.26 Serum levels of IL1RL1 are elevated in atopic asthmatic patients during acute exacerbations.27 IL18R1 encodes the alpha chain of the ILR18 receptor (IL18R).28 IL18R binds IL18 and enhances T helper type1 (TH1) cell-driven immune responses in synergy with interleukin 12 (IL12).28 IL18 can also induce the development of TH2 cells, stimulate TH2 cytokine release, and plays a complicated role in atopic asthma depending on its immunological environment.28
SNPs in IL1RL1 and IL18 have been associated with asthma-related phenotypes in only three previous studies conducted in several European populations and one Korean population.29–31 IL1RL1 and IL18R1 are located together in a LD block in Europeans29, 30 and Mexicans. We examined 11 SNPs in IL1RL1 and 9 SNPs in IL18R1. Eleven of the 20 SNPs were associated with asthma in the Mexican population (p < 0.01 for 6 SNPs and 0.01 ≤ p < 0.05 for 5 SNPs). There is little overlap between the SNPs genotyped across studies.29–31 Two (rs1041973 and rs10206753) of the 4 coding non-synonymous IL1RL1 SNPs associated with asthma in our Mexican population were also examined in a Dutch population, where they showed no associations.29 An intronic IL1RL1 SNP rs1420101 or its tightly linked SNP rs950880 (r2 = 0.96 in European HapMap samples) has been significantly associated with blood eosinophil count and asthma in European and Korean populations,31 but not in our Mexican population. The rs1420094 SNP in IL18R1 was significantly associated with atopic asthma in Europeans30 and our Mexican population.
DPP10 was identified as an asthma candidate gene by positional cloning,32 but its definitive function is still unclear. DPP10 is a member of the dipeptidyl peptidase family that can remove N-terminal dipeptides from chemokines and cytokines, and thus might modify their functional activities.32, 33 Alternative transcriptional spliced variants of DPP10 are expressed in many tissues including airways (trachea), and are abundant in T-cells.32 SNPs in a LD island across the first 60 kb region of DPP10 intron1 were associated with asthma in British and German populations.32, 34 Of note, only SNPs in the first 200 kb of the DPP10 genomic DNA were examined for association with asthma-related phenotypes in the original report and the study of Blakey et al.32, 34 A previous examination of DPP10 within a GWAS evaluated 252 SNPs and found that 25 SNPs gave p values smaller than 0.05 for association with asthma in a non-Hispanic white North American population (smallest P = 0.001).6 Among the 253 SNPs we studied, 36 SNPs spreading over a 900 kb genomic region encompassing intron1 to intron3 of DPP10 all gave p values < 0.05 for association with asthma in the Mexican population. To our knowledge, no functional DPP10 SNPs have been reported yet. Allen et al32 identified several alternative splicing sites located in an 850 kb region across exon1, intron1 and exon2, which can lead to the production of membrane-bound and other isoforms of DPP10. Polymorphisms in regulatory elements resulting in alternative splicing of DPP10 may explain effects on asthma susceptibility from this region.32
ORMDL3 was the first asthma candidate gene identified using the GWAS approach.1 We previously examined rs4378650 in ORMDL3 and rs7216389 in the neighboring GSDML in 615 nuclear families.35 Rs7216389 in GSDML was also on the Illumina 550K array used in the present analysis. Although rs4378650 in ORMDL3 was not on the Illumina 550K array, it can be tagged by rs7216389 (r2 = 0.92) in Mexicans. 35 The results for rs7216389 from our two analyses were consistent [RR (95% CI) = 1.20 (1.01–1.43), p = 0.043 in the previous report with 615 families; RR (95% CI) = 1.22 (1.01–1.49), p = 0.042 in the present analysis of 492 trios; a log-additive risk model with C as the reference allele specified for both analyses].35
Our study has several strengths. The triad design and analysis protects against population stratification, a potential source of bias in an admixed population such as the Mexican population.7 The demographic and clinical characteristics of our asthmatic children are well characterized. Our asthma cases were diagnosed by pediatric allergists at a pediatric allergy specialty clinic of a large public referral hospital. Consultation with this pediatric allergy clinic is a tertiary referral in Mexico, and thus the children in our study had already been seen by a generalist and a pediatrician over time for recurrent asthma symptoms. Diagnoses were made on clinical grounds according to previous guidelines.9 We did not test for bronchial hyperreactivity (BHR). However, physician diagnosis of asthma is a valid outcome compared to objective measurements.36 We had objective data on atopy; skin prick tests revealed the vast majority of these children with asthma (92%) to be skin test positive to common environmental aeroallergens. Thus all findings may apply primarily to atopic asthma.
We comprehensively evaluated the relationship between SNPs in 237 previously published candidate genes and childhood asthma within the context of a GWAS. Our single SNP and multimarker analysis results suggest that SNPs in multiple genes including TGFB1, IL1RL1, IL18R1 and DPP10 may contribute to childhood asthma susceptibility in a Mexican population.
ACKNOWLEDGMENTS
We thank the children and parents who participated in this study; Dr. Deborah Nickerson and Joshua Smith, University of Washington, for their genotyping services; Kevin Jacobs, National Cancer Institute (NCI), for technical assistance with the GLU software; Drs. Douglas Bell, Xuting Wang, and Lauranell Burch, National Institute of Environmental Health Sciences (NIEHS); Dr. Patrick Sullivan, University of North Carolina – Chapel Hill, for bioinformatics support; Stephanie Holmgren, NIEHS, for reference services; and Dr. Stephan Chanock, NCI, for determination of short tandem repeats for parentage testing.
Declaration of all sources of funding: This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES49019). Subject enrollment was supported in part by the National Council of Science and Technology (grant 26206-M), Mexico. Dr. Romieu was supported in part by the National Center for Environmental Health at the Centers for Disease Control.
Abbreviations used
- CI
confidence interval
- CYFIP2
cytoplasmic FMR1 interacting protein 2
- DPP10
dipeptidyl-peptidase 10
- ESR1
estrogen receptor 1
- FDR
false discovery rate
- FEV1
forced expiratory volume in one second
- GWAS
genome-wide association study
- IL1RL1
interleukin 1 receptor-like 1
- IL18R1
interleukin 18 receptor 1
- LD
linkage disequilibrium
- MAF
minor allele frequency
- RR
relative risk
- SNP
single nucleotide polymorphism
- TDT
transmission disequilibrium test
- TGFB1
transforming growth factor, beta 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
Clinical implications
The associations between asthma and polymorphisms in multiple candidate genes, including TGFB1, IL1RL1, IL18R1 and DPP10, provide insights to disease pathogenesis and suggest potential therapeutic targets.
Capsule summary
Findings from this study suggest that genetic variants in multiple candidate genes including TGFB1, IL1RL1, IL18R1 and DPP10 may play roles in childhood asthma susceptibility.
REFERENCES
- 1.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Li M, Lange EM, Watanabe RM. Prioritized subset analysis: improving power in genome-wide association studies. Hum Hered. 2008;65:129–141. doi: 10.1159/000109730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Pare PD, Sandford AJ. Recent advances in asthma genetics. Respir Res. 2008;9:4. doi: 10.1186/1465-9921-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 6.Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, et al. Assessing the Reproducibility of Asthma Candidate Gene Associations Using Genome-wide Data. Am J Respir Crit Care Med. 2009;179:1084–1090. doi: 10.1164/rccm.200812-1860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcox AJ, Weinberg CR, Lie RT. Distinguishing the effects of maternal and offspring genes through studies of "case-parent triads". Am J Epidemiol. 1998;148:893–901. doi: 10.1093/oxfordjournals.aje.a009715. [DOI] [PubMed] [Google Scholar]
- 9.BTS/SIGN. British guideline on the management of asthma. Thorax. 2003;58 Suppl 1:i1–i94. doi: 10.1136/thorax.58.suppl_1.1i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NHLBI. NIH Publication: National Institute of Health: Bethesda, MD; Pocket Guide for Asthma Management and Prevention: Global Initiative for Asthma. 1998
- 11.ATS. Standardization of Spirometry, 1994 Update. American Thoracic Society. Am J Respir Crit Care Med. 1995;152:1107–1136. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635–643. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- 16.van Den Oord EJ, Vermunt JK. Testing for linkage disequilibrium, maternal effects, and imprinting with (In)complete case-parent triads, by use of the computer program LEM. Am J Hum Genet. 2000;66:335–338. doi: 10.1086/302708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Oord EJ, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–542. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Shi M, Umbach DM, Weinberg CR. Identification of risk-related haplotypes with the use of multiple SNPs from nuclear families. Am J Hum Genet. 2007;81:53–66. doi: 10.1086/518670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–182. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Romieu I, Wu H, Sienra-Monge JJ, Ramirez-Aguilar M, del Rio-Navarro BE, et al. Genetic polymorphisms in transforming growth factor beta-1 (TGFB1) and childhood asthma and atopy. Hum Genet. 2007;121:529–538. doi: 10.1007/s00439-007-0337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dale M, Nicklin MJ. Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics. 1999;57:177–179. doi: 10.1006/geno.1999.5767. [DOI] [PubMed] [Google Scholar]
- 23.Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Mangan NE, Dasvarma A, McKenzie AN, Fallon PG. T1/ST2 expression on Th2 cells negatively regulates allergic pulmonary inflammation. Eur J Immunol. 2007;37:1302–1312. doi: 10.1002/eji.200636520. [DOI] [PubMed] [Google Scholar]
- 26.Cherry WB, Yoon J, Bartemes KR, Iijima K, Kita H. A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J Allergy Clin Immunol. 2008;121:1484–1490. doi: 10.1016/j.jaci.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshikawa K, Kuroiwa K, Tago K, Iwahana H, Yanagisawa K, Ohno S, et al. Elevated soluble ST2 protein levels in sera of patients with asthma with an acute exacerbation. Am J Respir Crit Care Med. 2001;164:277–281. doi: 10.1164/ajrccm.164.2.2008120. [DOI] [PubMed] [Google Scholar]
- 28.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 is a unique cytokine that stimulates both Th1 and Th2 responses depending on its cytokine milieu. Cytokine Growth Factor Rev. 2001;12:53–72. doi: 10.1016/s1359-6101(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 29.Reijmerink NE, Postma DS, Bruinenberg M, Nolte IM, Meyers DA, Bleecker ER. Association of IL1RL1, IL18R1, and IL18RAP gene cluster polymorphisms with asthma and atopy. J Allergy Clin Immunol. 2008;122:651–654. doi: 10.1016/j.jaci.2008.06.030. e8. [DOI] [PubMed] [Google Scholar]
- 30.Zhu G, Whyte MK, Vestbo J, Carlsen K, Carlsen KH, Lenney W, et al. Interleukin 18 receptor 1 gene polymorphisms are associated with asthma. Eur J Hum Genet. 2008;16:1083–1090. doi: 10.1038/ejhg.2008.67. [DOI] [PubMed] [Google Scholar]
- 31.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 32.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003;35:258–263. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 33.Kere J, Laitinen T. Positionally cloned susceptibility genes in allergy and asthma. Curr Opin Immunol. 2004;16:689–694. doi: 10.1016/j.coi.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Blakey JD, Sayers I, Ring S, Strachan D, Hall I. Positionally cloned Asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth Cohort. Thorax. 2009;64:381–387. doi: 10.1136/thx.2008.102053. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Romieu I, Sienra-Monge JJ, Li H, Del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins MA, Clarke JR, Carlin JB, Robertson CF, Hopper JL, Dalton MF, et al. Validation of questionnaire and bronchial hyperresponsiveness against respiratory physician assessment in the diagnosis of asthma. Int J Epidemiol. 1996;25:609–616. doi: 10.1093/ije/25.3.609. [DOI] [PubMed] [Google Scholar]