Abstract
Reactive oxygen species (ROS) are products of normal metabolism and xenobiotic exposure, and depending on concentrations, ROS can be beneficial or harmful to cells and tissues. At physiological low levels, ROS function as “redox messengers” in intracellular signaling and regulation while excess ROS induce oxidative modification of cellular macromolecules, inhibit protein function and promote cell death. Additionally, various redox systems, such as the glutathione, thioredoxin, and pyridine nucleotide redox couples, participate in cell signaling and modulation of cell function, including apoptotic cell death. Cell apoptosis is initiated by extracellular and intracellular signals via two main pathways, the death receptor- or mitochondria-mediated pathways. Various pathologies can result from oxidative stress induced apoptotic signaling that is consequent to ROS increases and/or antioxidant decreases, disruption of intracellular redox homeostasis, and irreversible oxidative modifications of lipid, protein or DNA. In the current review, we focused on several key aspects of ROS and redox mechanisms in apoptotic signaling, and highlighted the gaps in knowledge and potential avenues for further investigation. A full understanding of redox control of apoptotic initiation and execution could underpin the development of therapeutic interventions targeted at oxidative stress associated disorders.
1. OVERVIEW OF REACTIVE OXYGEN SPECIES AND INTRACELLULAR SOURCES
Reactive oxygen species (ROS) is a collective term that broadly describes O2-derived free radicals such as superoxide anions (O2•−), hydroxyl radicals (HO•), peroxyl (RO2•), alkoxyl (RO•), as well as O2-derived non-radical species such as hydrogen peroxide (H2O2) [1]. The mitochondrion is a major intracellular source of ROS. Of total mitochondrial O2 consumed, 1–2% is diverted to the formation of ROS, mainly at the level of complex I and complex III of the respiratory chain, and is believed to be tissue and species dependent [2, 3]. Mitochondria-derived O2•− is dismutated to H2O2 by manganese superoxide dismutase, and, in the presence of metal ions, highly reactive HO• is generated via Fenton and/or Haber-Weiss reactions, inflicting significant damage to cellular proteins, lipids, and DNA. To date ~10 potential mitochondrial ROS-generating systems have been identified [4]. Among these, Krebs cycle enzyme complexes, such as α-ketoglutarate dehydrogenase (α-KGDH) and pyruvate dehydrogenase have been implicated as significant mitochondrial O2•− and H2O2 sources [5]. Notably, increased nicotinamide adenine dinucleotide (NADH) is linked to elevated H2O2 production by mitochondrial α-KGDH, and this elevated oxidant burden elicits further ROS production from mitochondrial complex I and accelerates cell death [6]. Other interesting mitochondrial ROS sources include p66Shc, an intermembrane space enzyme [7], and monoamine oxidase, an outer membrane enzyme [4], and altered mitochondrial membrane potential [8] or matrix pH [9]. As major ROS generators, mitochondria are often targets of high ROS exposure with deleterious consequences, such as oxidative damage to mitochondrial DNA [10, 11]. While elevated O2•− and HO• associated with mtDNA damage have been implicated in cell apoptosis [12], the precise mechanism whereby mtDNA damage mediates apoptotic signaling is incompletely understood and should provide a fruitful avenue for future investigation.
Peroxisomes are sources of cytosolic H2O2 under physiological conditions, and peroxisomal H2O2 elimination is compartmentalized given its structural organization. Matrix H2O2 is removed by catalase [13] while urate oxidase-catalyzed H2O2 generation within the core is directly released into the cytosol via cristaloid core tubules [13]. Endoplasmic reticular (ER) monoxygenases (e.g., cytochrome P450) contributes to increased cellular H2O2 and O2•− that promotes lipid peroxidation, altered calcium homeostasis, mitochondrial dysfunction and cell apoptosis [14, 15], while NADPH oxidase (Nox)-derived ROS at the plasma membrane function in cellular signaling [16]. In the extrinsic apoptotic pathway, ligand-death receptor engagement (Fas ligand-Fas receptor, TNF-α-TNF receptor1) induced lipid raft formation and Nox recruitment/activation and ROS generation that signal acid sphingomyelinase activation, ceramide production and receptor clustering. These combined processes constituted lipid raft-derived signaling platforms that mediated death receptor activation and induction of apoptosis [17, 18]. The physiological relevance and quantitative significance of ROS-dependent receptor-mediated apoptosis as compared to the classical receptor/ligand-induced apoptotic signaling is, at present incompletely understood and warrant further investigation.
2. CELLULAR REDOX SYSTEMS
2.1 The glutathione redox system and its cellular compartmentation
The centrality of glutathione (L-γ-glutamyl-L-cysteinyl-glycine, GSH) in cellular redox homeostasis is well recognized, and collective studies from our laboratory have established the importance of GSH redox in cell apoptosis in a variety of cell types [19–22]. GSH, as the most abundant free thiol in eukaryotic cells maintains an optimal intracellular redox environment for proper function of cellular proteins. Reduced GSH is the biological active form that is oxidized to glutathione disulfide (GSSG) during oxidative stress, and the ratio of GSH-to-GSSG thus offers a simple and convenient expression of cellular oxidative stress [23]. Typically, cells exhibit a highly reduced GSH-to-GSSG ratio, and greater than 90% of total GSH is maintained in the reduced form through cytosolic de novo GSH synthesis, enzymatic reduction of GSSG, and exogenous GSH uptake [24]. Since GSH participates in numerous redox reactions, an oft-asked question that remains unresolved is that of specificity of GSH redox in the control of cell signaling. Current evidence suggests that in part, specificity and targeted redox control is achieved through the existence of distinct intracellular redox compartments that exhibit unique distribution of GSH and other redox couples [25].
Intracellular GSH is compartmentalized as distinct redox pools within the cellular compartments of cytosol, mitochondria, endoplasmic reticulum and nucleus. Cytosolic GSH is highly reduced, with GSH concentrations ranging between 2–10mM in most cell types. Cellular ratio of GSH-to-GSSG under physiological conditions highly favor reduced GSH (around 100-to-1 in liver) and is decreased during oxidative stress and apoptosis [26]. GSH within the ER is between 2–10mM, and it maintains catalytic site thiols of protein disulfide isomerase (PDI) for protein folding [27, 28] and buffers against ER-generated ROS [27, 28]. Altered GSH redox state triggers the unfolding protein response (UPR) and apoptosis [29]. Recent evidence suggests that the distribution of GSH and GSSG was closer to 5-to-1 [30], that is more reduced that previously thought (ratio of ~ 1-to1). The full implication of a more reducing redox environment for PDI–catalyzed protein folding is unclear at present and warrants reevaluation in the light of this new observation. An independent nuclear GSH pool functions to preserve nuclear proteins in a reducing environment and protects against oxidative and ionizing radiation-induced DNA damage [31]. Cytosol-to-nuclear GSH distribution is reportedly a dynamic process that correlates with cell cycle progression [32] with nuclear GSH being 4-fold greater than cytosolic GSH during cell proliferation, and equally distributed between the two compartments when cells reached confluency [31, 32]. Cytosol-to-nuclear GSH import was suggested to occur by passive diffusion via nuclear pores [33] that is facilitated by the anti-apoptotic protein, Bcl-2 [34]. A distinct mitochondrial GSH (mtGSH) pool preserves the integrity of mitochondrial proteins and lipids and controls mitochondrial ROS generation. The pool size of mtGSH is cell-type specific, varying from 10–15% of the total GSH in the liver [35] to 15–30% of total GSH in the renal proximal tubule [36]. Matrix GSH is maintained through active transport from the cytosolic compartment via inner membrane dicarboxylate and 2-oxoglutarate GSH carriers [37]. Bcl-2 reportedly functions in the preservation of GSH in the intermembrane space through interactions with GSH via its BH2 groove [38], thus contributing to a localized source of mtGSH at this site. Disruption of this interaction by apoptotic stimuli inhibits cytosol-to-mitochondria GSH transport, inducing mtGSH efflux and the apoptotic cascade [38].
2.2 The thioredoxin redox system
The function of the GSH/GSSG redox couple in cellular redox homeostasis occurs in conjunction with redox proteins. Thioredoxins (Trxs), which represent a pivotal partner with GSH/GSSG in redox regulation, are small ubiquitous proteins that possess two catalytic site redox active cysteines (Cys-XX-Cys) [39]. Trxs catalyze the reversible reduction of protein disulfide bonds, and Trx active site cysteines are regenerated by Trx reductase and NADPH [39]. The Trx system collaborates with the glutaredoxin (Grx) system in the reduction of protein mixed disulfides. Like GSH, mammalian forms of Trx are compartmentalized within cells. Trx1 localizes within the cytosol and during oxidative stress, translocates to the nucleus. However, cytosolic and nuclear Trx1 are regulated independently of each other and exhibits functional differences [40, 41]. Trx2 resides exclusively in the mitochondria and functions in mitochondrial redox homeostasis [40, 41]. The regulation of mitochondrial Trx2 is distinct from that of cytosolic Trx1 and mitochondrial GSH/GSSG [40, 42]; thus, reduced and oxidized Trx (Trx-SH/Trx-SS) as well as GSH and GSSG are distinct and independently controlled redox systems.
Described as unique “redox control nodes or circuitry”, the GSH and Trx redox systems reportedly function as rheostat on/off switches in the redox regulation of cellular proteins [43]. Based on a simple concept that the GSH/GSSG, Trx/TrSSG, and cysteine/cystine redox couples are not at equilibrium in biologic systems that is in part supported by experiemental data, Jones and co-workers have presented compelling argument that the existence of such unique compartments of redox pools could, in fact, afford an elegant mechanism for redox control of specific protein sets [41, 44]. Thus, the mode for independent regulation of the redox status of specific protein sets could represent a crucial and generalized redox signaling mechanism in the control of redox-sensitive biological processes within mammalian cells. However, the full extent of the biological importance of metabolically distinct redox pools in redox regulation and the universality of such a regulatory redox mechanism in all tissue types remains to be established. In addition, much remains unknown about precise interactions of compartmental regulatory redox mechanisms, the extent of GSH/Trx cross-talk and communication among the various redox compartments, and the quantitative impact of altered GSH/GSSG and Trx/TrxSS status within one compartment such as the mitochondria on the threshold for redox signaling and gene expression in another compartment such as the nucleus. A full discussion of the integration of redox compartments and communications and implications for redox biological processes is beyond the scope of current review. The reader is referred to several excellent reviews by Jones and coworkers [43, 44] and our recent review [25] for a more in-depth coverage of the concept of redox compartmentation in mammalian cells, non equilibrium thiol/disulfide redox systems and implications for redox signaling and regulation. Given the conceptual novelty that distinct redox nodes/circuitry could serve as a general paradigm of redox control of cell fate, vis-à-vis proliferation, differentiation, and apoptosis, it is anticipated that future research into this area of redox regulation will continue to grow.
Peroxiredoxins (Prx) is a group of non-seleno thiol-specific peroxidases that also contribute to cellular redox control via their ability to eliminate of organic hydroperoxides and H2O2 [45]. All Prxs possess within their catalytic site, peroxidatic cysteines (fast reactive cysteines) that are oxidizable to sulfenic acids (Cys-SOH) that rapidly form disulfide bonds with another cysteine at the C-terminal subunit. Cysteine regeneration is catalyzed by the Trx/TrxR) system [46]. The six isoforms of Prx (PrxI to PrxVI) in mammalian cells are classified into typical 2-Cys Prx, atypical 2-Cys Prx and 1-Cys Prx. Among these, Prx I-II and VI are cytosolic, Prx II is mitochondrial, and Prx IV is extracellular. Prx V is localized in the mitochondria and peroxisomes. The functional significance of redundancies of multiple Prx isoforms in the control of cellular oxidative stress, whether Prxs integrate with the GSH and Trx redox nodes, and whether Prxs, directly or indirectly, participate in redox regulation of apoptotic signaling, are important unresolved questions that need to be addressed.
2.3 The pyridine nucleotide redox system
The redox state of pyridine nucleotides is intricately tied to that of GSH and Trx in redox-dependent cellular processes. Indeed, NAPDH, with a redox potential of ~400mV is the electron donor in the rejuvenation of GSH and Trx redox status. Pyridine nucleotides collectively comprise of reduced and oxidized nicotinamide adenine dinucleotide (NADH/NAD+) and reduced and oxidized nicotinamide adenine dinucleotide phosphate (NADPH/NADP+) that are classically associated with ATP production and reductive biosynthesis, respectively. NADPH/NADP+ is also linked to oxidative stress defense and redox regulation. However, more versatile biological functions have been attributed to these molecules. For instance, NADH/NAD+ was shown to modulate gene transcription via the carboxy-terminal binding protein (CtBP) [47], Ca2+ signaling via cyclic ADP-ribose [48], cell death via poly(ADP-ribose) polymerase-1 (PARP) [49] and sirtuin protein function (see section 2.3.2). NADPH/NADP+ was found to influence cellular signaling via nicotinic acid adenine dinucleotide phosphate (NAADP) [50], and cellular ROS production by electron transport chain or NADPH oxidases [51]. The specific contribution of pyridine nucleotides to redox regulation of cell apoptosis and their interactions with the GSH and Trx redox circuitry in cell signaling are poorly understood and should provide exciting and novel avenues for future research. Of relevance, NADPH/NADP+ participates in antioxidant defense in the control of cellular oxidative stress and GSH/GSSG redox balance. In addition to its classical role in mitochondrial energy production, novel roles of NADH/NAD+ in oxidative stress and apoptosis are underscored by its function in modulating activities of sirtuin proteins, a class of NAD+-dependent deacetylases and mono-ADP-ribosyl transferases. Aspects of NADPH/NADP+ and NADH/NAD+ redox functions pertinent to detoxication reactions and sirtuin activity are discussed in the following sections.
2.3.1 NADPH and antioxidant defense
NADPH is pivotal in GSSG and Trx-SS reduction. NADPH generation during oxidative stress is accomplished by mechanisms that involve either NADP+ reduction or NAD(H)-to-NADP(H) conversion. Consistent with a role for the pentose phosphate shunt in NADPH production, ROS-mediated increase in glucose-6-phosphate dehydrogenase (G6PDH) activity was associated with increased protection against oxidative stress-induced apoptosis [52, 53]. Moreover, although dispensable for pentose synthesis, G6PDH was essential for NADPH-mediated antioxidant protection in mouse embryonic stem cells [54]. NADP+-linked dehydrogenases such as isocitrate dehydrogenase (ICDH) are major contributors to NADPH maintenance, and ICDH-mediated production of NADPH was correlated with increased protection against oxidative stress-induced cellular damage and apoptosis [55]. The two mammalian ICDH isoforms are localized to the mitochondria (mICDH) and cytosol (cICDH), the latter is capable of translocating to the peroxisomes [56]. Overexpression of mICDH improved mitochondrial NADPH and GSH levels and protected human neuroblastoma cells SH-SY5Y against cytochrome c release and ROS-mediated mitochondrial apoptotic signaling [57], while siRNA silencing of mICDH compromised mitochondrial redox status and enhanced HeLa cell apoptotic susceptibility to TNF-α, anticancer drugs or heat shock [58, 59]. Interestingly, ICDH-mediated NADPH production did not appear to compensate for loss of NADPH supply by G6PDH deficiency; G6PDH-deficient mouse cells remained highly sensitive to oxidative stress [60]. Malic enzyme (ME) is another important enzymatic source of NADPH; of the three known mammalian MEs, NADP+ and NAD+-specific ME are mitochondrial, and NADP+-dependent ME is cytosolic [61]. In addition to NADPH generation, mitochondrial NADP+-specific ME reportedly participates in pyruvate-recycling and maintenance of mtGSH [62]. The existence of multienzyme systems for NADPH production is consistent with the importance of its dual functions in reductive biosynthesis as well as detoxication and redox reactions. However, the biological significance of site-specific NADPH generation within the cytosolic and mitochondrial compartments in redox maintenance and drug detoxication remains to be better defined. Indeed, as compared to the GSH and Trx redox systems, the questions of integration of compartmental pyridine nucleotide pools, communication among cellular compartments, and implications for redox biological processes are relatively unexplored.
An important aspect of antioxidant defense by pyridine nucleotides is underscored by the enzymatic NADH-to-NADPH conversion. Located in the inner membrane, the mitochondrial nicotinamide nucleotide transhydrogenase (NNT) coordinates proton translocation across the mitochondrial membrane and transfer of reducing equivalents between NADH and NADPH, representing a key mechanism for mitochondrial NADH-to-NADPH conversion. Under physiological conditions, NNT favors NADPH production that maintains intramitochondrial homeostatic redox state while during oxidative stress, NNT-derived NADPH mediates the regeneration of GSH and protein thiols [63]. Within the cytosol, NAD(H)-to-NADP(H) conversion is mediated by cytosolic NAD kinases (NADK), a class of ubiquitous enzymes that catalyze the phosphorylation of NAD+. To date only a cytosolic mammalian NADK has been identified [64], and its importance in maintaining cellular NADPH for reductive biosynthesis in organogenesis is documented [64, 65]. The relevance of NADK to NADPH-dependent protection against oxidative challenge is, however, less well understood. Pollak et al found that NADK overexpression conferred only moderate protection against oxidative stress, and that NADK deficient cells exhibited similar sensitivity to oxidative stress as NADK sufficient cells [64], implying a relatively minor role for NADK in NADPH biogenesis. Moreover, whereas NADP+-linked dehydrogenases were activated by oxidative stress, NADK activity and expression remained unchanged [64], suggesting a lack of sensitivity to oxidative challenge. This not-withstanding, the importance of enzymatic conversion of NADH-to-NADPH and its relevance in antioxidant defense is underscored by recent studies demonstrating that during oxidative challenge, gluconeogenic and tricarboxylic acid cycle enzymes in NADH production were downregulated in favor of upregulation of NADPH-producing enzymes of the glyoxalate and glycolytic [66, 67]. Consequently, substrate flow was channeled away from NADH production towards increased NADPH generation. Significant diversion of reducing equivalents from mitochondrial energy production to NADPH generation for detoxication reactions is likely to markedly impact mitochondrial respiratory integrity. Furthermore, sustained compromise in mitochondrial respiration during oxidative challenge, particularly under conditions of decreased glucose, would have important biological implications for cell survival.
2.3.2 NAD+ and function of sirtuin proteins
In recent years, much interest has focused on the function of sirtuin proteins, a conserved family of NAD+-dependent deacetylases and mono-ADP-ribosyl transferases in cellular processes such as gene silencing, DNA repair, life span extension and cell apoptosis [68–71]. Sirtuins (Sirts) catalyze protein deacetylation, functioning as cellular rheostats that sense changes in the energy and redox status. For instance, an increase in the NAD+/NADH ratio enhanced protein deacetylation, while elevated NADH or nicotinamide inhibited Sirt activity [72], suggesting a close association between the cellular redox status of pyridine nucleotides and Sirt function. To date, seven members of mammalian Sirts (Sirt1-7) have been identified, each exhibiting distinct subcellular localization. While Sirt members reportedly control function of specific protein sets, the biological roles of most Sirt proteins have not been fully explored.
Sirt 1 is among the best studied member, an NAD+-deacetylase with a predominant nuclear localization that is capable of nuclear-to-cytosol shuttling [73]. Sirt1 acts on numerous substrates that controls cell senescence, proliferation, and apoptosis [74, 75]. The association of Sirt1 with the tumor suppressor p53 and the transcription factor, Forkhead box O (FOXO) is of relevance to oxidative stress-induced apoptosis. At low H2O2, Sirt1-mediated p53 deacetylation promoted p53 destruction via MDM2-dependent ubiquitination [76, 77], and Sirt1 overexpression inhibited p53-mediated nuclear transactivation and blocked oxidant-induced apoptosis [78]. However, Sirt1 could induce proapoptotic effects in certain cell types under oxidative conditions; at high ROS, hyperacetylated nuclear p53 promoted apoptosis through transactivation of proapoptotic PUMA and Bax genes [79, 80]. Interestingly, physiological levels of ROS can promote mitochondria-induced apoptosis in mouse embryonic cells via Sirt1-dependent mitochondrial translocation of p53 [81]. Sirt1 interacts with FOXO-1, -3a and -4 wherein FOXO deacetylation conferred cell resistance to oxidative stress and apoptosis [82]. During oxidative stress, Sirt1-FOXO3a interaction increased transcription of stress-resistant genes and decreased expression of FOXO3a-dependent proapoptotic genes [82]. In renal tubular cells, Sirt1 induced catalase downregulation and maintained steady state ROS levels appropriate for cell signaling in the absence of oxidative stress [83], while increased H2O2 promoted Sirt1/FOXO3a-dependent catalase expression that protected against apoptosis [83]. Thus, it appears that Sirt1 controls renal tubular cell apoptosis by modulating intracellular ROS levels through bidirectional regulation of nuclear catalase expression, and thus affords cytoprotection under physiological as well as oxidative conditions. In other examples, increased human cardiac fibroblast resistance to oxidative stress was associated with elevated FOXO3a and mitochondrial Prx III expression [84] and blockade of caspase-3 and -7 induced apoptosis in cancer epithelial cells was linked to Sirt1-mediated FOXO4 deacetylation [85]. A suggestion that Sirt1 may potentially be a key cellular target for prostate cancer therapy was evidenced by the observation that Sirt1-dependent deacetylation of FOXO1 was associated with uncontrolled growth and proliferation of prostate cancer cells [86]. Much of current Sirt1 research is in the area of cell proliferation and differentiation of cancer relevance, but a link to redox control is unknown. The findings that Sirt1 is sensitive to H2O2 and responsive to NAD+/NADH suggests a role for redox in the regulation of Sirt1 function, an exciting possibility which hitherto, has not been rigorously explored, and one that warrants detailed study.
Functions of other Sirt members (Sirt 2 to 6) are less studied. To date, Sirt 3, 4, and 5 have been identified as mitochondrial proteins possessing NAD+-deacetylase (Sirt4), ADP-ribosyl transferase (Sirt5) or both enzyme activities (Sirt3) [87–89] suggesting that mitochondrial Sirts may control a broad spectrum of mitochondrial functions. In several studies, mitochondrial Sirt proteins are implicated in energy metabolism such as ATP generation, membrane potential regulation, and ROS production. For instance, Sirt3-mediated deacetylation of mitochondrial complex I engaged ATP production under normal homeostatic conditions, while decreased ATP generation during oxidant exposure resulted from Sirt3 dissociation and complex I acetylation [90]. Sustained expression of Sirt3 induced thermogenesis in brown adipocytes through a decrease in mitochondrial membrane potential and enhanced ROS production [91]. Deacetylation by Sirt3 under caloric restriction activated glutamate dehydrogenase (GDH) [92] while ADP-ribosylation by Sirt4 repressed GDH activity and decreased ATP production due to reduced trafficking through tricarboxylic acid cycle intermediates [88]. Evidence of the contribution of mitochondrial Sirt proteins to redox modulation and cell apoptosis is sketchy. However, the notion that Sirts can contribute to mitochondrial redox control and apoptosis is supported by evidence that Sirt3 and -5 exert roles in mitochondrial antioxidant defense and apoptotic signaling. NADP+-dependent ICDH (ICDH2) is a mitochondrial target of Sirt3, and protein deacetylation stimulated enzyme activity and NADPH regeneration [89]. Moreover, Sirt5 reportedly can translocate to the mitochondrial intermembrane space and deacetylate cytochrome c [89], but the precise impact of cytochrome c deacetylation on mitochondrial respiration and/or apoptosis is unknown. Current paradigm proposes an oxidant-induced disruption of the cardiolipin-cytochrome c complex in mitochondria-to-cytosol translocation of cytochrome c during apoptosis initiation (Section 4.1). Demonstration that cytochrome c deacetylation is mechanistically associated with mitochondrial cytochrome c release could constitute a novel paradigm shift.
Current research on Sirt2, -6, and -7 is scanty, and avenues for future investigations into the functions and redox control of these Sirt members are wide open. Evidence to date identifies Sirt2 as a cytosolic enzyme that exhibits deacetylase and ADP-ribosyl transferase activities and colocalizes with the microtubule network within the cytosol, causing the deacetylation of α-tubulin. Nuclear translocation of Sirt2 during G2/M phase of the cell cycle suggests a role in the control of cell cycle check-point [93], but the mechanism of Sirt2-mediated cell cycle regulation is unclear. Beyond a reported involvement in maintaining genome integrity through activating base excision repair of oxidative damaged DNA or single-strand DNA breaks [94], the biological importance of Sirt6, a nucleolus-located deacetylase [95, 96] is relatively unexplored. Similarly, despite documentation that Sirt7 can bind to heterochromatin regions within the nucleolus [95, 96], a possible role for Sirt7 in promoting RNA polymerase 1-catalyzed gene transcription [96] remains to be defined. At present, the involvement of members of sirtuin proteins in the apoptotic process is unknown, and validation of an epigenetic control of apoptosis by sirtuin proteins should represent a fertile area for future research. One intriguing hypothesis is that acetylation/deacetylation functions in post-translational regulation of cell apoptosis in a manner akin to glutathiolation/deglutathiolation and/or nitrosation/denitrosation.
ROS AND REDOX INVOLVEMENT IN APOPTOSIS
3.1 Overview of death receptor and mitochondrial apoptotic pathways
Death receptor pathway
The extrinsic pathway of apoptosis is mediated by death receptors in which ligand-receptor binding initiates protein-protein interactions at cell membranes that activate initiator caspases. Major known receptors include Fas (also called CD95 or APO-1), TNF receptor 1 (TNFR1) and TNF-related apoptosis-inducing ligand (TRAIL) receptor 1 (TRAIL-R1; also called DR4) and TRAIL receptor 2 (TRAIL-R2; also called DR5) [97]. TRAIL-R3, TRAIL-R4 and soluble receptor, osteoprotegerin lack functional cytosolic domains and are decoy receptors where ligand binding does not transmit an apoptotic signal [98]. The death receptor is comprised of three functional extracellular ligand-binding, transmembrane, and intracellular domains. Ligands that activate death receptors belong to the TNF superfamily of cytokines; these include TNFα, Fas ligand (FasL), and TRAIL. Ligand binding induces receptor trimerization and cross-linking via disulfide bond formation, a step that is necessary for receptor stabilization and activity [99]. Typically, apoptotic signaling is initiated by the association of death domain-containing adaptor proteins within the death domain located at the C-terminal domain of the receptor. As discussed in Section 1, newer evidence suggests possible direct roles for ROS in mediating death receptor activation and apoptotic induction through ROS-induced receptor clustering and formation of lipid raft-derived signaling platforms. The pathophysiological scenarios whereby this would be a major mechanism of apoptotic signaling remains to be defined.
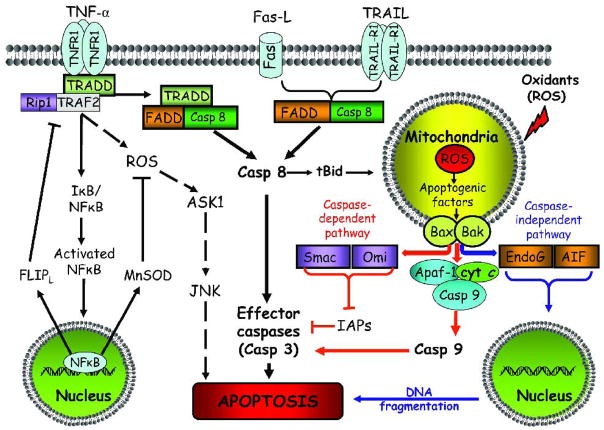
Figure 1 summarizes the key players and mechanistic differences in the three major classical death receptor signaling pathways in cell apoptosis. The Fas/FasL system is one of the best studied systems in death receptor-mediated apoptosis. Within minutes of Fas/FasL binding, Fas associated DD (FADD) and pro-caspase-8 are recruited, and the resultant death-inducing signaling complex (DISC) is endocytosed [100]. The release of endosomal DISC from the receptor accumulates as cytosolic DISC to which additional FADD and pro-caspase-8 are recruited, resulting in activation of the initiator caspase-8 [101, 102]. The extent of activated caspase-8 at the DISC determines Type 1 or Type 2 mechanisms; significant caspase-8 activation directly activate caspase-3 (Type 1), while low caspase-8 activation mediates caspase -3 activation through an amplification loop involving the mitochondria (Type 2) [103]. In Type 2 apoptosis, activated caspase-8 cleaves pro-apoptotic Bid that induces outer mitochondrial membrane permeabilization through the interactions of truncated tBid with Bax/Bak, resulting in the mitochondrial release of apoptogenic cytochorme c, second mitochondria-derived activator of caspases/direct IAP binding protein with low pI (Smac/Diablo) or apoptosis inducing factor (AIF). Additionally, Fas-induced NADPH oxidase-dependent H2O2 and O2.− generation further down-regulates the anti-apoptotic FLIPL through FLIPL ubiquitination/proteasomal degradation or through NO scavenging that prevented FLIPL S-nitrosation and cytoprotection [104]. This newly described ROS-NO interaction in controlling FLIPL down-regulation was considered a key regulatory mechanism of Fas-induced apoptosis [104] which is likely to be significantly impacted under pathophysiological conditions of altered ROS and NO availability, such as occurs during ischemia-reperfusion or chronic inflammation.
Figure 1. Death receptor-mediated and mitochondrial pathways of cell apoptosis.
Death receptor extrinsic pathway. The major death receptor pathways include Fas/FasL, TNF-R1/TNFα, and TRAIL-R1/TRAIL. Binding of ligands to respective receptors activates downstream signaling and the formation of death-inducing signaling complex. Activation of the NF-κB survival pathway enhances transcription of the anti-apoptotic proteins like FLIPL or MnSOD and apoptosis blockade. At high ROS and failure to activate NF-κB promotes ASK1/JNK activation that triggers cell apoptosis. Death receptor signaling is associated with caspase-8 activation that promotes apoptosis via activation of effector caspases (e.g. caspase-3) or engages mitochondrial apoptotic signaling via truncated Bid, leading to the release of apoptogenic factors such as cytochrome c into the cytosol. ASK1, apoptosis signal-regulating kinase 1; Apaf-1: apoptotic protease-activating factor-1; Bid, BH3-only pro-apoptotic protein; tBid, truncated form of Bid; casp 8, 9, active forms of caspases-8 and -9; cyt c: cytochrome c; FLIPL: FLICE inhibitory protein; FADD: Fas-associated death domain; FasL: Fas ligand; JNK: c-Jun N-terminal kinase; NF-κB: nuclear transcription factor kappa B; I B/NF-κB: the inactive form of NF-κB associated with its inhibitor; MnSOD: manganese superoxide dismutase; RIP1: receptor-interacting kinase1; ROS: reactive oxygen species; TNF-α: tumor necrosis factor-α; TNFR1: TNF receptor-1; TRAIL: TNF-related apoptosis-inducing ligand; TRAIL-R1: Trail receptor-1; TRADD: TNF receptor-associated death domain; TRAF-2: TNF receptor-associated factor-2.
Mitochondrial intrinsic pathway. Various apoptotic stimuli (e.g. ROS) mediate permeabilization of the mitochondrial outer membrane and the release of pro-apoptotic proteins. Within the cytosol, cytochrome c together with Apaf-1, and dATP form the apoptosome complex to which the initiator procaspase-9 is recruited and activated. Caspase-9-catalyzed activation of the effector caspase-3 executes the final steps of apoptosis. Caspase activation is further enhanced through neutralization of caspase inhibitors by apoptogenic proteins like Smac/Diablo and Omi/HtrA2 that are released from the mitochondria. In addition, mitochondrial proteins such as AIF and endoG promote caspase-independent apoptosis through nuclear translocation and mediating genomic DNA fragmentation. Cyt c: cytochrome c; casp 9: activated caspase-9; Apaf-1: apoptotic protease activation factor-1; Smac/Diablo, second mitochondria-derived activator of caspases/direct IAP binding protein of low pI; Omi/HtrA2: high temperature requirement A2 serine protease; IAP: inhibitors of apoptosis proteins; AIF: apoptosis inducing factor; endoG, endonuclease G; Bax/Bak, pro-apoptotic proteins.
TNF-R1 is a death receptor that mediates the major biological functions of TNFα. TNFα-TNF-R1 binding elicits receptor trimerization, release of inhibitory silencer of death domain (SODD) [105], and recruitment of TNF-R1-associated death domain (TRADD) that results in complexes I and II formation that activate distinct downstream survival or apoptotic signaling pathways [106]. At complex I, TRADD serves as a scaffold for the receptor interacting protein 1 (RIP1) and TNF receptor-associated factor 2 (TRAF2) in the recruitment of TGF-β activated kinase 1 and activation of NF-κB and p38 and Jun N-terminal protein kinase (JNK) [107]. NF-κB activation is associated with induction of anti-apoptotic proteins, FLIPL, Bcl-xL, A1/Bfl-1, X-linked inhibitor of apoptosis (XIAP) and cellular inhibitor of apoptosis (c-IAP)1 and 2 [108–110], while JNK activation is associated with ROS-induced activation of apoptosis signal-regulating kinase 1 (ASK1) and proteasomal degradation of FLIPL [111, 112]. Complex II comprises of TRAAD, FAAD, and caspase-8, that is formed within the cytosol following TNF-R1 receptosome endocytosis [113]. The cellular status of FLIPL and RIP1 appears to be important check points in determining whether TNF-R1 induces apoptotic or survival signaling. High concentrations of FLIPL competitively inhibited caspase-8 binding at complex II and prevented DISC formation [114], and complex I dissolution mediated by caspase-8 cleavage of RIP1 promoted complex II formation [115]. Given the similarity between the Fas and TNF-R1 systems, the activation of TNF-R1-mediated apoptosis could similarly subscribe to modulation by ROS through clustering of receptors, signaling via lipid raft platforms, and interaction with NO, a suggestion that remains to be tested.
The widely expressed TRAIL receptors (DR4 and DR5), unlike Fas and TNF-R1, signal through Type II mechanisms [116, 117], and are not dependent on internalization of the ligand-receptor complex for full activation of initiator caspases [118]. In addition, TRAIL receptor signaling partners with decoy receptors that competitively bind TRAIL ligands. Since decoy receptor expression was reportedly lower in tumor cells, cancer cell susceptibility to TRAIL would likely be enhanced, a notion that requires validation and could be capitalized in cancer therapy [119].
Mitochondrial pathway
Stimulation of intrinsic mitochondrial apoptotic pathway by ROS and mitochondrial DNA damage promotes outer membrane permeabilization and mitochondria-to-cytosol translocation of cytochrome c, AIF, or Smac/Diablo that trigger caspase-dependent or caspase-independent cytosolic signaling events [120] (Figure 1). In caspase-dependent signaling, cytochrome c forms the apoptosome complex with apoptotic protease-activating factor-1 (Apaf-1) and recruited pro-caspase-9 that induces cleavage of downstream effector caspases-3 and -7. Additionally, Smac/Diablo antagonizes the inhibitory effects of IAPs which enhances caspase activation. AIF mediates caspase-independent signaling through cytosol-to-nuclear translocation and induction of nuclear chromatin condensation and DNA fragmentation [121].
Central to mitochondrial permeabilization and mitochondrial release of apoptogenic factors is the permeability transition pore (PTP), a megapore spanning the inner and outer mitochondrial membrane. Of the three PTP component proteins, viz., cyclophylin D (cypD), voltage-dependent anion channel (VDAC), and the adenine nucleotide translocase (ANT), only cypD is a permanent constituent and modulator of PTP [122–124]. Key members of the anti-apoptotic (Bcl-2, Bcl-XL, and Bcl-w) and pro-apoptotic (Bax, Bak, Bad, Bim, and Bid) BcL-2 superfamily of proteins are major players in mitochondrial outer membrane permeabilization and apoptotic susceptibility [125]. In the presence of an apoptotic stimulus, tBid promotes Bax/Bak oligomerization and membrane insertion that results in megapore formation, a highly orchestrated and active process [126, 127]. Additionally, tBid/Bax-induced mitochondrial permeabilization was shown to be mediated through interaction with TOM, a functional outer membrane translocase, the TOM complex [128]. The pivotal contribution of tBid to apoptotic signaling in both the mitochondrial and death receptor pathways is consistent with cross-talk between intrinsic and extrinsic apoptotic signaling. However, the intracellular regulatory events and/or mechanisms that would preferentially trigger the engagement of the mitochondrial cascade following exposure to extrinsic Fas or TNFα signals remain to be determined.
3.2 ROS and JNK-mediated apoptotic signaling
ROS activation of JNK can induce extrinsic or intrinsic apoptotic signaling [129]. Upstream of JNK is the redox sensitive MAPK kinase kinase, ASK1. ASK1 activity is inhibited by interactions with redox proteins (Grx, Trx1), heat shock proteins (Hsp90, Hsp72) and 14-3-3 [130–132], and is stimulated by TRAF proteins, ASK1-interacting protein 1(AIP1), Daxx, and JASP/JIP3 [133–135]. TNFα is a potent activator of MAPK cascade, and the ASK1-JNK pathway plays an important role in TNF-R1-mediated apoptotic signaling in various cell types [136]. Whether TNFα induces anti- or pro-apoptotic effects depends on the level and duration of JNK activation by ROS [137]. A transient and modest JNK activation mediated cell survival via NF-κB-induced anti-apoptotic gene expression, while a prolonged and robust JNK activation was associated with cell apoptosis via ASK1 signaling [138, 139]. Thus, development of a strategy to manipulate the degree and duration of cellular JNK activation could provide a reasonable approach to specifically target cell survival or cell death such as in cancer therapy.
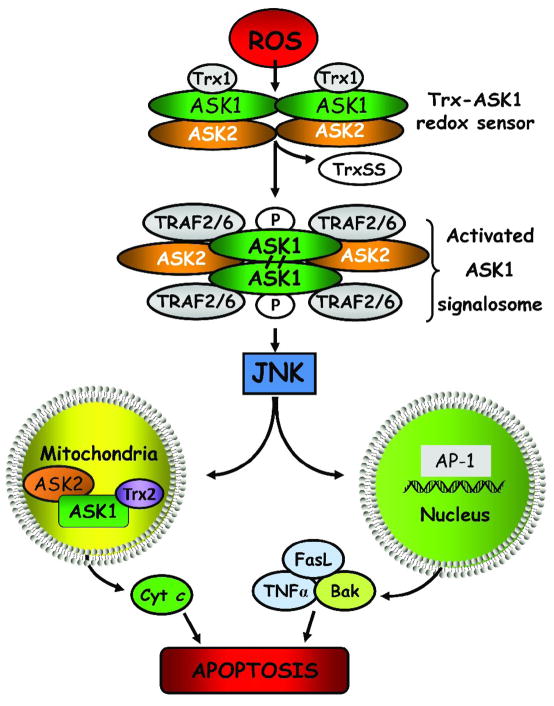
Figure 2 summarizes our current understanding of the mechanism of ROS and redox modulation of ASK1/JNK signaling in cell apoptosis. This model proposes that ROS mediates the interaction of Trx1 and the N-terminal domain of ASK1, preventing ASK1 activation and downstream propagation of an apoptotic signal [130]. Only reduced Trx1 binds ASK1; the resultant Trx1/ASK1 complex, termed “ASK1 signalosome” functions as a redox switch that senses cellular ROS and is activated under oxidizing conditions [140]. Elevated cellular ROS induce the dissociation of oxidized Trx1 from the complex, and permits ASK1 oligomerization through N-terminal coiled-coil domains and a gain of full ASK1 kinase activity [141]. Formation of a functional ASK1 signalosome complex is linked to the recruitment of TRAF2/6 that promotes ASK1 autophosphorylation and JNK activation [130, 141]. ASK2, another member of the ASK family, binds and stabilizes ASK1 in the cytosol, nucleus and mitochondria [142] and thus, play an important role in the regulation of ASK1/JNK signaling and cell apoptosis [143]. Current evidence show that heterodimeric complexes of ASK1 and 2 stimulate JNK and p38 activity while the absence of ASK2 diminishes apoptotic signaling [143]. ROS was shown to promote ASK1 activation by inducing the dissociation of the docking protein 14-3-3 [144] or blocking the inhibitory effects of protein phosphatases, PP5 and PP2A [145]. Interestingly, mitochondrial ASK1-dependent apoptotic signaling pathway reportedly activated both JNK-dependent and -independent apoptosis [146]. Nuclear translocation of activated JNK promoted activator protein-1 (AP-1)-mediated expression of pro-apoptotic TNFα, Fas-L and Bak [147], while mitochondrial JNK translocation promoted cytochrome c release [148]. While these latter observations suggest cross-talk among ASK1/JNK signaling and the classical mitochondrial and death receptor pathways in cell apoptosis, the extent of interaction and integration among these various apoptotic pathways is unclear as is the universality and quantitative importance of ASK1/ASK2-JNK signaling in apoptosis of various cell types induced by intrinsic versus extrinsic signals.
Figure 2. ROS-induced ASK1/JNK signaling and apoptosis.
Reactive oxygen species initiate Trx dissociation from the ASK1-Trx complex, the ASK1 signalosome, through oxidation of Trx redox active site. ASK1 undergoes auto-phosphorylation and covalent binding between its subunits, leading to the formation of “activated signalosome”, and recruitment of tumor necrosis factor receptor-associated factor 2 and 6 to the complex. Activated ASK1 signals downstream JNK activation and induce apoptosis either via mitochondrial signaling or via transcription of AP-1-dependent pro-apoptotic genes. Additionally, ROS-mediated disruption of mitochondrial ASK1/ASK2/Trx2 complex induces cytochrome c release. ROS: reactive oxygen species; TRAF2/6: TNFα receptor-associated factor 2, 6; ASK1, -2: apoptosis signal-regulating kinase 1 and 2; JNK: c-Jun N-terminal kinase, Trx1: thioredoxin 1, reduced form; TrxSS: thioredoxin, oxidized form; Trx2: thioredoxin 2, mitochondrial enzyme; Bak: pro-apoptotic protein; cyt c, cytochrome c; TNFα: tumor necrosis factor alpha; FasL: Fas ligand.
3.3 GSH redox status and apoptotic signaling
ROS-mediated apoptotic signaling is associated with decreased cellular GSH levels and the loss of cellular redox balance [149, 150]. Decreased cell GSH can occur through ROS-induced GSH oxidation or GSH export from cells; the resultant GSH reduction would enhance further ROS production during oxidative challenge [151]. It was demonstrated that GSH loss due to decreased de novo GSH synthesis triggered redox activation of protein kinase C (PKCδ) [152] and through GSH efflux, induced JNK-dependent apoptosis [153]. The initiation of apoptosis through GSH efflux was a ROS-independent mechanism since apoptosis was attenuated by blockade of GSH export but not by antioxidants [154]. This suggestion is supported by our recent studies, demonstrating that intestinal cell apoptosis induced by staurosporine (STP) was linked to GSH efflux without accompanying GSH/GSSG redox changes. This STP-induced export of cellular GSH was driven by γ-glutamyl transferase-catalyzed extracellular GSH hydrolysis [155] and was associated with caspase-3 activation independent of caspases-8 or 9 functions. In other studies, FasL-induced GSH export was shown to be essential for the development of apoptosis in lymphoid cells; ROS production was merely a bystander phenomenon [154]. Collectively, the evidence to date has ruled out a major role for ROS signaling in GSH efflux-mediated cell apoptosis, and key challenges for future research will be delineating the mechanism(s) that couples the export of GSH to the triggering of apoptotic signals at cell membranes, and the nature of these signals.
GSH oxidation is a major contributor to cell apoptosis mediated by oxidants. Accumulated evidence from our laboratory has consistently shown that an early spike in GSSG formation, typically within minutes of oxidant exposure, preceded oxidant-induced activation of mitochondrial apoptotic signaling and cell apoptosis hours later [20, 21, 156, 157]. Importantly, post-oxidant recovery of cellular GSH/GSSG redox status did not influence the apoptotic outcome, indicating that oxidant-induced apoptotic initiation occurred within an early and narrow window of GSH/GSSG redox shift. Indeed, while cell apoptosis was effectively blocked by NAC pretreatment, apoptosis was not prevented when the thiol antioxidant was administered at a time post oxidant-induced GSH oxidation [21]. Collectively, our studies establish that this mode of early induction of cellular GSH redox imbalance in apoptotic redox signaling is common among diverse oxidants such as hydroperoxides (tert-butyl, lipid hydroperoxides), redox cycling quinones (MQ), and methylglyoxal, and in different intestinal (CaCo2, HT-29, NCM460) and neuronal (naïve and differentiated PC12) cell types [19, 21, 156, 158–160]. We further show that oxidant-mediated apoptotic susceptibility is related to a cell’s phenotype. The induction of PC12 cell differentiation generated a phenotype that was more resistant to tert-butylhydroperoxide; this oxidative resistance was associated with a reduced intracellular GSH redox environment and decreased Apaf-1 expression [20]. Attenuated cellular susceptibility of the differentiated PC12 phenotype was also observed for carbonyl stress [160] and hyperglycemic stress [22, 159]. If validated, the paradigm of enhanced cellular vulnerability associated with a proliferative state should provide a strong cellular basis for targeting actively growing tumor cells for oxidative stress induced cell killing.
The mitochondrial GSH/GSSG redox status is critical for preserving mitochondrial function during oxidative stress. Studies from our laboratory and others have documented a relationship between mitochondrial GSH (mtGSH) loss and cell apoptosis. Earlier studies demonstrated that decreased mtGSH correlated with apoptosis induced by exposure to aromatic hydrocarbons [161], hypoxia [162], tert-butylhydroperoxide [163], and ethanol [164–166]. Associated with decreased mtGSH were mitochondrial ROS production, loss of mitochondrial membrane potential, and mitochondria-to-cytosol release of cytochrome c [167]. Interestingly, moderate mtGSH decrease was insufficient to elicit apoptosis in hepatocytes during hypoxia, suggesting that achievement of a critical threshold of mtGSH loss is necessary to trigger mitochondrial apoptotic signaling [162]. Moreover, Ghosh et al demonstrated that oxidation of mtGSH was a requisite for mitochondrial ROS generation, membrane potential collapse and caspases-9 and -3 activation in cardiomyocyte apoptosis caused by short-term diabetes [168]. We similarly showed that early ROS production and mtGSSG formation preceded mitochondrial dysfunction and apoptosis in intestinal cells exposed to menadione [19]. Given the oxidative vulnerability of the mtGSH pool and the dynamics of its maintenance, manipulation of the mitochondrial redox compartment can be capitalized to selectively sensitize cells to oxidative damage.
4. MODULATORS OF INTIATION AND EXECUTION OF APOPTOSIS
4.1 Mitochondrial modulators of apoptotic initiation
ROS and mitochondrial permeability transition
ROS are known triggers of the intrinsic apoptotic cascade via interactions with proteins of the mitochondrial permeability transition complex [169]. Components of PTP, viz., VDAC [170], ANT [171] and cypD [172] are targets of ROS, and oxidative modifications of PTP proteins will significantly impact mitochondrial anion fluxes. Indeed, a mere transient increase in mitochondrial membrane hyperpolarization following exposure to H2O2 initiated collapse of the mitochondrial membrane potential (Δψm) [173], mitochondrial translocation of Bax and Bad, and cytochrome c release [174]. Even non oxidants such as cadmium and staurosporine can trigger intrinsic apoptotic signaling through induction of ROS production and associated mitochondrial permeability transition and cytochrome c translocation [175, 176]. Significant mitochondrial loss of cytochrome c will lead to further ROS increase due a disrupted electron transport chain [177].
Oxidative mitochondrial DNA damage
Mitochondrial DNA (mtDNA) is a circular double-stranded DNA organized in nucleoids in proximity to the electron transport chain. mtDNA lacks introns and, being close to an ROS source, is prone to oxidative damage. Since mtDNA encodes 13 polypeptides of the respiratory chain, impaired mtRNA transcription would compromise mitochondrial ATP production [178]. mtDNA damage-induced decreased respiratory function enhances ROS generation, thus eliciting a vicious cycle of ROS-mtDNA damage that ultimately trigger apoptosis [179, 180]. A limited nucleotide excision DNA repair capacity coupled to a high mitochondrial ROS load, further contributes to oxidative damage to mtDNA. Single strand breaks and abasic sites formed during enhanced ROS generation can induce apoptotic signaling [181]. Interestingly, of the two sources of O2•− production elicited by angiotensin II exposure, viz., NADPH oxidase and mitochondria, only the latter resulted from mtDNA damage-induced impaired mitochondrial complex I activity. The subsequent collapse of Δψm collapse, release of cytochrome c and cell apoptosis, were all prevented by inhibiting mtDNA damage [12]. Thus, the protection of mtDNA intergrity is critical not only to bioenergetic homeostasis, but to cell survival as well. Recent findings by Rachek et al (2009) revealed that mtDNA damage was an initiating event in mitochondrial dysfunction and hepatotoxicity induced by pharmacologic levels of the diabetic drug, troglitazone, thus ascribing potential clinical importance of mtDNA damage in drug toxicity [11].
Our recent studies demonstrated that mtGSH is a determinant of the extent of oxidant-induced DNA damage [10]. MQ-induced oxidative mtDNA damage paralleled the formation of mitochondrial GSSG and protein disulfide which was blunted by NAC and exacerbated by inhibition of GSH synthesis in accordance with increased and decreased cellular GSH, respectively [182]. Significantly, mtDNA damage was potentiated by blockade of mtGSH transport and prevented by overexpression of the oxoglutarate mtGSH carrier, validating the link between mtGSH and mtDNA integrity. Moreover, post-MQ recovery of mtDNA was preceded by restored cellular GSH, suggesting that DNA repair may also be GSH-dependent [182]. In previous studies, an inverse relationship between GSH and basal oxidative DNA damage [183], and an association between ROS-induced DNA deletions and genomic rearrangements with GSH depletion/oxidation have been documented [184–187]. Moreover, age-derived ROS-induced mtDNA damage was linked to mtGSH oxidation [188]. It remains to be established as to whether GSH functions in attenuating mtDNA damage or in stimulating mtDNA repair. Post-translational phosphorylation of Ogg1 [189] and acetylation/deacetylation of Ape1 [190] have been implicated in the control of DNA repair; the possibility that glutathiolation is another key post-translational mechanism in the regulation of base-excision repair enzymes is an exciting notion that warrants investigation.
Cytochrome c and cardiolipin interaction
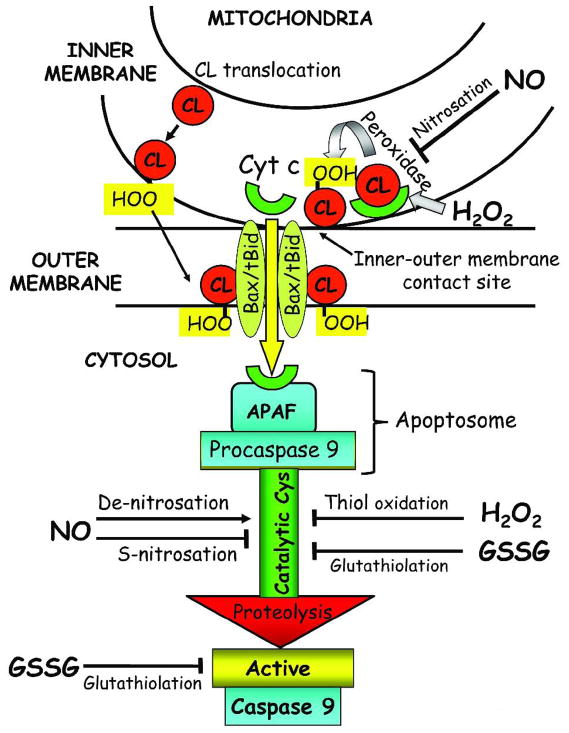
Cytochrome c is a water-soluble heme-containing protein bound to the outer leaflet of the mitochondrial inner membrane through interactions with the anionic phospholipid, cardiolipin. Normally, cytochrome c participates in shuttling electrons between Complex III and Complex IV of the mitochondrial electron transport chain; its release from the mitochondria initiates the apoptotic cascade (Figure 3). Mitochondria-to-cytosol release of cytochrome c sequentially occurs via detachment from cardiolipin and translocation through the mitochondrial outer membrane [191]. At low mitochondrial ROS, tightly bound cytochrome c exhibits increased peroxidase activity [192] that oxidizes cardiolipin and facilitates its detachment [193, 194]. Oxidized cardiolipin is distributed to the outer leaflet of the mitochondrial membrane where it functions as a docking platform for tBid, enabling mitochondrial membrane permeabilization and cytochrome c movement across the outer membrane into the cytosol [195]. Since oxidative modification of cardiolipin is pivotal in mitochondrial cytochrome c loss and cell commitment to apoptosis, cardiolipin-bound cytochrome c could be viewed as a mitochondrial oxidative stress sensor and redox regulator of apoptosis. Therefore, the extent of cardiolipin peroxidation would likely be an important determinant of apoptotic susceptibility of different cell types possessing various cardiolipin species and fatty acyl side chains. A possible quantitative relationship between cardiolipin oxidation products and propensity for cell apoptosis has yet to be rigorously tested.
Figure 3. Redox mediation of mitochondria-to-cytosol release of cytochrome c and activation/inactivation of caspase-9.
Interaction with mitochondria-specific cardiolipin sequesters cytochrome c in the mitochondrial inter-membrane space. Enhanced mitochondrial generation of H2O2 activates cytochrome c peroxidase activity and induces cardiolipin peroxidation and cytochrome c detachment. Oxidized cardiolipin is translocated to the mitochondrial outer membrane, and together with the proapoptotic proteins, Bid and Bax, forms a megapore channel that enables the mitochondria-to-cytosol transit of cytochrome c. Through a nitrosation reaction, the peroxidase activity of cytochrome c is inhibited by NO. Within the cytosol, cytochrome c interacts with Apaf-1 and dATP, forming the apoptosome complex to which pro-caspase-9 is recruited. Proteolytic activation of pro-caspase-9 is mediated by post-translational modification of the catalytic site cysteines through thiol oxidation, nitrosation or glutathiolation. H2O2- and GSSG-mediated cysteine oxidation or S-glutathiolation, respectively results in pro-caspase inactivation. NO-mediated S-nitrosation similarly inhibits caspase-9 activation whereas de-nitrosation promotes proenzyme proteolysis and activation. Additionally, GSSG-dependent glutathiolation of active caspase-9 results in direct inhibition of enzyme activity. Apaf-1: apoptotic protease activation factor-1; CL: cardiolipin; CL-OOH: peroxidized cardiolipin; cyt c: cytochrome c; H2O2: hydrogen peroxide; NO.: nitric oxide; Bid/Bax: proapoptotic proteins.
Nitric oxide (NO) reportedly modulate cytochrome c peroxidase function at the early stage of apoptotic initiation [196]. Physiological NO levels inhibited peroxidase activity and prevented cardiolipin oxidation, while elevated NO induced peroxynitrite-mediated nitration of tyr-67 and enhanced cytochrome c peroxidase activity [197, 198]. However, the nitration of tyr-74 following continuous peroxynitrite exposure prevented cytochrome c/apoptosome complex formation [199]. Interestingly, the oxidation/reduction state of cytochrome c per se has been implicated in mitochondrial apoptotic signaling [200], and cytochrome c-containing oxidized, but not reduced, heme was capable of caspase activation [201]. The redox state of cytochrome c within cells appears to be dependent a non-apoptotic or apoptotic phenotype; generally, non-apoptotic cells favored cytochrome c reduction and cell survival, while apoptotic cells favored cytochrome c oxidation and apoptosis [202]. Precisely how the redox state of cytochrome c contributes to the initiation of apoptosis is unknown, but it does not appear to involve direct effects on apoptosome formation or caspase-9 activation [200].
4.2 Redox modulation of apoptotic execution: control of caspase activity
Cellular caspases belong to a highly conserved family of cysteine proteases that cleave aspartate residues of caspase substrates and are the main players in the execution phase of apoptosis. The mammalian caspase family contains at least 14 members that are divided into initiator and executioner caspases [203]. Initiator or apical caspases are recruited at the death receptor via the death effector domain (caspases 8 and -10) or in the cytosol via the recruiting domain (caspases 2 and -9). Activated initiator caspases cleave executioner caspases such as caspases-3, and -7 which execute apoptosis through cleavage of protein substrates that include mediators and regulators of apoptosis, structural proteins, and DNA repair and cell-cycle related proteins [204]. Additionally, activated caspase-3 promotes caspases-2 and -6 activation in an amplification loop that enhances caspase-9 processing [205]. Caspases are constitutively expressed as monomers in the cytosol as inactive zymogen, and are activated by apoptotic signals such as ROS via proteolysis at internal sites [206]. Proteolytic cleavage of caspases at N-terminal prodomains results in the generation of small p10 and large p20 active subunits, forming active p10/p20 tetramers. The activation of caspases is prevented by specific inhibitors belonging to members of the IAP family; members like XIAP, c-IAP1, c-IAP2 and survivin bind and suppress enzyme catalytic activity [207]. During apoptotic signaling IAPs are antagonized by mitochondria-derived Smac/Diablo and Omi/HtrA2 proteins, allowing caspase-mediated execution [208, 209].
Our present understanding of the redox control of apoptotic execution is sketchy, but is a growing area of research. Post-translational modification of catalytic site cysteine residues has gained recognition as a potentially important redox mechanism in the control of caspase activity. Redox active catalytic site cysteines of caspases are prone to oxidation, nitrosation or glutathiolation (Figure 3) [203]. Direct ROS effects on caspase activation have been documented; for instance, H2O2 derived from endogenous and exogenous sources was shown to induce reversible inactivation of caspase 3 and caspase 8 through oxidation of their catalytic site cysteines [210]. For caspase-9, H2O2-induced enzyme inactivation was specifically mediated through iron-catalyzed oxidation of procaspase 9 catalytic site cysteine [211]. Additionally, H2O2-mediated redox-dependent intra-mitochondrial autoactivation of caspase-9 has been demonstrated in U937 cells in which procaspase-9 dimerization was induced by thiol-disulfide bond formation, a process that was inhibited by Trx [212]. Since mitochondrial procaspase-9 activation occurred during the pre-apoptotic phase prior to cytochrome c release, it was suggested that this mechanism could amplify the pro-apoptotic effect of cytochrome c [212].
Low levels of NO have been shown to exert anti-apoptotic effects via S-nitrosation of single cysteine residue at catalytic sites of caspases [213, 214]. To date, enzyme activities of seven members of the caspase family have been shown to be inhibited by redox-modulation through this mechanism [215]. Evidence that caspase-3 is nitrosated with resultant inhibition of enzyme activity comes from studies in human umbilical vein endothelial cells. Using electron spin resonance spectroscopy of Myc-tagged p17 (a subunit of caspase 3), Rossig et al found that S-nitrosation of cys-163 prevented caspase-3 mediated apoptotic cascade [216]. In hepatocytes, NO blocked Bid activation through S-nitrosation of caspase-8 and prevented TNFα-induced mitochondrial apoptotic signaling [217–219]. Additionally, NO donors were found to inhibit proper assembly of the Apaf-1/caspase-9 apoptosome complex and caspase-9 activation [217–219]. De-nitrosation is reportedly pro-apoptotic. For instance, de-nitrosation of procaspase-3 and procaspase-9 has been shown to be associated with proteolytic enzyme activation in Fas-mediated [220] and cytokine-induced apoptosis [221]. A role for Trx1 in procaspase-3 S-nitrosation has been documented, an interaction that involved transnitrosation reaction between the procaspase-3 and Trx1 [222]. The cellular sites of caspase nitrosation/denitrosation have not been fully investigated, but mitochondria are reportedly key locations for S-nitrosation reactions, judging by the preferred mitochondrial distribution of nitrosated caspases [223]. If this is the case, the intriguing question of how nitrosation/activation of matrix caspases mediates downstream cytosolic events in apoptotic execution warrants further study.
Apart from ROS and NO, other recent evidence implicates a direct role for cellular GSH in redox regulation of caspase activity, mediated by S-glutathiolation. Reportedly, S-glutathiolation of cysteine contributes to caspase stability and decreased accessibility for proteolytic cleavage, consistent with apoptotic resistance. Conversely, recent work of Pan and Berk demonstrated that de-glutathiolation of caspase-3 increased caspase-3 activity and TNFα-induced endothelial cell apoptosis [224]. Grx-catalyzed reversible glutathiolation of caspase-3 has been suggested to represent a novel redox signaling mechanism in TNFα-mediated cell apoptosis; specifically, TNFα-induced Grx assisted in thiol transfer in caspase-3 de-glutathiolation [225]. Recent findings in HL60 cells show that GSSG at physiological levels mediated cysteine glutathiolation of both caspase-3 subunits and inhibition of enzyme activity [226], linking S-glutathiolation with direct inhibition of caspase activity. Additional findings that procaspases-9 and -3 were targets of glutathiolation further suggests that proteolytic activation of caspases may also be under GSH redox control [226]. While caspases are subject to individual modification by nitrosation and glutathiolation, unanswered questions remain as to whether or how individual post-translational mechanisms interact and integrate with one another that collectively optimize caspase activities during cell apoptosis.
5. CONCLUDING REMARKS
Apoptosis has long been appreciated as an important form of cell death in biological processes and various pathologies. Our current understanding of the regulation of apoptosis is incomplete despite decades of research. The recognition that ROS play a central role in cell signaling has spurred much recent interest in the role of redox mechanisms in apoptotic signaling and control. Evidence to date has implicated redox-dependent mechanisms in mitochondria-to-cytosol release of cytochrome c, a central event in apoptotic initiation. Additionally, S-nitrosation and S-glutathiolation of catalytic site cysteines were reportedly important post-translational redox mechanisms in the reversible activation/inactivation of caspases in the control of apoptotic execution. Cellular redox systems, most notably, the GSH/GSSG redox couple, often functioning in conjunction with the thioredoxin system, are central in redox regulation and cell apoptosis. Less well understood is the contribution of the pyridine nucleotide couples of NAD+/NADH and NADP+/NADPH. Apart from their classical roles in bioenergetic homeostasis and reductive biosynthesis, respectively, recent evidence suggests that pyridine nucleotides have broader biological functions, including controlling cell death. At present, the precise contribution of pyridine nucleotides and the extent to which they interact with the thiol redox systems of GSH/GSSG and thioredoxin in redox regulation of apoptosis is unclear and should provide fruitful avenues for future research.
Acknowledgments
Research in the authors’ laboratory was supported by a grant from the National Institutes of Health, DK44510.
ABBREVIATION LIST
- AIF
apoptosis inducing factor
- ANT
adenine nucleotide translocase
- Apaf-1
apoptotic protease activation factor-1
- ASK-1
-2, apoptosis signal-regulating kinase 1, -2
- Bax
pro-apoptotic protein Bax
- Bcl-2
anti-apoptotic protein
- Bid
BH3-only pro-apoptotic protein Bid
- BIR
Baculovirus IAP repeat
- BSO
L-buthionine-S,R-sulfoximine
- CARD
caspase activation and recruiting domain
- caspase- 8
-9,-3, active form of caspase-8,-9,-3
- CL
cardiolipin
- CL-OOH
peroxidized cardiolipin
- cypD
cyclophylin D
- cys
cysteine
- cyt c
cytochrome c
- DD
death domain
- DIC
dicarboxylate carrier
- DISC
death-inducing signaling complex
- ER
endoplasmic reticulum
- FADD
Fas-associated death domain
- FasL
Fas ligand
- FLIPL
FLICE inhibitory protein
- FOXO
Forkhead box O
- G6PD
glucose-6-phosphate dehydrogenase
- GAPDH
glyceraldehyde-3-phospate dehydrogenase
- G6PDH
glucose-6-phosphate dehydrogenase
- 6PGlcDH
6-phosphogluconate dehydrogenase
- GCS
glutamate-cysteine syntethase
- Glu
glutamic acid
- Gly
glycine
- GPx
GSH peroxidase
- GR
GSH reductase
- GS
glutathione synthase
- GSH
glutathione
- GSSG
glutathione disulfide
- Grx
glutaredoxin
- GST
GSH-S-transferase
- HO•
hydroxyl radical
- H2O2
hydrogen peroxide
- IAP
inhibitors of apoptosis proteins
- ICDH
isocitrate dehydrogenase
- JNK
c-Jun N-terminal kinase
- ME
malic enzyme
- MnSOD
manganese superoxide dismutase
- MPT
mitochondrial permeability transition
- mtDNA
mitochondrial DNA
- mtGSH/GSSG
mitochondrial GSH/GSSG
- NAC
N-acetylcysteine
- NADH/NAD+
reduced and oxidized nicotinamide adenine dinucleotide
- NADPH/NADP+
reduced and oxidized nicotinamide adenine dinucleotide phosphate
- NNT
nicotine amide nucleotide transhydrogenase
- NADK
NAD kinase
- NO•
nitric oxide
- NO2•
nitrogen dioxide
- NOS
nitric oxid synthase
- Nox
NADPH oxidase
- OGC
oxo-glutarate carrier
- OGG1
8-oxodG glycosylase
- O2.−
superoxide anion
- OPA1
optic atrophya 1 protein
- Prx
peroxiredoxin
- PTP
permeability transition pore
- RIP1
receptor interacting protein 1
- ROS
reactive oxygen species
- RNO
reactive nitrogen species
- Sirts
sirtuin proteins
- Smac/Diablo
second mitochondria-derived activator of caspases/direct IAP binding protein with low pI
- tBH
tert-butyl hydroperoxide
- tBid
truncated form of Bid
- TRAF2/6
TNFα receptor-associated factor 2/6
- TRADD
TNFR-associated death domain
- TRAIL
TNF-related apoptosis-inducing ligand
- Trx1
thioredoxin 1, reduced form
- Trx2
thioredoxin 2, mitochondrial enzyme
- TRX-S-S
thioredoxin, oxidized form
- UPR
unfolding protein response
- VDAC
voltage-dependent anion channel
- XIAP
X-linked inhibitor of apoptosis protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Halliwell B, Cross CE. Oxygen-derived species: their relation to human disease and environmental stress. Environ Health Perspect. 1994;102(Suppl 10):5–12. doi: 10.1289/ehp.94102s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 2005;70:200–214. doi: 10.1007/s10541-005-0102-7. [DOI] [PubMed] [Google Scholar]
- 5.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migliaccio E, Giorgio M, Pelicci PG. Apoptosis and aging: role of p66Shc redox protein. Antioxid Redox Signal. 2006;8:600–608. doi: 10.1089/ars.2006.8.600. [DOI] [PubMed] [Google Scholar]
- 8.Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 9.Lambert AJ, Brand MD. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem J. 2004;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Circu ML, Moyer MP, Harrison L, Aw TY. Contribution of glutathione status to oxidant-induced mitochondrial DNA damage in colonic epithelial cells. Free Radic Biol Med. 2009;47:1190–1198. doi: 10.1016/j.freeradbiomed.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rachek LI, Yuzefovych LV, Ledoux SP, Julie NL, Wilson GL. Troglitazone, but not rosiglitazone, damages mitochondrial DNA and induces mitochondrial dysfunction and cell death in human hepatocytes. Toxicol Appl Pharmacol. 2009;240:348–354. doi: 10.1016/j.taap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- 13.Fritz R, Bol J, Hebling U, Angermuller S, Volkl A, Fahimi HD, Mueller S. Compartment-dependent management of H(2)O(2) by peroxisomes. Free Radic Biol Med. 2007;42:1119–1129. doi: 10.1016/j.freeradbiomed.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharmacol. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 15.Caro AA, Cederbaum AI. Role of cytochrome P450 in phospholipase A2- and arachidonic acid-mediated cytotoxicity. Free Radic Biol Med. 2006;40:364–375. doi: 10.1016/j.freeradbiomed.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 16.Dumitru CA, Zhang Y, Li X, Gulbins E. Ceramide: a novel player in reactive oxygen species-induced signaling? Antioxid Redox Signal. 2007;9:1535–1540. doi: 10.1089/ars.2007.1692. [DOI] [PubMed] [Google Scholar]
- 17.Zhang AY, Yi F, Zhang G, Gulbins E, Li PL. Lipid raft clustering and redox signaling platform formation in coronary arterial endothelial cells. Hypertension. 2006;47:74–80. doi: 10.1161/10.1161/01.HYP.0000196727.53300.62. [DOI] [PubMed] [Google Scholar]
- 18.Zhang AY, Yi F, Jin S, Xia M, Chen QZ, Gulbins E, Li PL. Acid sphingomyelinase and its redox amplification in formation of lipid raft redox signaling platforms in endothelial cells. Antioxid Redox Signal. 2007;9:817–828. doi: 10.1089/ars.2007.1509. [DOI] [PubMed] [Google Scholar]
- 19.Circu ML, Rodriguez C, Maloney R, Moyer MP, Aw TY. Contribution of mitochondrial GSH transport to matrix GSH status and colonic epithelial cell apoptosis. Free Radic Biol Med. 2008;44:768–778. doi: 10.1016/j.freeradbiomed.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekshyyan O, Aw TY. Decreased susceptibility of differentiated PC12 cells to oxidative challenge: relationship to cellular redox and expression of apoptotic protease activator factor-1. Cell Death Differ. 2005;12:1066–1077. doi: 10.1038/sj.cdd.4401650. [DOI] [PubMed] [Google Scholar]
- 21.Pias EK, Ekshyyan OY, Rhoads CA, Fuseler J, Harrison L, Aw TY. Differential effects of superoxide dismutase isoform expression on hydroperoxide-induced apoptosis in PC-12 cells. J Biol Chem. 2003;278:13294–13301. doi: 10.1074/jbc.M208670200. [DOI] [PubMed] [Google Scholar]
- 22.Okouchi M, Okayama N, Alexander JS, Aw TY. NRF2-dependent glutamate-L-cysteine ligase catalytic subunit expression mediates insulin protection against hyperglycemia- induced brain endothelial cell apoptosis. Curr Neurovasc Res. 2006;3:249–261. doi: 10.2174/156720206778792876. [DOI] [PubMed] [Google Scholar]
- 23.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 24.Meister A, Tate SS. Glutathione and related gamma-glutamyl compounds: biosynthesis and utilization. Annu Rev Biochem. 1976;45:559–604. doi: 10.1146/annurev.bi.45.070176.003015. [DOI] [PubMed] [Google Scholar]
- 25.Circu ML, Aw TY. Glutathione and apoptosis. Free Radic Res. 2008;42:689–706. doi: 10.1080/10715760802317663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 27.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessop CE, Bulleid NJ. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 29.Frand AR, Kaiser CA. Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2833–2843. doi: 10.1091/mbc.11.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon BM, Heath SH, Kim R, Suh JH, Hagen TM. Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid Redox Signal. 2008;10:963–972. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Delannoy M, Odwin S, He P, Trush MA, Yager JD. Enhanced mitochondrial gene transcript, ATP, bcl-2 protein levels, and altered glutathione distribution in ethinyl estradiol-treated cultured female rat hepatocytes. Toxicol Sci. 2003;75:271–278. doi: 10.1093/toxsci/kfg183. [DOI] [PubMed] [Google Scholar]
- 32.Markovic J, Borras C, Ortega A, Sastre J, Vina J, Pallardo FV. Glutathione is recruited into the nucleus in early phases of cell proliferation. J Biol Chem. 2007;282:20416–20424. doi: 10.1074/jbc.M609582200. [DOI] [PubMed] [Google Scholar]
- 33.Ho YF, Guenthner TM. Isolation of liver nuclei that retain functional trans-membrane transport. J Pharmacol Toxicol Methods. 1997;38:163–168. doi: 10.1016/s1056-8719(97)00082-8. [DOI] [PubMed] [Google Scholar]
- 34.Voehringer DW, McConkey DJ, McDonnell TJ, Brisbay S, Meyn RE. Bcl-2 expression causes redistribution of glutathione to the nucleus. Proc Natl Acad Sci U S A. 1998;95:2956–2960. doi: 10.1073/pnas.95.6.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jocelyn PC, Kamminga A. The non-protein thiol of rat liver mitochondria. Biochim Biophys Acta. 1974;343:356–362. doi: 10.1016/0304-4165(74)90099-3. [DOI] [PubMed] [Google Scholar]
- 36.Schnellmann RG. Renal mitochondrial glutathione transport. Life Sci. 1991;49:393–398. doi: 10.1016/0024-3205(91)90447-j. [DOI] [PubMed] [Google Scholar]
- 37.Soderdahl T, Enoksson M, Lundberg M, Holmgren A, Ottersen OP, Orrenius S, Bolcsfoldi G, Cotgreave IA. Visualization of the compartmentalization of glutathione and protein-glutathione mixed disulfides in cultured cells. Faseb J. 2003;17:124–126. doi: 10.1096/fj.02-0259fje. [DOI] [PubMed] [Google Scholar]
- 38.Zimmermann AK, Loucks FA, Schroeder EK, Bouchard RJ, Tyler KL, Linseman DA. Glutathione binding to the Bcl-2 homology-3 domain groove: a molecular basis for Bcl-2 antioxidant function at mitochondria. J Biol Chem. 2007;282:29296–29304. doi: 10.1074/jbc.M702853200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura H, Nakamura K, Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 40.Watson WH, Jones DP. Oxidation of nuclear thioredoxin during oxidative stress. FEBS Lett. 2003;543:144–147. doi: 10.1016/s0014-5793(03)00430-7. [DOI] [PubMed] [Google Scholar]
- 41.Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- 42.Hansen JM, Zhang H, Jones DP. Differential oxidation of thioredoxin-1, thioredoxin-2, and glutathione by metal ions. Free Radic Biol Med. 2006;40:138–145. doi: 10.1016/j.freeradbiomed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 46.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 48.Guse AH. Second messenger function and the structure-activity relationship of cyclic adenosine diphosphoribose (cADPR) Febs J. 2005;272:4590–4597. doi: 10.1111/j.1742-4658.2005.04863.x. [DOI] [PubMed] [Google Scholar]
- 49.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 50.Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol. 2001;41:317–345. doi: 10.1146/annurev.pharmtox.41.1.317. [DOI] [PubMed] [Google Scholar]
- 51.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 52.Bolanos JP, Delgado-Esteban M, Herrero-Mendez A, Fernandez-Fernandez S, Almeida A. Regulation of glycolysis and pentose-phosphate pathway by nitric oxide: impact on neuronal survival. Biochim Biophys Acta. 2008;1777:789–793. doi: 10.1016/j.bbabio.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 53.Scott MD, Zuo L, Lubin BH, Chiu DT. NADPH, not glutathione, status modulates oxidant sensitivity in normal and glucose-6-phosphate dehydrogenase-deficient erythrocytes. Blood. 1991;77:2059–2064. [PubMed] [Google Scholar]
- 54.Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. Embo J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HJ, Kang BS, Park JW. Cellular defense against heat shock-induced oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. Free Radic Res. 2005;39:441–448. doi: 10.1080/10715760500066265. [DOI] [PubMed] [Google Scholar]
- 56.Yoshihara T, Hamamoto T, Munakata R, Tajiri R, Ohsumi M, Yokota S. Localization of cytosolic NADP-dependent isocitrate dehydrogenase in the peroxisomes of rat liver cells: biochemical and immunocytochemical studies. J Histochem Cytochem. 2001;49:1123–1131. doi: 10.1177/002215540104900906. [DOI] [PubMed] [Google Scholar]
- 57.Kim SJ, Yune TY, Han CT, Kim YC, Oh YJ, Markelonis GJ, Oh TH. Mitochondrial isocitrate dehydrogenase protects human neuroblastoma SH-SY5Y cells against oxidative stress. J Neurosci Res. 2007;85:139–152. doi: 10.1002/jnr.21106. [DOI] [PubMed] [Google Scholar]
- 58.Shin SW, Kil IS, Park JW. Silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase by small interfering RNA enhances heat shock-induced apoptosis. Biochem Biophys Res Commun. 2008;366:1012–1018. doi: 10.1016/j.bbrc.2007.12.067. [DOI] [PubMed] [Google Scholar]
- 59.Kil IS, Kim SY, Lee SJ, Park JW. Small interfering RNA-mediated silencing of mitochondrial NADP+-dependent isocitrate dehydrogenase enhances the sensitivity of HeLa cells toward tumor necrosis factor-alpha and anticancer drugs. Free Radic Biol Med. 2007;43:1197–1207. doi: 10.1016/j.freeradbiomed.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 60.Filosa S, Fico A, Paglialunga F, Balestrieri M, Crooke A, Verde P, Abrescia P, Bautista JM, Martini G. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDonald MJ. Evidence for the malate aspartate shuttle in pancreatic islets. Arch Biochem Biophys. 1982;213:643–649. doi: 10.1016/0003-9861(82)90594-x. [DOI] [PubMed] [Google Scholar]
- 62.McKenna MC, Stevenson JH, Huang X, Tildon JT, Zielke CL, Hopkins IB. Mitochondrial malic enzyme activity is much higher in mitochondria from cortical synaptic terminals compared with mitochondria from primary cultures of cortical neurons or cerebellar granule cells. Neurochem Int. 2000;36:451–459. doi: 10.1016/s0197-0186(99)00148-5. [DOI] [PubMed] [Google Scholar]
- 63.Vogel R, Wiesinger H, Hamprecht B, Dringen R. The regeneration of reduced glutathione in rat forebrain mitochondria identifies metabolic pathways providing the NADPH required. Neurosci Lett. 1999;275:97–100. doi: 10.1016/s0304-3940(99)00748-x. [DOI] [PubMed] [Google Scholar]
- 64.Pollak N, Niere M, Ziegler M. NAD kinase levels control the NADPH concentration in human cells. J Biol Chem. 2007;282:33562–33571. doi: 10.1074/jbc.M704442200. [DOI] [PubMed] [Google Scholar]
- 65.Akella SS, Harris C. Developmental ontogeny of NAD+ kinase in the rat conceptus. Toxicol Appl Pharmacol. 2001;170:124–129. doi: 10.1006/taap.2000.9093. [DOI] [PubMed] [Google Scholar]
- 66.Singh R, Lemire J, Mailloux RJ, Appanna VD. A novel strategy involved anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS ONE. 2008;3:e2682. doi: 10.1371/journal.pone.0002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh R, Mailloux RJ, Puiseux-Dao S, Appanna VD. Oxidative stress evokes a metabolic adaptation that favors increased NADPH synthesis and decreased NADH production in Pseudomonas fluorescens. J Bacteriol. 2007;189:6665–6675. doi: 10.1128/JB.00555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 69.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 71.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 73.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 74.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 75.Smith BC, Hallows WC, Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isono M, Isshiki K, Uzu T, Kashiwagi A, Koya D. Silent information regulator 2 (SIRT1) attenuates oxidative stress-induced mesangial cell apoptosis via p53 deacetylation. Free Radic Biol Med. 2006;40:2175–2182. doi: 10.1016/j.freeradbiomed.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 77.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 78.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 79.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 80.Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol Cell Biol. 2003;23:8576–8585. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 83.Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Washida N, Tokuyama H, Hayashi K, Itoh H. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem Biophys Res Commun. 2008;372:51–56. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- 84.Chiribau CB, Cheng L, Cucoranu IC, Yu YS, Clempus RE, Sorescu D. FOXO3A regulates peroxiredoxin III expression in human cardiac fibroblasts. J Biol Chem. 2008;283:8211–8217. doi: 10.1074/jbc.M710610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 86.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase Sirt1 in prostate cancer: A target for prostate cancer management via its inhibition? J Biol Chem. 2008;284:3823–3832. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 89.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 90.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 92.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 93.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 94.Lombard DB, Schwer B, Alt FW, Mostoslavsky R. SIRT6 in DNA repair, metabolism and ageing. J Intern Med. 2008;263:128–141. doi: 10.1111/j.1365-2796.2007.01902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 96.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 98.LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 99.Berg D, Lehne M, Muller N, Siegmund D, Munkel S, Sebald W, Pfizenmaier K, Wajant H. Enforced covalent trimerization increases the activity of the TNF ligand family members TRAIL and CD95L. Cell Death Differ. 2007;14:2021–2034. doi: 10.1038/sj.cdd.4402213. [DOI] [PubMed] [Google Scholar]
- 100.Watanabe N, Kuriyama H, Sone H, Neda H, Yamauchi N, Maeda M, Niitsu Y. Continuous internalization of tumor necrosis factor receptors in a human myosarcoma cell line. J Biol Chem. 1988;263:10262–10266. [PubMed] [Google Scholar]
- 101.Lee KH, Feig C, Tchikov V, Schickel R, Hallas C, Schutze S, Peter ME, Chan AC. The role of receptor internalization in CD95 signaling. Embo J. 2006;25:1009–1023. doi: 10.1038/sj.emboj.7601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang X, Masselli A, Frisch SM, Hunton IC, Jiang Y, Wang JY. Blockade of tumor necrosis factor-induced Bid cleavage by caspase-resistant Rb. J Biol Chem. 2007;282:29401–29413. doi: 10.1074/jbc.M702261200. [DOI] [PubMed] [Google Scholar]
- 103.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 104.Wang L, Azad N, Kongkaneramit L, Chen F, Lu Y, Jiang BH, Rojanasakul Y. The Fas Death Signaling Pathway Connecting Reactive Oxygen Species Generation and FLICE Inhibitory Protein Down-Regulation. J Immunol. 2008;180:3072–3080. doi: 10.4049/jimmunol.180.5.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- 106.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 107.Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, Yamada G, Akira S, Matsumoto K, Ghosh S. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stehlik C, de Martin R, Kumabashiri I, Schmid JA, Binder BR, Lipp J. Nuclear factor (NF)-kappaB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J Exp Med. 1998;188:211–216. doi: 10.1084/jem.188.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 112.Pantano C, Shrivastava P, McElhinney B, Janssen-Heininger Y. Hydrogen peroxide signaling through tumor necrosis factor receptor 1 leads to selective activation of c-Jun N-terminal kinase. J Biol Chem. 2003;278:44091–44096. doi: 10.1074/jbc.M308487200. [DOI] [PubMed] [Google Scholar]
- 113.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwender M, Adam D, Mentlein R, Kabelitz D, Schutze S. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 114.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 115.Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.LeBlanc H, Lawrence D, Varfolomeev E, Totpal K, Morlan J, Schow P, Fong S, Schwall R, Sinicropi D, Ashkenazi A. Tumor-cell resistance to death receptor--induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat Med. 2002;8:274–281. doi: 10.1038/nm0302-274. [DOI] [PubMed] [Google Scholar]
- 117.Deng Y, Lin Y, Wu X. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 2002;16:33–45. doi: 10.1101/gad.949602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jin Z, El-Deiry WS. Distinct signaling pathways in TRAIL- versus tumor necrosis factor-induced apoptosis. Mol Cell Biol. 2006;26:8136–8148. doi: 10.1128/MCB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 120.Ryter SW, Kim HP, Hoetzel A, Park JW, Nakahira K, Wang X, Choi AM. Mechanisms of cell death in oxidative stress. Antioxid Redox Signal. 2007;9:49–89. doi: 10.1089/ars.2007.9.49. [DOI] [PubMed] [Google Scholar]
- 121.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 122.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, MacGregor GR, Wallace DC. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc Natl Acad Sci U S A. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kelekar A, Thompson CB. Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- 126.Eskes R, Desagher S, Antonsson B, Martinou JC. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol Cell Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou JC. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ott M, Norberg E, Walter KM, Schreiner P, Kemper C, Rapaport D, Zhivotovsky B, Orrenius S. The mitochondrial TOM complex is required for tBid/Bax-induced cytochrome c release. J Biol Chem. 2007;282:27633–27639. doi: 10.1074/jbc.M703155200. [DOI] [PubMed] [Google Scholar]
- 129.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. Embo J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song JJ, Rhee JG, Suntharalingam M, Walsh SA, Spitz DR, Lee YJ. Role of glutaredoxin in metabolic oxidative stress. Glutaredoxin as a sensor of oxidative stress mediated by H2O2. J Biol Chem. 2002;277:46566–46575. doi: 10.1074/jbc.M206826200. [DOI] [PubMed] [Google Scholar]
- 132.Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci U S A. 1999;96:8511–8515. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang H, Zhang R, Luo Y, D’Alessio A, Pober JS, Min W. AIP1/DAB2IP, a novel member of the Ras-GAP family, transduces TRAF2-induced ASK1-JNK activation. J Biol Chem. 2004;279:44955–44965. doi: 10.1074/jbc.M407617200. [DOI] [PubMed] [Google Scholar]
- 134.Chang HY, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 135.Matsuura H, Nishitoh H, Takeda K, Matsuzawa A, Amagasa T, Ito M, Yoshioka K, Ichijo H. Phosphorylation-dependent scaffolding role of JSAP1/JIP3 in the ASK1-JNK signaling pathway. A new mode of regulation of the MAP kinase cascade. J Biol Chem. 2002;277:40703–40709. doi: 10.1074/jbc.M202004200. [DOI] [PubMed] [Google Scholar]
- 136.Matsuzawa A, Ichijo H. Redox control of cell fate by MAP kinase: physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochim Biophys Acta. 2008:1708. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 137.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 138.Liu H, Lo CR, Czaja MJ. NF-kappaB inhibition sensitizes hepatocytes to TNF-induced apoptosis through a sustained activation of JNK and c-Jun. Hepatology. 2002;35:772–778. doi: 10.1053/jhep.2002.32534. [DOI] [PubMed] [Google Scholar]
- 139.Deng Y, Ren X, Yang L, Lin Y, Wu X. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell. 2003;115:61–70. doi: 10.1016/s0092-8674(03)00757-8. [DOI] [PubMed] [Google Scholar]
- 140.Fujino G, Noguchi T, Takeda K, Ichijo H. Thioredoxin and protein kinases in redox signaling. Semin Cancer Biol. 2006;16:427–435. doi: 10.1016/j.semcancer.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 141.Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27:8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang XS, Diener K, Tan TH, Yao Z. MAPKKK6, a novel mitogen-activated protein kinase kinase kinase, that associates with MAPKKK5. Biochem Biophys Res Commun. 1998;253:33–37. doi: 10.1006/bbrc.1998.9749. [DOI] [PubMed] [Google Scholar]
- 143.Ortner E, Moelling K. Heteromeric complex formation of ASK2 and ASK1 regulates stress-induced signaling. Biochem Biophys Res Commun. 2007;362:454–459. doi: 10.1016/j.bbrc.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 144.Goldman EH, Chen L, Fu H. Activation of apoptosis signal-regulating kinase 1 by reactive oxygen species through dephosphorylation at serine 967 and 14-3-3 dissociation. J Biol Chem. 2004;279:10442–10449. doi: 10.1074/jbc.M311129200. [DOI] [PubMed] [Google Scholar]
- 145.Chen L, Liu L, Huang S. Cadmium activates the mitogen-activated protein kinase (MAPK) pathway via induction of reactive oxygen species and inhibition of protein phosphatases 2A and 5. Free Radic Biol Med. 2008;45:1035–1044. doi: 10.1016/j.freeradbiomed.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 146.Zhang R, Al-Lamki R, Bai L, Streb JW, Miano JM, Bradley J, Min W. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res. 2004;94:1483–1491. doi: 10.1161/01.RES.0000130525.37646.a7. [DOI] [PubMed] [Google Scholar]
- 147.Fan M, Goodwin ME, Birrer MJ, Chambers TC. The c-Jun NH(2)-terminal protein kinase/AP-1 pathway is required for efficient apoptosis induced by vinblastine. Cancer Res. 2001;61:4450–4458. [PubMed] [Google Scholar]
- 148.Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, Cheng K, Chen YN, Campbell A, Sudha T, Yuan ZM, Narula J, Weichselbaum R, Nalin C, Kufe D. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J Biol Chem. 2000;275:322–327. doi: 10.1074/jbc.275.1.322. [DOI] [PubMed] [Google Scholar]
- 149.Merad-Boudia M, Nicole A, Santiard-Baron D, Saille C, Ceballos-Picot I. Mitochondrial impairment as an early event in the process of apoptosis induced by glutathione depletion in neuronal cells: relevance to Parkinson’s disease. Biochem Pharmacol. 1998;56:645–655. doi: 10.1016/s0006-2952(97)00647-3. [DOI] [PubMed] [Google Scholar]
- 150.Lu C, Armstrong JS. Role of calcium and cyclophilin D in the regulation of mitochondrial permeabilization induced by glutathione depletion. Biochem Biophys Res Commun. 2007;363:572–577. doi: 10.1016/j.bbrc.2007.08.196. [DOI] [PubMed] [Google Scholar]
- 151.D’Alessio M, Cerella C, De Nicola M, Bergamaschi A, Magrini A, Gualandi G, Alfonsi AM, Ghibelli L. Apoptotic GSH extrusion is associated with free radical generation. Ann N Y Acad Sci. 2003;1010:449–452. doi: 10.1196/annals.1299.082. [DOI] [PubMed] [Google Scholar]
- 152.Marengo B, De Ciucis C, Verzola D, Pistoia V, Raffaghello L, Patriarca S, Balbis E, Traverso N, Cottalasso D, Pronzato MA, Marinari UM, Domenicotti C. Mechanisms of BSO (L-buthionine-S,R-sulfoximine)-induced cytotoxic effects in neuroblastoma. Free Radic Biol Med. 2008;44:474–482. doi: 10.1016/j.freeradbiomed.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 153.Meurette O, Lefeuvre-Orfila L, Rebillard A, Lagadic-Gossmann D, Dimanche-Boitrel MT. Role of intracellular glutathione in cell sensitivity to the apoptosis induced by tumor necrosis factor {alpha}-related apoptosis-inducing ligand/anticancer drug combinations. Clin Cancer Res. 2005;11:3075–3083. doi: 10.1158/1078-0432.CCR-04-1764. [DOI] [PubMed] [Google Scholar]
- 154.Franco R, Panayiotidis MI, Cidlowski JA. Glutathione depletion is necessary for apoptosis in lymphoid cells independent of reactive oxygen species formation. J Biol Chem. 2007;282:30452–30465. doi: 10.1074/jbc.M703091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Circu ML, Stringer S, Rhoads CA, Moyer MP, Aw TY. The role of GSH efflux in staurosporine-induced apoptosis in colonic epithelial cells. Biochem Pharmacol. 2009;77:76–85. doi: 10.1016/j.bcp.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang TG, Gotoh Y, Jennings MH, Rhoads CA, Aw TY. Lipid hydroperoxide-induced apoptosis in human colonic CaCo-2 cells is associated with an early loss of cellular redox balance. Faseb J. 2000;14:1567–1576. doi: 10.1096/fj.14.11.1567. [DOI] [PubMed] [Google Scholar]
- 157.Pias EK, Aw TY. Early redox imbalance mediates hydroperoxide-induced apoptosis in mitotic competent undifferentiated PC-12 cells. Cell Death Differ. 2002;9:1007–1016. doi: 10.1038/sj.cdd.4401064. [DOI] [PubMed] [Google Scholar]
- 158.Pias EK, Aw TY. Apoptosis in mitotic competent undifferentiated cells is induced by cellular redox imbalance independent of reactive oxygen species production. Faseb J. 2002;16:781–790. doi: 10.1096/fj.01-0784com. [DOI] [PubMed] [Google Scholar]
- 159.Okouchi M, Okayama N, Aw TY. Hyperglycemia potentiates carbonyl stress-induced apoptosis in naive PC-12 cells: relationship to cellular redox and activator protease factor-1 expression. Curr Neurovasc Res. 2005;2:375–386. doi: 10.2174/156720205774962665. [DOI] [PubMed] [Google Scholar]
- 160.Okouchi M, Okayama N, Aw TY. Differential susceptibility of naive and differentiated PC-12 cells to methylglyoxal-induced apoptosis: influence of cellular redox. Curr Neurovasc Res. 2005;2:13–22. doi: 10.2174/1567202052773535. [DOI] [PubMed] [Google Scholar]
- 161.Hallberg E, Rydstrom J. Selective oxidation of mitochondrial glutathione in cultured rat adrenal cells and its relation to polycyclic aromatic hydrocarbon-induced cytotoxicity. Arch Biochem Biophys. 1989;270:662–671. doi: 10.1016/0003-9861(89)90549-3. [DOI] [PubMed] [Google Scholar]
- 162.Lluis JM, Morales A, Blasco C, Colell A, Mari M, Garcia-Ruiz C, Fernandez-Checa JC. Critical role of mitochondrial glutathione in the survival of hepatocytes during hypoxia. J Biol Chem. 2005;280:3224–3232. doi: 10.1074/jbc.M408244200. [DOI] [PubMed] [Google Scholar]
- 163.Olafsdottir K, Reed DJ. Retention of oxidized glutathione by isolated rat liver mitochondria during hydroperoxide treatment. Biochim Biophys Acta. 1988;964:377–382. doi: 10.1016/0304-4165(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 164.Fernandez-Checa JC, Garcia-Ruiz C, Ookhtens M, Kaplowitz N. Impaired uptake of glutathione by hepatic mitochondria from chronic ethanol-fed rats. Tracer kinetic studies in vitro and in vivo and susceptibility to oxidant stress. J Clin Invest. 1991;87:397–405. doi: 10.1172/JCI115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Garcia-Ruiz C, Fernandez-Checa JC. Mitochondrial glutathione: hepatocellular survival-death switch. J Gastroenterol Hepatol. 2006;21(Suppl 3):S3–6. doi: 10.1111/j.1440-1746.2006.04570.x. [DOI] [PubMed] [Google Scholar]
- 166.Lluis JM, Colell A, Garcia-Ruiz C, Kaplowitz N, Fernandez-Checa JC. Acetaldehyde impairs mitochondrial glutathione transport in HepG2 cells through endoplasmic reticulum stress. Gastroenterology. 2003;124:708–724. doi: 10.1053/gast.2003.50089. [DOI] [PubMed] [Google Scholar]
- 167.Lluis JM, Buricchi F, Chiarugi P, Morales A, Fernandez-Checa JC. Dual Role of Mitochondrial Reactive Oxygen Species in Hypoxia Signaling: Activation of Nuclear Factor-{kappa}B via c-SRC and Oxidant-Dependent Cell Death. Cancer Res. 2007;67:7368–7377. doi: 10.1158/0008-5472.CAN-07-0515. [DOI] [PubMed] [Google Scholar]
- 168.Ghosh S, Pulinilkunnil T, Yuen G, Kewalramani G, An D, Qi D, Abrahani A, Rodrigues B. Cardiomyocyte apoptosis induced by short-term diabetes requires mitochondrial GSH depletion. Am J Physiol Heart Circ Physiol. 2005;289:H768–776. doi: 10.1152/ajpheart.00038.2005. [DOI] [PubMed] [Google Scholar]
- 169.Tsujimoto Y, Shimizu S. Role of the mitochondrial membrane permeability transition in cell death. Apoptosis. 2007;12:835–840. doi: 10.1007/s10495-006-0525-7. [DOI] [PubMed] [Google Scholar]
- 170.Madesh M, Hajnoczky G. VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release. J Cell Biol. 2001;155:1003–1015. doi: 10.1083/jcb.200105057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Giron-Calle J, Zwizinski CW, Schmid HH. Peroxidative damage to cardiac mitochondria. II. Immunological analysis of modified adenine nucleotide translocase. Arch Biochem Biophys. 1994;315:1–7. doi: 10.1006/abbi.1994.1463. [DOI] [PubMed] [Google Scholar]
- 172.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 173.Skulachev VP. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis. 2006;11:473–485. doi: 10.1007/s10495-006-5881-9. [DOI] [PubMed] [Google Scholar]
- 174.Cook SA, Sugden PH, Clerk A. Regulation of bcl-2 family proteins during development and in response to oxidative stress in cardiac myocytes: association with changes in mitochondrial membrane potential. Circ Res. 1999;85:940–949. doi: 10.1161/01.res.85.10.940. [DOI] [PubMed] [Google Scholar]
- 175.Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 176.Oh SH, Lim SC. A rapid and transient ROS generation by cadmium triggers apoptosis via caspase-dependent pathway in HepG2 cells and this is inhibited through N-acetylcysteine-mediated catalase upregulation. Toxicol Appl Pharmacol. 2006;212:212–223. doi: 10.1016/j.taap.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 177.Chen Q, Lesnefsky EJ. Depletion of cardiolipin and cytochrome c during ischemia increases hydrogen peroxide production from the electron transport chain. Free Radic Biol Med. 2006;40:976–982. doi: 10.1016/j.freeradbiomed.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 178.Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 179.Lenaz G, Bovina C, D’Aurelio M, Fato R, Formiggini G, Genova ML, Giuliano G, Merlo Pich M, Paolucci U, Parenti Castelli G, Ventura B. Role of mitochondria in oxidative stress and aging. Ann N Y Acad Sci. 2002;959:199–213. doi: 10.1111/j.1749-6632.2002.tb02094.x. [DOI] [PubMed] [Google Scholar]
- 180.Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 181.Grishko V, Pastukh V, Solodushko V, Gillespie M, Azuma J, Schaffer S. Apoptotic cascade initiated by angiotensin II in neonatal cardiomyocytes: role of DNA damage. Am J Physiol Heart Circ Physiol. 2003;285:H2364–2372. doi: 10.1152/ajpheart.00408.2003. [DOI] [PubMed] [Google Scholar]
- 182.Circu MLHL, Aw YY. Menadione-induced mitochondrial DNA damage in colonic cells: influence of cellular GSH redox status and mitochondrial base excision repair (BER) capacity. In: Med FRB, editor. SFRBM’s 14-th Annual Meeting; Washington DC: 2007. p. S148. [Google Scholar]
- 183.Will O, Mahler HC, Arrigo AP, Epe B. Influence of glutathione levels and heat-shock on the steady-state levels of oxidative DNA base modifications in mammalian cells. Carcinogenesis. 1999;20:333–337. doi: 10.1093/carcin/20.2.333. [DOI] [PubMed] [Google Scholar]
- 184.Reliene R, Schiestl RH. Glutathione depletion by buthionine sulfoximine induces DNA deletions in mice. Carcinogenesis. 2006;27:240–244. doi: 10.1093/carcin/bgi222. [DOI] [PubMed] [Google Scholar]
- 185.Suliman HB, Carraway MS, Velsor LW, Day BJ, Ghio AJ, Piantadosi CA. Rapid mtDNA deletion by oxidants in rat liver mitochondria after hemin exposure. Free Radic Biol Med. 2002;32:246–256. doi: 10.1016/s0891-5849(01)00797-3. [DOI] [PubMed] [Google Scholar]
- 186.Esteve JM, Mompo J, Garcia de la Asuncion J, Sastre J, Asensi M, Boix J, Vina JR, Vina J, Pallardo FV. Oxidative damage to mitochondrial DNA and glutathione oxidation in apoptosis: studies in vivo and in vitro. Faseb J. 1999;13:1055–1064. doi: 10.1096/fasebj.13.9.1055. [DOI] [PubMed] [Google Scholar]
- 187.Hollins DL, Suliman HB, Piantadosi CA, Carraway MS. Glutathione regulates susceptibility to oxidant-induced mitochondrial DNA damage in human lymphocytes. Free Radic Biol Med. 2006;40:1220–1226. doi: 10.1016/j.freeradbiomed.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 188.de la Asuncion JG, Millan A, Pla R, Bruseghini L, Esteras A, Pallardo FV, Sastre J, Vina J. Mitochondrial glutathione oxidation correlates with age-associated oxidative damage to mitochondrial DNA. Faseb J. 1996;10:333–338. doi: 10.1096/fasebj.10.2.8641567. [DOI] [PubMed] [Google Scholar]
- 189.Dantzer F, Luna L, Bjoras M, Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic acids research. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yamamori T, Dericco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic acids research. 2009 doi: 10.1093/nar/gkp1039. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc Natl Acad Sci U S A. 2002;99:1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 193.Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, Fujii Y. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic Biol Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 194.Dejean LM, Martinez-Caballero S, Kinnally KW. Is MAC the knife that cuts cytochrome c from mitochondria during apoptosis? Cell Death Differ. 2006;13:1387–1395. doi: 10.1038/sj.cdd.4401949. [DOI] [PubMed] [Google Scholar]
- 195.Gonzalvez F, Pariselli F, Dupaigne P, Budihardjo I, Lutter M, Antonsson B, Diolez P, Manon S, Martinou JC, Goubern M, Wang X, Bernard S, Petit PX. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 2005;12:614–626. doi: 10.1038/sj.cdd.4401571. [DOI] [PubMed] [Google Scholar]
- 196.Vlasova II, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- 197.Jang B, Han S. Biochemical properties of cytochrome c nitrated by peroxynitrite. Biochimie. 2006;88:53–58. doi: 10.1016/j.biochi.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 198.Cassina AM, Hodara R, Souza JM, Thomson L, Castro L, Ischiropoulos H, Freeman BA, Radi R. Cytochrome c nitration by peroxynitrite. J Biol Chem. 2000;275:21409–21415. doi: 10.1074/jbc.M909978199. [DOI] [PubMed] [Google Scholar]
- 199.Nakagawa H, Komai N, Takusagawa M, Miura Y, Toda T, Miyata N, Ozawa T, Ikota N. Nitration of specific tyrosine residues of cytochrome C is associated with caspase-cascade inactivation. Biol Pharm Bull. 2007;30:15–20. doi: 10.1248/bpb.30.15. [DOI] [PubMed] [Google Scholar]
- 200.Brown GC, Borutaite V. Regulation of apoptosis by the redox state of cytochrome c. Biochim Biophys Acta. 2008;1777:877–881. doi: 10.1016/j.bbabio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 201.Suto D, Sato K, Ohba Y, Yoshimura T, Fujii J. Suppression of the pro-apoptotic function of cytochrome c by singlet oxygen via a haem redox state-independent mechanism. Biochem J. 2005;392:399–406. doi: 10.1042/BJ20050580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Borutaite V, Brown GC. Mitochondrial regulation of caspase activation by cytochrome oxidase and tetramethylphenylenediamine via cytosolic cytochrome c redox state. J Biol Chem. 2007;282:31124–31130. doi: 10.1074/jbc.M700322200. [DOI] [PubMed] [Google Scholar]
- 203.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 204.Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 205.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Salvesen GS, Abrams JM. Caspase activation - stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 207.Verhagen AM, Coulson EJ, Vaux DL. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2001;2:REVIEWS3009. doi: 10.1186/gb-2001-2-7-reviews3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 209.Suzuki Y, Takahashi-Niki K, Akagi T, Hashikawa T, Takahashi R. Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways. Cell Death Differ. 2004;11:208–216. doi: 10.1038/sj.cdd.4401343. [DOI] [PubMed] [Google Scholar]
- 210.Borutaite V, Brown GC. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Lett. 2001;500:114–118. doi: 10.1016/s0014-5793(01)02593-5. [DOI] [PubMed] [Google Scholar]
- 211.Barbouti A, Amorgianiotis C, Kolettas E, Kanavaros P, Galaris D. Hydrogen peroxide inhibits caspase-dependent apoptosis by inactivating procaspase-9 in an iron-dependent manner. Free Radic Biol Med. 2007;43:1377–1387. doi: 10.1016/j.freeradbiomed.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 212.Katoh I, Tomimori Y, Ikawa Y, Kurata S. Dimerization and processing of procaspase-9 by redox stress in mitochondria. J Biol Chem. 2004;279:15515–15523. doi: 10.1074/jbc.M311819200. [DOI] [PubMed] [Google Scholar]
- 213.Chung HT, Pae HO, Choi BM, Billiar TR, Kim YM. Nitric oxide as a bioregulator of apoptosis. Biochem Biophys Res Commun. 2001;282:1075–1079. doi: 10.1006/bbrc.2001.4670. [DOI] [PubMed] [Google Scholar]
- 214.Kim KM, Kim PK, Kwon YG, Bai SK, Nam WD, Kim YM. Regulation of apoptosis by nitrosative stress. J Biochem Mol Biol. 2002;35:127–133. doi: 10.5483/bmbrep.2002.35.1.127. [DOI] [PubMed] [Google Scholar]
- 215.Li J, Billiar TR, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 216.Rossig L, Fichtlscherer B, Breitschopf K, Haendeler J, Zeiher AM, Mulsch A, Dimmeler S. Nitric oxide inhibits caspase-3 by S-nitrosation in vivo. J Biol Chem. 1999;274:6823–6826. doi: 10.1074/jbc.274.11.6823. [DOI] [PubMed] [Google Scholar]
- 217.Kim YM, Kim TH, Chung HT, Talanian RV, Yin XM, Billiar TR. Nitric oxide prevents tumor necrosis factor alpha-induced rat hepatocyte apoptosis by the interruption of mitochondrial apoptotic signaling through S-nitrosylation of caspase-8. Hepatology. 2000;32:770–778. doi: 10.1053/jhep.2000.18291. [DOI] [PubMed] [Google Scholar]
- 218.Tzeng E, Kim YM, Pitt BR, Lizonova A, Kovesdi I, Billiar TR. Adenoviral transfer of the inducible nitric oxide synthase gene blocks endothelial cell apoptosis. Surgery. 1997;122:255–263. doi: 10.1016/s0039-6060(97)90016-7. [DOI] [PubMed] [Google Scholar]
- 219.Zech B, Kohl R, von Knethen A, Brune B. Nitric oxide donors inhibit formation of the Apaf-1/caspase-9 apoptosome and activation of caspases. Biochem J. 2003;371:1055–1064. doi: 10.1042/BJ20021720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 221.Kim JE, Tannenbaum SR. S-Nitrosation regulates the activation of endogenous procaspase-9 in HT-29 human colon carcinoma cells. J Biol Chem. 2004;279:9758–9764. doi: 10.1074/jbc.M312722200. [DOI] [PubMed] [Google Scholar]
- 222.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci U S A. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 225.Sykes MC, Mowbray AL, Jo H. Reversible glutathiolation of caspase-3 by glutaredoxin as a novel redox signaling mechanism in tumor necrosis factor-alpha-induced cell death. Circ Res. 2007;100:152–154. doi: 10.1161/01.RES.0000258171.08020.72. [DOI] [PubMed] [Google Scholar]
- 226.Huang Z, Pinto JT, Deng H, Richie JP., Jr Inhibition of caspase-3 activity and activation by protein glutathionylation. Biochem Pharmacol. 2008;75:2234–2244. doi: 10.1016/j.bcp.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]