Summary
AF4 and ENL family proteins are frequently fused with MLL, and comprise a higher order complex (designated AEP) containing the P-TEFb transcription elongation factor. Here, we show that AEP is normally recruited to MLL-target chromatin to facilitate transcription. By contrast, MLL oncoproteins fused with AEP components constitutively form MLL/AEP hybrid complexes to cause sustained target gene expression, which leads to transformation of hematopoietic progenitors. Furthermore, MLL-AF6, an MLL fusion with a cytoplasmic protein, does not form such hybrid complexes, but nevertheless constitutively recruits AEP to target chromatin via unknown alternative mechanisms. Thus, AEP recruitment is an integral part of both physiological and pathological MLL-dependent transcriptional pathways. Bypass of its normal recruitment mechanisms is the strategy most frequently employed by MLL oncoproteins.
Keywords: AF4, MLL, ENL, P-TEFb, DOT1L, leukemia
Introduction
Leukemia is a heterogeneous disease with distinctive biological and clinical properties that are conferred by a variety of acquired genetic mutations (Gilliland et al. 2002). Chromosomal translocations of the MLL gene account for 5-10% of acute leukemias and are generally associated with poor prognosis (Daser and Rabbitts, 2004; Krivtsov and Armstrong, 2007; Pui et al., 2004). MLL gene rearrangements create fusion genes that contain the 5′ portion of MLL and the 3′ portion of its fusion partner, whose products cause sustained expression of MLL target genes and consequent enhanced proliferation of hematopoietic progenitors (Ayton and Cleary, 2003; Lavau et al., 1997; Cozzio et al., 2003). The amino-terminal portion of MLL serves as a targeting unit to direct MLL oncoprotein complexes to their target loci through DNA binding (Ayton et al., 2004; Slany et al., 1998) and association with menin and LEDGF (Yokoyama et al., 2005; Yokoyama and Cleary, 2008), while the fusion partner portion serves as an effecter unit that causes sustained transactivation (Cheung et al., 2007; Lavau et al., 2000; DiMartino et al., 2000; 2002; Slany et al., 1998; So and Cleary 2002; 2003). To date, approximately 50 different fusion partners have been reported to form chimeric MLL oncoproteins (Huret et al., 2001). However, the mechanisms underlying this molecular diversity have not been revealed.
The AF4 and ENL protein families are the most frequent MLL fusion partners, accounting for two-thirds of MLL-associated leukemia incidence (Huret et al., 2001). The AF4 family is comprised of four paralogous proteins including AF4, AF5q31, LAF4 and FMR2. The ENL family includes ENL and AF9 and has structural homology to the yeast Anc1 protein. The members of both protein families possess transactivation domains and therefore are thought to be involved in transcriptional regulation (Prasad et al., 1995; Ma and Staudt, 1996; Morrissey et al., 1997; Slany et al., 1998). All but FMR2 have been reported to form fusion genes with MLL in leukemia (Domer et al., 1993; Taki et al., 1999; von Bergh et al., 2002; Iida et al., 1993; Nakamura et al., 1993; Tkachuk et al., 1992). AF4 family proteins associate with ENL family proteins and P-TEFb (Positive Transcription Elongation Factor b) (Erfurth et al., 2004; Zeisig et al., 2005; Bitoun et al., 2007; Mueller et al., 2007). P-TEFb is composed of CDK9 and cyclin T1 (or cyclin T2) and capable of phosphorylating the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII) and DSIF to facilitate transcriptional elongation (Saunders et al. 2006; Peterlin and Price, 2006). AF4 functions as a positive regulator of P-TEFb kinase (Bitoun et al., 2007), which in turn controls the transactivation activity and/or stability of AF4 and ENL family proteins. ENL family proteins also associate with DOT1L (Bitoun et al., 2007; Mueller et al., 2007; Zhang et al., 2006), the major histone methyltransferase responsible for the H3K79 methylation mark (Jones et al. 2008), which is predominantly associated with actively transcribed genes (Steger et al. 2008). It has been reported that DOT1L also associates with MLL-AF10 and plays a critical role in its oncogenic transformation (Okada et al., 2005). However, the molecular roles of these components in MLL-dependent leukemogenesis have not been clearly defined.
In this study we investigated the contributions of a higher order complex containing AF4- and ENL-family proteins with P-TEFb in physiologic and pathologic MLL-dependent transcription.
Results
AF4 forms a higher order complex with AF5q31, ENL and P-TEFb in hematopoietic cells
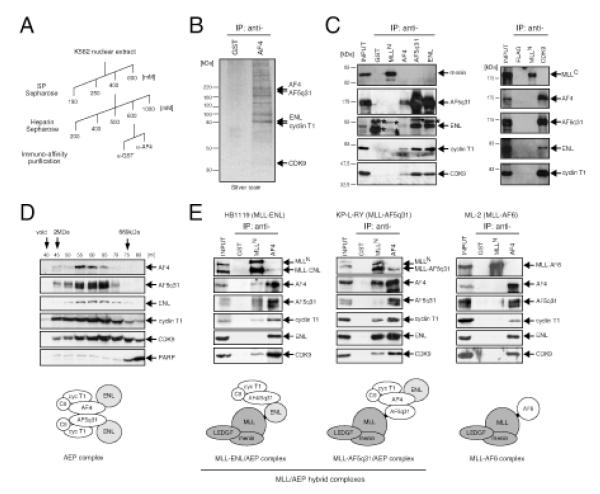
To identify AF4-associated proteins in vivo, we biochemically purified AF4 complexes from K562 cells using column chromatography followed by an immuno-affinity purification with a highly specific anti-AF4 monoclonal antibody (Figure 1A). Mass spectrometry identified AF5q31, ENL, CDK9 and cyclin T1 in the purified materials (Figure 1B). Reciprocal immunoprecipitation (IP) further confirmed that all five proteins comprise an endogenous bona fide complex (Figure 1C) consistent with previous observations (Erfurth et al., 2004, Zeisig et al., 2005; Bitoun et al., 2007; Mueller et al., 2007). In gel filtration analysis, the AF4 complex components co-distributed in fractions that eluted at an average mass of ~0.8 MDa (Figure 1D). A similar complex was obtained using a monoclonal antibody specific for AF5q31 in the immuno-affinity step (Figure S1A). However, neither purification yielded other proteins previously reported to interact with ENL (e.g. DOT1L, AF10) (Zeisig et al., 2005; Bitoun et al., 2007; Mueller et al., 2007). These data demonstrate that AF4, AF5q31 and ENL associate in an endogenous higher-order complex (hereafter referred to as AEP for the AF4 family/ENL family/P-TEFb complex) containing P-TEFb in hematopoietic lineage cells.
Figure 1.
Heterologous associations of wild type and oncogenic AF4 and ENL family proteins
(A) The scheme employed for purification of the AF4 complex.
(B) A silver-stained image shows the proteins immuno-purified using anti-AF4 antibody, and subsequently identified by mass spectrometry as indicated by arrows on the right. Anti-GST antibody served as a negative control.
(C) K562 nuclear extracts were analyzed by IP western blotting. IP was performed with the antibodies indicated on the top and the precipitates were immunoblotted with the antibodies indicated on the right. Anti-GST and anti-FLAG antibodies served as negative controls. Asterisks indicate signals from IgG used for IP.
(D) Selected fractions from gel filtration analysis of K562 nuclear extracts were analyzed by western blotting for AF4-associated factors (PARP served as a negative control). Molecular weight standards are shown on the top. A cartoon of a putative AEP complex is depicted. C9, CDK9; cyc T1, cyclin T1.
(E) IP western blot analysis was performed as in (C) on human leukemia cell lines that harbor MLL chromosomal translocations and express MLL chimeric oncoproteins (indicated at tops). Cartoons of putative MLL fusion complexes are depicted below.
See also Figure S1.
Leukemogenic fusion proteins inappropriately tether AEP components with MLL
Co-IP analyses were performed to determine whether MLL chimeric oncoproteins participate in higher-order associations that recapitulate the composition of AEP. Reciprocal IP using human leukemia cell lines that express MLL-ENL, MLL-AF4 or MLL-AF5q31 showed that the respective fusion proteins form similar AEP-like complexes (Figures 1E and S1B). Conversely, MLL-AF6, an MLL fusion with a cytoplasmic protein that was not co-purified with AF4 or AF5q31, did not co-precipitate any of the AEP components in ML-2 cells (Figure 1E). Similarly, wild type (wt) MLL did not pull down AEP components in K562 cells while co-precipitating menin, a component of the MLL complex (Yokoyama et al., 2004) (Figure 1C). Therefore, the MLL and AEP complexes are separate biochemical entities that are inappropriately tethered to form MLL/AEP hybrid complexes by a subset of covalent fusions of MLL in human leukemia cells.
MLL-ENL and MLL-AF4 consistently recruit AEP components to MLL target genes
Genomic localizations of MLL chimeric proteins and AEP components were analyzed by chromatin immunoprecipitation (ChIP) in human leukemia cell lines. Histone marks indicative of open chromatin states (tri-methyl H3K4 and acetyl H3K9) (Li et al., 2007) were associated with transcriptionally active loci, while histone marks indicative of closed chromatin (di-methyl H3K9 and high levels of histone H3) were associated with transcriptionally inactive loci (Figures 2A-2C), verifying the integrity of ChIP assays. In HB1119 cells, MLL-ENL specifically co-localized with AF4 and AF5q31 at promoter-adjacent regions of the HOXA9 and MEIS1 genes, which are known to serve critical roles in MLL-associated leukemogenesis (Ayton and Cleary, 2003; Nakamura et al., 1996; Wong et al., 2007), whereas the presence of AEP at non-MLL target loci such as β-ACTIN and GAPDH was minimal or negligible (Figures 2B and S2A). Similarly, ChIP analysis showed that AF5q31 and ENL co-localized with endogenous MLL-AF4 on the HOXA9 and MEIS1 promoters in MV4-11 cells (Figures 2C and S2B). Co-localization of AEP components with MLL oncoproteins was also observed on other MLL target genes such as CDKN1B and CDKN2C (Milne et al., 2005) and the transcribed regions of HOXA9 and MEIS1 (Figures 2B and 2C), suggesting that MLL/AEP hybrid complexes may function in transcriptional elongation. Therefore, a subset of MLL oncoproteins results in consistent recruitment of AEP components at MLL target chromatin in leukemia cells.
Figure 2.
Co-localization of MLL fusion proteins and AEP components on chromatin
(A) Relative expression of various genes (indicated on the right) in seven human cell lines was analyzed by quantitative RT-PCR. Expression levels were normalized to GAPDH and depicted relative to the highest value among the seven cell lines arbitrarily set as 100. Error bars represent standard deviations of triplicate PCRs.
(B) Genomic localizations of various proteins in HB1119 cells were determined by ChIP assay. Cross-linked chromatin was immunoprecipitated with antibodies specific for the indicated proteins and analyzed by quantitative PCR using primer/probe sets that target promoter-adjacent regions or other genomic regions indicated at the bottom. Occupancies are displayed relative to the highest value in the group arbitrarily set as 100. Error bars represent standard deviations of triplicate PCRs. Genes expressed more than 20% of the highest levels in panel A are defined as active genes.
(C) A comparable analysis as (B) was performed for MV4-11 cells, which harbor a t(4;11) translocation and express MLL-AF4 proteins. The purple rectangle highlights a locus on which di-methyl H3K79 marks were absent, but the MLL-AF4/AEP complex was present.
See also Figure S2.
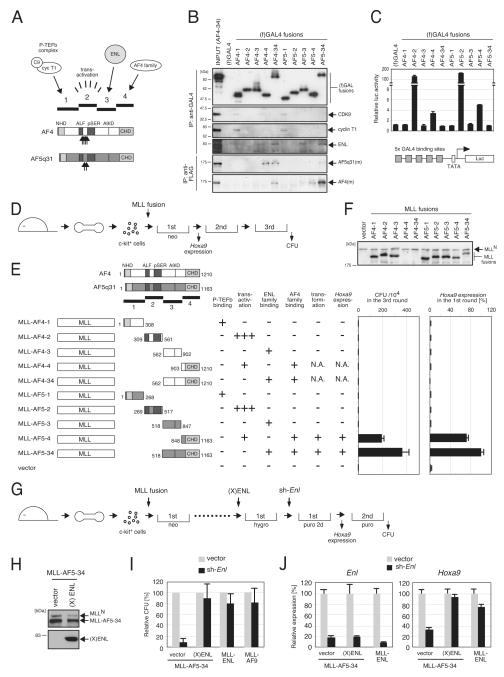
Formation of a higher order MLL-AF5q31/AEP hybrid complex is required for sustained transcription of target genes and transformation
AF4 and AF5q31 share extensive sequence similarity that resides in four sub-regions of the respective proteins (Figure 3A). A structure/function analysis (Figures 3B and 3C) revealed that: (1) P-TEFb interacts with AF4 and AF5q31 via sub-region 1, which contains the N-terminal homology domain (NHD) (Nilson et al., 1997); (2) strong transactivation activity is conferred by sub-region 2, consistent with previous observations (Prasad et al., 1995; Ma and Staudt, 1996; Morrissey et al., 1997); (3) ENL interacts with AF4 and AF5q31 through sub-region 3 that encompasses the AF9 interaction domain (Srinivasan et al., 2004; Zeisig et al. 2005); and (4) the C-terminal homology domain (CHD) within sub-region 4 mediates hetero-association of AF4 and AF5q31, which appears to be highly preferred over their respective homo-dimerization (Figure 3B). Preferential hetero-dimerization was also observed in co-IP experiments of endogenous or transfected MLL-AF5q31 (Figures 1E and S3A), as well as an interaction assay based on GAL4-dependent transactivation (Figure S3B).
Figure 3.
Formation of an AEP-like complex is required for MLL-AF5q31-dependent myeloid transformation
(A) The structures of AF4 and AF5q31 are schematically illustrated. Subregions (1-4) of AF4 and AF5q31 are indicated with associated functions. Upward arrows indicate the sites of fusion with MLL in human leukemia oncoproteins (Jansen et al. 2005). A9ID, AF9 interaction domain (Srinivasan et al. 2004).
(B) The four subregions fused to GAL4 DNA binding domain were expressed in 293T cells (upper four panels) or co-expressed with myc-tagged AF4 or AF5q31 [AF4(m) or AF5q31(m)] (lower two panels) and analyzed by IP western blotting. IP antibodies are indicated on the left and proteins detected by western blotting are indicated on the right. (f)GAL4 fusions and myc-tagged AF4 family proteins were visualized with anti-FLAG and anti-myc antibodies, respectively.
(C) Transactivation activity of respective GAL4 fusions was analyzed using the reporter gene shown below. Error bars represent standard deviations from triplicate analyses.
(D) The experimental scheme of myeloid progenitor transformation assays to evaluate the oncogenic potentials of various MLL mutants shows the time points at which CFU (colony forming unit) activity or Hoxa9 expression was examined.
(E) The structures of various MLL-AF4/AF5q31 mutants and their associated functions are summarized schematically. Hoxa9 levels were normalized to Gapdh and displayed relative to MLL-AF5-34-transduced cells arbitrarily set at 100 %. Error bars represent standard deviations of three independent analyses (left) or triplicate PCRs (right). N.A., not applicable due to unstable expression of MLL fusion proteins.
(F) Protein levels of respective MLL mutants in virus packaging cells were examined by western blotting with anti-MLLN antibody. MLL-AF4-4 and MLL-AF4-34 proteins were not stably expressed.
(G) The experimental scheme to evaluate the effect of Enl knockdown on MLL transformation is shown schematically. (X)ENL, Xpress-tagged human ENL.
(H) Transduced myeloid progenitors were analyzed by western blotting with anti-MLLN (top) and anti-Xpress (bottom) antibodies to detect exogenous MLL-AF5q31 and human (X)ENL, respectively
(I) The clonogenic potentials of MLL-AF5-34-transformed cells transduced with or without (X)ENL are shown at the second plating after sh-RNA transduction (vector or sh-Enl). MLL-ENL- or MLL-AF9-transformed cells were also subjected to sh-RNA transduction for comparison. CFUs are expressed relative to the vector control arbitrarily set as 100. Error bars represent standard deviations of three independent analyses.
(J) Cells from first round colonies following sh-RNA transduction (vector or sh-Enl) were analyzed by RT-PCR for expression of endogenous Enl or Hoxa9. Expression levels were normalized to Gapdh and displayed relative to the vector/vector control cells arbitrarily set at 100. Error bars represent standard deviations of triplicate PCRs.
See also Figure S3.
MLL fusion proteins containing the respective sub-regions of AF4 or AF5q31 were assessed for their oncogenic potentials in a myeloid progenitor transformation assay (Figure 3D) (Lavau et al., 1997). Only MLL-AF5q31 constructs containing sub-region 4 (MLL-AF5-4 and MLL-AF5-34) induced serial replating activity and up-regulation of Hoxa9 transcription (Figures 3E and 3F). This result indicates that none of the single functions (i.e. P-TEFb recruitment, transactivation, or association with ENL) is sufficient for transformation but rather CHD-mediated association with endogenous AEP is required. The corresponding MLL-AF4-4 and MLL-AF4-34 proteins were not stably expressed and thus unable to be evaluated (Figure 3F). Although recruitment of ENL was not sufficient for MLL-AF5q31-dependent transformation, ENL was required since its knockdown by sh-RNA substantially decreased the clonogenicity and Hoxa9 expression of MLL-AF5q31-transformed cells (Figures 3G-3J). This phenotype was rescued by exogenous expression of human ENL, thus verifying the target specificity of the sh-RNA. Hence, formation of a higher order MLL/AEP hybrid complex on target genes is necessary for MLL-AF5q31-dependent transformation.
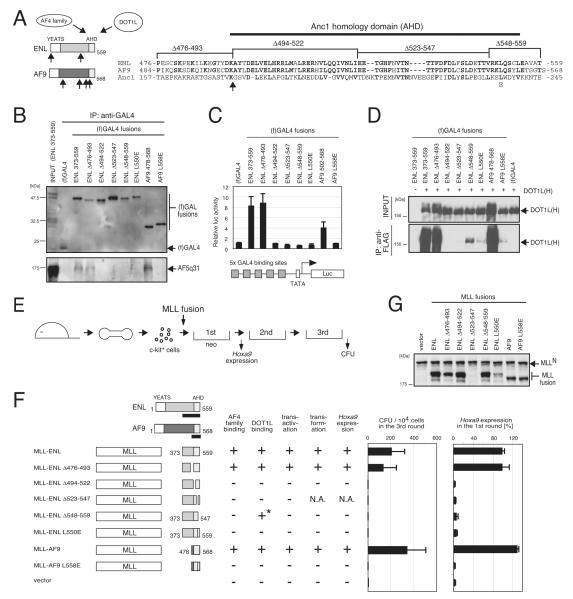
Transforming properties of MLL-ENL and MLL-AF9 correlate with association with AF4 family proteins and DOT1L
A similar structure/function analysis of MLL-ENL demonstrated that C-terminal ENL residues (494-559) are required for the interaction with AF5q31 (Figures 4A and 4B). This region, which is evolutionally conserved with AF9 and S. cerevisiae Anc1 (designated AHD: Anc1 homology domain), displayed transactivation potential that correlated with association with AF4 family proteins (Figure 4C). The AHD of ENL also mediated association with DOT1L (Figure 4D) consistent with previous studies (Muller et al., 2007). Mutations of MLL-ENL that abolished AF5q31 and DOT1L interaction (including a single L550E point mutation) resulted in failure to upregulate Hoxa9 transcription and transform myeloid progenitors (Figures 4E-4G). Similarly, the portion of AF9 retained in MLL-AF9 oncoproteins, which includes AHD (residues 502-568) (Figure 4A), mediated AF5q31 and DOT1L association, and conferred GAL4-dependent transactivation, MLL-dependent Hoxa9 expression, and myeloid transformation (Figures 4B-4G). Unlike MLL-AF5q31-transformed cells, MLL-ENL and MLL-AF9 transformed cells did not require wt ENL since their clonogenicities were unaffected by its knockdown (Figures 3I and 3J), consistent with the observation that MLL-ENL did not form a complex with wt ENL in HB1119 cells (Figure 1E). These results suggest that association with AF4 family proteins and/or DOT1L is required for the oncogenic properties of MLL-ENL and MLL-AF9.
Figure 4.
MLL-ENL and MLL-AF9 transform myeloid progenitors via the AHD, which is responsible for association with AF4 family proteins and DOT1L
(A) The structures of ENL and AF9 are schematically illustrated with associated functions (Zeisig et al. 2005). Aligned amino acid sequences for the minimum transformation domain are also shown with the positions of deletion or substitution mutations and AHD. Upward arrows indicate the sites of fusion with MLL in human leukemia oncoproteins (Jansen et al., 2005).
(B) Domain mapping of ENL family proteins for association with AF5q31 was performed with FLAG-tagged GAL4 fusion constructs of ENL (372-559 aa) and AF9 (478-568 aa). IP was performed with anti-GAL4 antibody and the precipitates were immunoblotted with anti-FLAG antibody for (f)GAL4 fusions or anti-AF5q31 antibody for endogenous AF5q31.
(C) Transactivation activity of indicated GAL4 constructs was analyzed by luciferase assay as in Figure 3C.
(D) The same set of GAL4 fusion proteins used in (B) and HA-tagged DOT1L [DOT1L(H)] were co-expressed in 293T cells and analyzed by IP western blotting. IP was performed with anti-FLAG antibody and the precipitates were immunoblotted with anti-HA antibody.
(E) The experimental scheme is shown for myeloid progenitor transformation assays to evaluate the oncogenic potentials of MLL mutants.
(F) The structures of MLL-ENL and MLL-AF9 mutants and their associated functions are summarized with schematic representations. Hoxa9 expression levels were normalized to Gapdh and displayed relative to the MLL-ENL-transduced cells arbitrarily set at 100 %. Error bars represent standard deviations of three independent analyses (left) or triplicate PCRs (right). N.A., not applicable due to unstable expression of MLL fusion proteins. The asterisk indicates that association of ENL Δ548-559 mutant with DOT1L was detected but reduced substantially compared to wt ENL.
(G) Protein levels of respective MLL mutants in virus packaging cells were examined by western blotting with anti-MLLN antibody. MLL-ENL Δ523-547 was not stably expressed.
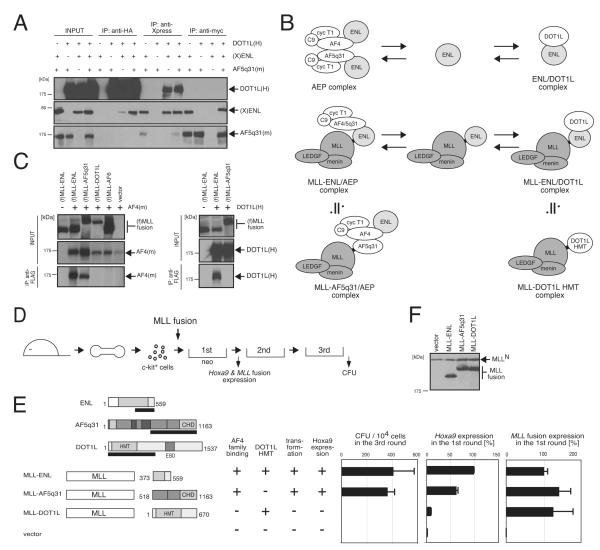
Interactions of ENL with DOT1L or AF4 family proteins are mutually exclusive
To determine whether ENL can simultaneously co-associate with AF4-family proteins and DOT1L, IP analysis was performed on cells transiently expressing ENL, AF5q31 and DOT1L. Although ENL co-precipitated both AF5q31 and DOT1L, the latter two did not pull down each other (Figure 5A), indicating that the three proteins do not form a trimeric complex. Similarly, GAL4-AF5-3 effectively co-precipitated ENL but not DOT1L under conditions where GAL4-ENL successfully pulled down DOT1L (Figure S4). These data demonstrate that the associations of ENL family proteins with AF4 family proteins or DOT1L are mutually exclusive. Therefore, the ENL/DOT1L complex is a separate entity from AEP (Figure 5B).
Figure 5.
Associations of ENL family proteins with AF4 family proteins or DOT1L are mutually exclusive
(A) AF5q31(m), (X)ENL, and DOT1L(H) were co-expressed in 293T cells and analyzed by IP western blotting. IP was performed with antibodies indicated on the top and the precipitates were immunoblotted with anti-myc, anti-Xpress or anti-HA antibody.
(B) Putative conformations of various ENL complexes are shown schematically. ENL forms two distinct complexes: AEP and ENL/DOT1L. Similarly, MLL-ENL participates in two mutually exclusive associations to form the MLL-ENL/AEP and MLL-ENL/DOT1L complexes that are approximate to the MLL-AF5q31/AEP and MLL-DOT1L complexes, respectively.
(C) FLAG-tagged MLL fusion proteins [(f) MLL fusions] were co-expressed with AF4(m) or DOT1L(H) in 293T cells and analyzed by IP western blotting. IP was performed with anti-FLAG antibody and the precipitates were immunoblotted with anti-MLLN, anti-myc, or anti-HA antibody.
(D) The experimental scheme is shown for myeloid progenitor transformation assays to evaluate the oncogenic potentials of MLL mutants.
(E) The structures of MLL-fusion proteins and their associated functions are summarized. Expression of MLL fusion genes or Hoxa9 was examined by RT-PCR in first round colonies. Expression levels were normalized to Gapdh levels and displayed relative to the transcript levels in MLL-ENL transduced cells arbitrarily set at 100. Error bars represent standard deviations of three independent analyses (left) or triplicate PCRs (middle and right). HMT, histone methyltransferase catalytic domain; EBD, ENL binding domain (Okada et al. 2005; Mueller et al. 2007).
(F) Protein levels of MLL fusions in virus packaging cells were analyzed by western blotting with anti-MLLN antibody.
See also Figure S4.
Recruitment of AEP, versus DOT1L, plays a predominant role in MLL-dependent leukemogenesis
The ability of MLL-ENL to associate with AF4 family proteins or DOT1L raised the issue of which interaction (MLL-ENL/AEP versus MLL-ENL/DOT1L) is essential for leukemic transformation (Figure 5B). To address this, an artificial MLL fusion with DOT1L (MLL-DOT1L) that does not associate with AF4 (Figure 5C) but retains the HMT catalytic domain (thus mimics the MLL-ENL/DOT1L complex) was assessed for its transformation potential. MLL-DOT1L failed to sufficiently activate Hoxa9 expression to immortalize myeloid progenitors (Figures 5D and 5E) despite the comparable levels of protein expression in packaging cells (Figure 5F) and mRNA expression in first round colonies (Figure 5E). In the same experimental condition, MLL-AF5q31 successfully transformed myeloid progenitors (Figure 5E) without being able to directly associate with DOT1L (Figure 5C). These results, which contrast with previous studies (Okada et al., 2005), indicate that simple recruitment of DOT1L HMT activity alone to MLL target genes is not sufficient for transformation, and support a more predominant role for AEP recruitment.
Nevertheless, DOT1L-dependent H3K79 methylation co-localized with the presence of MLL-ENL at all target loci tested in HB1119 cells (Figure 2B), indicating that not only AEP components but also DOT1L is consistently recruited by MLL-ENL. In MV4-11 cells, H3K79 methylation marks also co-localized at most of the MLL-AF4-occupied loci consistent with previous observations (Krivtsov et al., 2008; Guenther et al., 2008) despite the apparent inability to directly recruit DOT1L (Figures 2C and 5C). However, the signal intensities of H3K79 di-methylation were relatively low at MLL-AF4-target loci compared to those at MLL-ENL-target loci (compare relative intensities to those of β-ACTIN and GAPDH which served as internal standards) (Figures 2B, 2C and S2) and were minimal at the CDKN2C promoter in spite of the localization of abundant AEP components (Figure 2C, purple rectangle). Thus, DOT1L-dependent H3K79 methylation is associated with the presence of the MLL-AF4/AEP-hybrid complex but the two distinct biochemical entities are not constitutively coupled. These results suggest that DOT1L is functionally linked to MLL-AF4 but normally recruited to target loci subsequent to AEP components.
AEP is indirectly recruited to MLL-AF6-occupied loci to sustain transcription and transformation
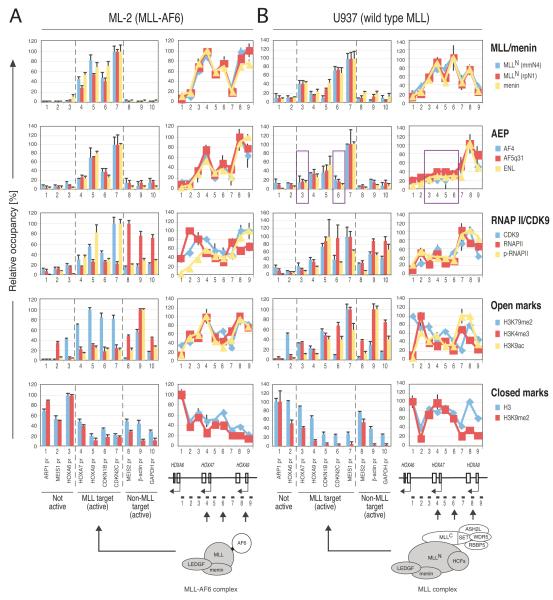
To investigate whether AEP involvement is restricted to MLL fusions with AF4 and ENL family proteins, ChIP analyses were performed on ML-2 cells. Surprisingly, MLL-AF6 co-localized with AEP at the chromatin of MLL target genes (HOXA7, HOXA9, CDKN1B and CDKN2C) (Figures 6A and S5A) despite its inability to directly associate with AEP (Figures 1E and 5C). The occupancies of CDK9 and phosphorylated RNAPII coincided with the presence of AEP on MLL-AF6 target genes (Figure 6A). Characteristically, high levels of dimethyl H3K79 were associated with the presence of AEP, corroborating the functional link between AEP and DOT1L. These results suggest that the AEP complex can be recruited to MLL target loci via an indirect mechanism potentially serving a role in MLL-AF6-dependent leukemogenesis.
Figure 6.
Indirect recruitment of AEP to MLL-AF6- or wt MLL-occupied loci
(A & B) Genomic localizations of indicated proteins in ML-2 (A) and U937 (B) cells were determined by ChIP assay as in Figure 2B. The purple rectangle highlights regions where AEP is absent while the MLL complexes are present. ChIP data using anti-MLLN (rpN1) and anti-menin antibodies are partially adapted from a previous report (Yokoyama and Cleary, 2008).
See also Figure S5.
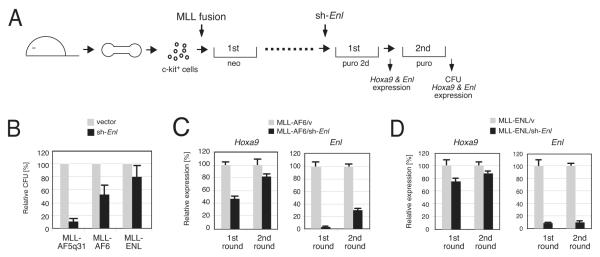
MLL-AF6 transformed cells were also dependent on ENL, since its knockdown reduced their clonogenicity and Hoxa9 expression by 50% (Figures 7A and 7C). This was less severe compared to MLL-AF5q31-transformed cells (Figure 7B), in part due to insufficient knockdown by the sh-RNA since secondary colonies expressed Hoxa9 at its normal levels accompanied with impaired knockdown of Enl (Figure 7C), indicating a selective proliferative advantage of cells in which Enl was incompletely knocked down (MLL-ENL served as a negative control in Figure 7D). Thus, transformation by MLL-AF6 is dependent on ENL, despite an inability to directly associate with AEP.
Figure 7.
ENL is required for MLL-AF6-dependent transactivation and transformaion
(A) The experimental scheme to evaluate the effect of Enl knockdown on MLL transformation is shown.
(B) Clonogenic potentials are shown for myeloid cells transformed by MLL oncogenes (indicated below) at the second plating after sh-RNA transduction (vector or sh-Enl). CFU numbers are displayed relative to the vector control arbitrarily set as 100. Error bars represent standard deviations of three independent analyses.
(C) MLL-AF6-transformed cells from first and second round colonies following sh-RNA transduction (vector or sh-Enl) were analyzed by RT-PCR for expression of endogenous Enl or Hoxa9. Expression levels were normalized to GAPDH levels and displayed relative to the transcript levels in vector control cells arbitrarily set as 100. Error bars represent standard deviations of triplicate PCRs.
(D) The same analysis as (C) was performed on MLL-ENL-transformed cells. Note that data in (B) and (D) are partially redundant with Figures 3I and 3J.
AEP facilitates the physiologic MLL-dependent transcriptional pathway
The foregoing results prompted studies of a potential relationship of AEP in physiologic transcriptional regulation by wt MLL. ChIP analyses of U937 cells, which lack an MLL chromosomal translocation (Dreying et al., 1996; Guenther et al., 2005), showed that AEP co-localized with wt MLL at the HOXA9, MEIS1 and CDKN1B promoters (Figures 6B and S5B). However, in contrast to MLL leukemia cell lines, co-localization was not observed at all of the MLL-occupied loci in U937 cells. For instance, the MLL complex occupied both the HOXA7 and HOXA9 loci, whereas AEP associated only with the latter (Figure 6B, purple rectangle). A similar disparity was observed at the CDKN2C promoter. These results suggest that AEP is recruited to wt MLL-occupied loci in a context dependent manner as opposed to its constitutive recruitment in MLL leukemia cells. The presence of AEP correlated more closely with active transcriptional marks like phospho-RNAPII and acetyl-histone H3K9 (e.g. the HOXA7-9 locus), suggesting that AEP recruitment to MLL-targeted chromatin facilitates transcription.
The role of ENL in physiologic MLL-dependent transcriptional maintenance was assessed by knocking down Enl in mouse embryonic fibroblasts (MEFs), in which Hoxc8 is a target gene of the MLL/menin complex (Figure 8A) (Hughes et al. 2004; Milne et al., 2002). Enl knockdown caused reduction of Hoxc8 expression, which could be prevented by antecedent expression of exogenous human ENL (Figure 8B). Thus, ENL is required for physiologic transcriptional regulation by the MLL/menin complex. Moreover, DOT1L-mediated histone methylation was decreased at the Hoxc8 promoter in Men1 null MEFs (Figure 8C), indicating that the MLL/menin complex functions upstream of ENL/DOT1L functions.
Figure 8.
ENL functions downstream of physiologic MLL-dependent transcriptional pathways
(A) Expression levels of Enl and Hoxc8 in wt or Men1-/- MEFs were determined by RT-PCR (normalized to β-actin levels and displayed relative to the vector control arbitrarily set as 100). Error bars represent standard deviations of triplicate PCRs.
(B) Expression of Enl and Hoxc8 with or without Enl knockdown/rescue was determined by RT-PCR. Expression levels were normalized to β-actin levels and expressed relative to the vector/vector control arbitrarily set as 100. Error bars represent standard deviations of triplicate PCRs. Protein levels of the exogenously expressed (X)ENL (right panels) were assessed by western blotting with an anti-Xpress antibody (actin immunoblot served as a loading control). nsp, non-specific band.
(C) ChIP assay was performed on wt or Men1-/- MEFs using anti-di-methyl H3K79 and histone H3 antibodies for the Hoxc8 promoter-adjacent region and results displayed as relative ratio (%) to the input DNA. Error bars represent standard deviations of triplicate PCRs.
(D) The effects of ENL-knockdown are shown for two different sh-RNAs in U937 cells. Expression of various genes was analyzed by RT-PCR 4 d after transduction/puromycin selection. Expression values were normalized to GAPDH levels and displayed relative to the vector control arbitrarily set as 100. Error bars represent standard deviations of triplicate PCRs.
(E) A three-step model of MLL-dependent transcription.
Furthermore, ENL knockdown in U937 cells caused down-regulation of HOXA9, CDKN1B and MEIS1, whose genomic loci were occupied by both MLL and AEP complexes, but did not affect expression of genes occupied by the MLL complex without AEP (HOXA7 and CDKN2C) (AF5q31, MLL or β-ACTIN served as negative controls) (Figures 6B and 8D). Thus, ENL is specifically required for the optimal transcription of genes occupied by both MLL and AEP complexes.
Discussion
Our biochemical purification of AF4 family proteins demonstrates that they normally associate with ENL and the P-TEFb elongation factor in an endogenous complex (AEP) in hematopoietic cells. MLL oncopoteins fused with AEP components (AF4 or ENL family proteins) nucleate formation of MLL/AEP hybrid complexes that constitutively occupy MLL-target chromatin. This aberrant recruitment of AEP components causes sustained activation of MLL target gene transcription and transformation of hematopoietic progenitors. Although the AEP and MLL complexes are normally separate biochemical entities, our studies support a dependent role for the AEP complex in physiologic MLL target gene expression pathways, whose conditional recruitment mechanisms are often bypassed by leukemic MLL fusion proteins.
The AEP complex purified from leukemia cell lines under our experimental conditions contained ENL as an integral component but lacked a number of previously reported ENL-associated proteins, most notably the DOT1L histone methyltransferase (Mueller et al., 2007). Our domain mapping analyses provide a molecular basis for its absence in that DOT1L and AF4 family proteins use the same binding surface within the AHD of ENL. Because of this physical constraint, DOT1L and AF4 family proteins are incapable of simultaneously associating with the AHD to form an AF4/ENL/DOT1L trimeric complex. Therefore, retention of DOT1L in the AF4 complex previously identified in thymus homogenates (Bitoun et al. 2008) is likely mediated by other proteins (e.g. AF10, RNAPII) but not by ENL/AF9. Our data suggest that an endogenous ENL/DOT1L complex and AEP normally exist as separate entities consistent with previous suggestions that ENL may participate in a mixture of different sub-complexes (Mueller et al., 2007).
A role for ENL in multiple sub-complexes raises the issue of which of its molecular interactions is essential for MLL leukemogenesis. This was addressed by assessing the oncogenic potential of MLL fused with the DOT1L catalytic domain, which effectively bypasses ENL. Contrary to a previous report (Okada et al., 2005), MLL-DOT1L was not sufficient for transactivation of MLL target genes and transformation of myeloid progenitors under our experimental conditions that readout the oncogenic properties of MLL-AF5q31 and MLL-ENL. This indicates that aberrant recruitment of AEP, not DOT1L, plays a primary rate-limiting role in transactivation and transformation by MLL fusion proteins, a conclusion further supported by structure/function analysis of MLL-AF5q31 showing that its CHD, which mediates hetero interactions with AF4 family members, was necessary and sufficient for transformation.
Nevertheless, ChIP analyses by us and others show that H3K79 methylation marks are present at most MLL-AF4-target loci (Figure 2B) (Krivtsov et al., 2008; Guenther et al., 2008), indicating that there is a strong functional interconnection between AEP and DOT1L. DOT1L-dependent H3K79 methylation is associated with transcribed regions and stimulated by histone H2B K120 mono-ubiquitination (a histone mark accompanied with transcription), but not required for transcription itself (Steger et al. 2008; McGinty et al. 2008). This suggests that DOT1L-dependent H3K79 methylation occurs after the traverse of RNAPII and may play roles in the maintenance of transcriptional memory rather than initiating transcription per se. In this context, our studies support dual roles for ENL, which is capable of interacting with AEP or DOT1L through its AHD to sequentially recruit them to the same target chromatin, possibly via its N-terminal YEATS domain that retains a chromatin binding property (Zeisig et al., 2005).
Our data demonstrate that AEP co-localizes with wt MLL on target promoters indicative of a role in physiologic as well as oncogenic MLL-dependent transcriptional pathways. Supporting this notion, knockdown of ENL impaired expression of MLL target genes in MEFs and U937 cells (Figures 8B and 8D), and Af9-deficient mice display homeotic transformations similar to those of Mll-deficient mice (Collins et al., 2002). The recruitment of AEP to MLL-target loci appears to be non-constitutive since some MLL-occupied loci do not contain AEP (Figure 6B). Because the presence of the MLL complex does not invariably correlate with occupancy by AEP, other factors/signals yet to be identified are likely required for AEP recruitment. Based on these observations/speculations, we propose a three-step model in which wt MLL first establishes/maintains the transcriptionally poised state (STEP 1), AEP is then recruited to facilitate onset of transcriptional initiation/elongation (STEP 2), which is followed by DOT1L-dependent H3K79 methylation post-transcription (STEP 3) (Figure 8E). In this model, ENL serves a key role in sequential recruitment of AEP and DOT1L, respectively.
To date, up to 50 different proteins have been reported to fuse with MLL in human leukemias. This promiscuity poses a question whether any common trait is shared among the fusion partners. We demonstrate here that AEP recruitment is a downstream event in physiologic MLL-dependent transcriptional pathways and regulated in a context-dependent manner. MLL-AF4 and MLL-ENL family fusions transform myeloid progenitors by constitutively recruiting AEP to MLL-target loci through direct association. Thus, one of the major mechanisms of MLL-dependent transformation is constitutive activation of MLL-dependent transcription by direct recruitment of AEP, which circumvents the regulatory mechanisms that normally control AEP recruitment (Figure 8E).
AEP does not physically interact with MLL-AF6, but nevertheless consistently co-localizes with MLL-AF6 at target chromatin to activate transcription (Figures 6A and 8E). Although the mechanism of this aberrant AEP recruitment is unknown, it indicates that AEP serves an even broader role in MLL leukemogenesis beyond the subset of fusions with AEP components. Determination of whether this role may extend to other MLL fusion proteins requires further investigation. Nevertheless, our studies show that most of the frequently occurring MLL fusions (e.g. MLL-AF4, MLL-AF9, MLL-ENL, and MLL-AF6) employ a similar strategy for leukemic transformation, in which AEP is constitutively recruited to MLL target genes either directly or indirectly.
A critical role for AEP in MLL-mediated leukemic transformation suggests that it may be an ideal target for molecular therapy of MLL-associated leukemias. In this regard, our results tentatively support the rationale for CDK9 inhibition as a potential therapeutic strategy, or inhibition of DOT1L whose activity appears to be functionally linked to AEP and possibly plays important roles in the maintenance of the epigenetic status of target genes. However, these molecules are likely to have more generalized roles other than AEP-dependent transcription (Jones et al. 2008; Peterlin and Price 2006), therefore serious side effects might occur if they are effectively inhibited. Thus, compounds that specifically target the function of AF4- and ENL-family proteins but not P-TEFb or DOT1L may selectively inhibit MLL-dependent transcription and benefit the treatment of MLL-associated leukemias.
Experimental procedures
Monoclonal antibodies
Highly specific monoclonal antibodies were generated against MBP fusion proteins containing portions of human AF4 (aa 782-979) (clone 2C.1), human AF5q31 (aa 489-680) (clone 1.3), and human ENL (414-472) (clone 3.1), respectively.
Cell culture
Human leukemia cell lines K562, HB1119, SEM-K2, KP-L-RY, ML-2, MV4-11, and U937 were cultured in RPMI 1640 medium supplemented with 15% fetal calf serum and non-essential amino acids. MEFs were prepared from E11.5 p53-null embryos. The 293T and plat-E cell lines, and MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal calf serum and non-essential amino acids.
Purification of the AEP complex, immunoprecipitation and western blotting
The purification procedure for AEP is described in the Supplemental Experimental Procedures. Immunoprecipitation and western blotting methods are described elsewhere (Yokoyama et al., 2004; 2005). Primary antibodies used in this study are summarized in the Supplemental Experimental Procedures.
Quantitative RT-PCR
Reverse transcription and quantitative PCR were performed as described previously (Yokoyama et al., 2005; Yokoyama and Cleary, 2008) using Taqman probes purchased from Applied Biosystems. The details of the probe set are summarized in the Supplemental Experimental Procedures. Expression levels (average values and standard deviations of triplicate determinations) normalized to housekeeping genes such as GAPDH and β-ACTIN were calculated using a standard curve and the relative quantification method as described in ABI User Bulletin #2.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitations were performed as previously described (Weinmann and Farnham, 2002; Yokoyama and Cleary, 2008). Primary antibodies used in ChIP assays are summarized in the Supplemental Experimental Procedures. Quantitative PCR was performed on the precipitated DNAs in triplicate using primers and probes described in the Supplemental Experimental Procedures. The values relative to input were determined using a standard curve and the relative quantification method as described in ABI User Bulletin #2.
Vector construction
cDNA fragments of AF4 and AF5q31 were cloned into pcDNA3.1/myc-His A (Invitrogen) for expressing c-myc tagged proteins, or pBICEP-CMV-2 (Sigma) for expressing FLAG-tagged proteins. pMSCV-neo constructs encoding MLL-ENL, MLL-AF9 and MLL-AF6 were described previously (Ayton and Cleary, 2003; Somervaille et al., 2006). pMSCV-hygro-Xpress tagged ENL and pMSCV-neo-Xpress tagged MLL-AF5q31-34 were generated by fusing the Xpress-tag sequence from pcDNA4 HisMax vector with the cDNAs of ENL or MLL-AF5q31, respectively. Other expression vectors for various MLL mutants were generated by restriction enzyme digestion or PCR-based mutagenesis. Various FLAG-tagged MLL fusions were also cloned into pCMV5 vector and used for IP analysis. The expression vectors for FLAG-tagged GAL4 fusion proteins were constructed by PCR using pM (Clontech) as template and cloned into pCMV5 vector. The sh-RNA expression vectors targeting murine Enl (TRCN0000084405,) and human ENL (TRCN000019291[#1], TRCN000019293[#2]) were purchased from Open Biosystems.
Virus production
Ecotropic retrovirus was produced using plat-E packaging cells (Morita et al., 2000). Lentivirus was produced by co-transfection of 293T cells with viral vectors and pCMV dR8.74 and pMD.G packaging constructs (Dull et al., 1998). Supernatant medium containing virus was harvested 48 h post transfection and used for transductions.
Myeloid progenitor transformation assay
Myeloid progenitor transformation was assessed as described elsewhere (Lavau et al., 1997; Yokoyama and Cleary, 2008) using cells harvested from the femurs of CD45.1 inbred C57BL/6 mice. C-kit positive cells were enriched by immuno-magnetic selection using an Auto MACS (Miltenyi Biotech), transduced with recombinant retrovirus by spinoculation, and plated in methylcellulose medium (M3231, Stemcell Technologies) containing SCF, IL-3, IL-6 and GM-CSF. The colony-forming units (CFUs) per 104 plated cells were quantified after 5-7 d of culture and expressed as the average and standard deviation of at least triplicate determinations. For secondary transductions, 105 cells were transduced with retrovirus by spinoculation, cultured in methylcellulose medium overnight, and selected for drug resistance (hygromycin 750 μg/ml, puromycin 4 μg/ml) for at least 2 days prior to CFC enumeration.
Transactivation assay
Transactivation assays were performed using 293T cells as described elsewhere (Yokoyama et al., 2002). Cells cultured in 24 well dishes were transfected with 25 ng of pRL-tk, 250 ng of pFR-luc, and 500 ng of pCMV5 FLAG-GAL4 fusion protein vector per well. Cells were lysed 24 h later and analyzed for luciferase activity using a dual luciferase assay kit according to the manufacturer’s instructions (Promega). Relative luciferase activities were normalized to renilla luciferase activities and expressed with the average values and standard deviations of triplicate determinations relative to the GAL4 DNA binding domain controls.
Significance.
MLL is fused by chromosomal translocations in 5-10% of acute leukemias to a variety of partner proteins (>50) of diverse molecular composition and function. Recent studies show that several of the more common MLL fusion partners (e.g. AF4, ENL, AF9) associate in a higher-order complex containing transcription elongation factors. Here we show that this complex is biochemically distinct from the MLL histone methyltransferase complex, but nevertheless normally present at MLL target genes during physiologic gene expression. In acute leukemias, the complex is constitutively recruited to target chromatin by covalent fusion of MLL with one of several complex components or non-covalent mechanisms employed by other MLL fusion proteins, thereby representing a unifying mechanism for MLL-mediated leukemogenesis that can be targeted by molecular therapy.
Highlights.
Proteins of the AF4 and ENL families form a complex (AEP) with P-TEFb on chromatin. The AEP complex facilitates physiologic MLL target gene expression.
Aberrant AEP recruitment is a unifying mechanism in MLL-mediated leukemogenesis.
Supplementary Material
Acknowledgments
We thank Dr. M. Meyerson for providing Men1 conditional knockout mice, Dr. T. Kitamura for the plat-E cell line, Dr. K. Yamagata for the DOT1L expression vector, and Dr. Y. Zhang for the MLL-DOT1L expression vector. We thank C. Nicolas, M. Ambrus, B. Rouse, C. Hatanaka and M. Kawaguchi for technical assistance. A.Y. was supported by a Special Fellow Award from the Leukemia and Lymphoma Society. These studies were supported by the Children’s Health Initiative of the Packard Foundation and grants from the National Institutes of Health (CA55029 and CA116606) and in part by Grants-in-Aid for Cancer Research (21-6-1) and for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labor of Japan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, Cleary ML, Shilatifard A. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;96:3887–3893. [PubMed] [Google Scholar]
- Domer PH, Fakharzadeh SS, Chen CS, Jockel J, Johansen L, Silverman GA, Kersey JH, Korsmeyer SJ. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci U S A. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfurth F, Hemenway CS, de Erkenez AC, Domer PH. MLL fusion partners AF4 and AF9 interact at subnuclear foci. Leukemia. 2004;18:92–102. doi: 10.1038/sj.leu.2403200. [DOI] [PubMed] [Google Scholar]
- Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39:6–11. doi: 10.1053/shem.2002.36921. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, Kamada N, Ariyoshi Y, Takahashi T, Ueda R. MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene. 1993;8:3085–3092. [PubMed] [Google Scholar]
- Jansen MW, van der Velden VH, van Dongen JJ. Efficient and easy detection of MLL-AF4, MLL-AF9 and MLL-ENL fusion gene transcripts by multiplex real-time quantitative RT-PCR in TaqMan and LightCycler. Leukemia. 2005;19:2016–2018. doi: 10.1038/sj.leu.2403939. [DOI] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. Embo J. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. Embo J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Ma C, Staudt LM. LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood. 1996;87:734–745. [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Morrissey JJ, Raney S, Cleary ML. The FEL (AF-4) protein donates transcriptional activation sequences to Hrx-Fel fusion proteins in leukemias containing T(4;11)(Q21;Q23) chromosomal translocations. Leuk Res. 1997;21:911–917. doi: 10.1016/s0145-2126(97)00012-x. [DOI] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale RP, Lange B, Crist WM, Nowell PC, et al. Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc Natl Acad Sci U S A. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Shaughnessy JD, Jr., Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- Nilson I, Reichel M, Ennas MG, Greim R, Knorr C, Siegler G, Greil J, Fey GH, Marschalek R. Exon/intron structure of the human AF-4 gene, a member of the AF-4/LAF-4/FMR-2 gene family coding for a nuclear protein with structural alterations in acute leukaemia. Br J Haematol. 1997;98:157–169. doi: 10.1046/j.1365-2141.1997.1522966.x. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce CM, Canaani E. Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci U S A. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Cleary ML. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol Cell Biol. 2002;22:6542–6552. doi: 10.1128/MCB.22.18.6542-6552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Cleary ML. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood. 2003;101:633–639. doi: 10.1182/blood-2002-06-1785. [DOI] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Srinivasan RS, Nesbit JB, Marrero L, Erfurth F, LaRussa VF, Hemenway CS. The synthetic peptide PFWT disrupts AF4-AF9 protein complexes and induces apoptosis in t(4;11) leukemia cells. Leukemia. 2004;18:1364–1372. doi: 10.1038/sj.leu.2403415. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki T, Kano H, Taniwaki M, Sako M, Yanagisawa M, Hayashi Y. AF5q31, a newly identified AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia with ins(5;11)(q31;q13q23) Proc Natl Acad Sci U S A. 1999;96:14535–14540. doi: 10.1073/pnas.96.25.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- von Bergh AR, Beverloo HB, Rombout P, van Wering ER, van Weel MH, Beverstock GC, Kluin PM, Slater RM, Schuuring E. LAF4, an AF4-related gene, is fused to MLL in infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2002;35:92–96. doi: 10.1002/gcc.10091. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–3718. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene. 2005;24:5525–5532. doi: 10.1038/sj.onc.1208699. [DOI] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.