Abstract
Two genes from Trypanosoma brucei brucei are predicted to encode FeII- and α-ketoglutarate-dependent enzymes related to fungal thymine 7-hydroxylase. Transcription of the thymine hydroxylase-like genes is up-regulated in the bloodstream-form of the parasite over the insect form, whereas Western blot analysis indicates more cross-reactive protein in the latter life stage. The genes were cloned, the proteins purified from Escherichia coli, and both proteins were shown to bind FeII and α-ketoglutarate, confirming proper folding. The isolated proteins were incubated with FeII and α-ketoglutarate plus thymine, thymidine, and other putative substrates, but no activity was detected. Furthermore, no thymine 7-hydroxylase activity was detected in extracts of procyclic or bloodstream form cells. Although the functions of these proteins remain unknown, we conclude they are unlikely to be involved in thymine salvage.
Keywords: Trypanosoma brucei, thymine 7-hydroxylase, EC 1.14.11.6, α-ketoglutarate, FeII
1. Introduction
Trypanosomes are the causative agents of several mammalian diseases, including African Sleeping Sickness and Chagas disease in humans and Nagana in cattle (Barrett et al., 2003). They are transmitted by a suite of insect vectors and infect the bloodstream of their mammalian hosts when the insect takes a blood meal. The genome of Trypanosoma brucei brucei was recently elucidated (Berriman et al., 2005), providing important insights into the physiology of this microorganism.
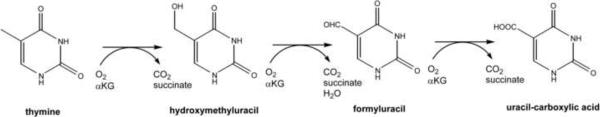
Two T. b. brucei genes, located on chromosomes 5 and 7 (Tb927.5.300 and Tb927.7.7500, respectively), encode full-length orthologues of thymine 7-hydroxylase (T7H) of Rhodotorula glutinis (Neidigh et al., 2009, Smiley et al., 2005) and are termed TLP5 and TLP7 for thymine hydroxylase-like proteins. T7H is a non-heme iron (FeII)- and α-ketoglutarate (αKG)-dependent hydroxylase that catalyzes sequential oxidations of thymine (Scheme 1) (Holme et al., 1970, Neidigh et al., 2009) as part of a pyrimidine salvage pathway in some fungi (Shaffer and Arst, 1984, Smiley et al., 2005). Hydroxylation of the unactivated C7 methyl group of thymine is driven by the oxidative decarboxylation of αKG, forming succinate and CO2. Significantly, trypanosomes have not been reported to contain a pyrimidine salvage pathway and show no mediated uptake of thymine or thymidine (Gudin et al., 2006), nor does the kinetoplastid genome contain an obvious candidate gene encoding uracil-5-carboxylate decarboxylase, the enzyme acting immediately downstream in the salvage pathway. Further, trypanosomes seem to have no direct need for pyrimidine salvage as they are known to be able to synthesize pyrimidine nucleotides (Randolph et al., 1995).
Scheme 1.
To date, three gene products in trypanosomes are predicted to belong to the FeII/αKG dependent hydroxylase family. One gene is suggested to encode AlkB, involved in oxidative repair of DNA damaged by alkylation (van den Born et al., 2008). Two others, encoding JBP1 and JBP2 are proposed to catalyze the hydroxylation of selected thymidine bases in trypanosomal DNA as the first step in formation of base J, β-D-glucosyl-hydroxymethyl-deoxyuracil (Cliffe et al., 2009, Iyer et al., 2009). Significantly, no direct evidence for hydroxylase activity has been reported for any of these proteins. Here, we examine whether the TLP5 and TLP7 genes encode T7H isozymes or have alternative functions.
2. Materials and methods
2.1. Candidate gene identification
To identify candidate thymine hydroxylase genes, the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990) was utilized to search the T. b. brucei genome with the gene sequence of R. glutinis thymine 7-hydroxylase (Neidigh et al., 2009) as the query, resulting in the identification of NCBI accession numbers XM_839611.1 and XM_841277.1. In addition, the available sequences of the kinetoplastid order were searched with the gene encoding uracil-5-carboxylate decarboxylase (Smiley et al., 2005), the next enzyme in the fungal pyrimidine salvage pathway, as a query.
2.2. Parasites and quantitative real-time PCR
Transcript levels were compared using T. b. brucei strain 427 bloodstream form (BF) paired with strain 427 (MITat-1) procyclic form (PF) and BF 427-2 (221) paired with PF 427, 29–13. Briefly, total RNA was treated with turbo DNAse (Ambion) according to the manufacturer's directions and 1 μg of RNA was then reverse transcribed (RT) in the presence of random hexamers (20 mM Tris 8.3, 50 mM KCl, 5 mM MgCl2, 2 mM mixed dNTPs, 0.25 units RNAsin and 2 units Seikagaku AMV reverse transcriptase). The RT reaction was stopped by heating at 70°C and diluted with 125 μl of TE. Real time PCR reactions (SYBR Green, Bio-Rad) were set up in triplicate (primers: chromosome 5 forward, 5'-GGT TGG GTA GAG TTG ATG AAC-3'; chromosome 5 reverse, 5'-TGG AGG ATA ATG TAG CAT ACG-3'; chromosome 7 forward, 5'-CAC ACT ATC GCG ATA TGC GGG AC-3'; chromosome 7 reverse, 5'-GCG GGG TAA TGC ACC ATG CG-3') using 2 μl of the diluted RT reaction. All reported data were normalized to β-tubulin gene internal control (primers: forward, 5' GAC GAA GGA GGT TGA TGA GCA GAT 3'; reverse, 5' TGA AGG TGA CAG CCA TCT TGA GTC 3') that was included in each run.
2.3. Construction of TLP5- and TLP7- expressing Escherichia coli strains
A 963-bp DNA fragment containing TLP5 was amplified by PCR using genomic T. b. brucei strain 427 DNA as a template with “forward” (5'-AGG ATA TAC CAT GGC TCA CGG CTC GAT T-3') and “reverse” (5'-GAG CAT CCT CGA GCA TCT TTG TTT TGC GAT G-3') primers which introduce NcoI and XhoI restriction sites (underlined), and Taq polymerase master mix kit (Promega) which leaves a single 3' adenine nucleotide overhang. The PCR product was treated with a PCR clean up kit (Qiagen, Inc.) and ligated into pGEM-T Easy (Promega). This was transformed into E. coli DH5α (Invitrogen), isolated from several transformants, and sequenced (Davis Sequencing). TLP5 was excised from the pGEM-T backbone by NcoI and XhoI restriction and ligated into pET28b (Novagen) which had been previously cut with the same enzymes, putting the coding sequence in frame with a sequence encoding a C-terminal 6-histidine tag. This plasmid was transformed into the expression strain E. coli BL21 (DE3).
Similarly, a 963-bp DNA fragment containing TLP7 was amplified from genomic T. b. brucei DNA using “forward” (5'-GGA TAT ACC ATG GTA ATG AAT CAC ACT TCG ATT CCA G-3') and “reverse” (5'-GAG CAT CCT CGA GCA TAT TTG CCT TGC G-3') primers, ligated into pGEM-T Easy, and transformed into E. coli DH5α. Plasmids isolated from several transformants were sequenced, one containing a minor A186V change was excised from the pGEM-T backbone by using NcoI and XhoI, and this fragment was ligated into pET28b to yield a sequence encoding TLP7 in frame with a C-terminal 6-histidine tag. The recombinant plasmid was transformed into E. coli BL21 (DE3) for expression.
2.4. Over-expression and purification of TLP5 and TLP7
E. coli BL21 (DE3) cells containing the pET28b derivatives encoding TLP5-His6 or TLP7-His6 (hereafter referred to simply as TLP5 and TLP7) were grown at 30 °C in LB medium supplemented with 100 μg/mL kanamycin while shaking at ~160 rpm to an optical density of 0.4 to 0.6 at 600 nm. Cultures were induced to overexpress the desired genes by addition of isopropyl-β-D-thiogalactopyranoside to 0.1 mM, and grown for an additional 4 h, then harvested at 4 °C by centrifugation at ~8,000 g for 8 min. The cell paste was either used immediately for protein purification or stored at −80 °C.
In a typical purification, 2.5 mL of binding buffer (30 mM imidazole, 10 mM Tris, 150 mM NaCl, pH 7.9) was added per gram of cell paste for resuspension. The protease inhibitor phenylmethylsulfonyl fluoride was added to 0.5 mM, cells were lysed by sonication (Branson Sonifier, 3 pulses of 1 min each, 3 W output power, duty cycle 50%, with cooling on ice), and the cell lysates were ultracentrifuged at 100,000 g for 1 h. Soluble cell extracts were loaded onto a Ni-bound nitrilotriacetic acid (NTA) column (Qiagen) pre-equilibrated with binding buffer. The column was washed with binding buffer until the baseline was reestablished, and proteins were released with elution buffer (150 mM imidazole, 10 mM Tris, 150 mM NaCl, pH 7.9). Fractions containing the purified proteins, as determined by denaturing polyacrylamide gel (12%) electrophoresis (Laemmli, 1970) and Coomassie staining, were pooled and exchanged back into binding buffer by using Amicon centrifugal filter units with a 10 kDa molecular weight cutoff. The concentrations of proteins were determined by using their calculated molar absorptivities at 280 nm (44,390 M−1 cm−1 for TLP5 and 49,890 M−1 cm−1 for TLP7 according to the ExPasy protein parameters prediction server (http://ca.expasy.org/tools/protparam.html). The proteins were stored at 4 °C and discarded after two weeks or if precipitation developed.
2.5. Antibody Production and Western Blotting
Purified recombinant TLP5 was provided to Lampire Biological Laboratories for polyclonal antibody production in a rabbit. The IgG fraction was isolated from the product antisera by ammonium sulfate precipitation and stored at 4 °C. Trypanosomes (107 cells in 20 μL cell wash buffer with 0.5 mM phenylmethylsulfonyl fluoride) were disrupted in a bath sonicator for 20 min. The lysate was centrifuged at ~16,000 g for 20 min at 4 °C, and soluble cell extracts were removed to a clean tube and used immediately or stored at −20 °C. Purified recombinant proteins or cell extracts of trypanosomes (both PF and BF) were subjected to 12% SDS-PAGE and Western blots were prepared by following standard protocols using Millipore ImmobilonP membrane. Protein bands were transferred at 4 °C either using 100 V for 1 h or 14 V overnight. Blots were probed with a 1:10,000 dilution of the rabbit anti-TLP5 antibody and a 1:30,000 dilution of goat anti-rabbit antibody conjugated to alkaline phosphatase from Sigma. Blots were developed using the BCIP/NBT liquid substrate system from Sigma.
2.6. Spectroscopy
UV/visible spectra were obtained by using an HP 8453 spectrophotometer (Hewlett Packard) equipped with circulating water bath and magnetic stirrer. TLP5 and TLP7 proteins were concentrated in Amicon centrifugal filter units with 10 kDa cutoff to 250 μM in binding buffer. Stock solutions of 10 mM FeII and 5 mM αKG were made by adding the dry reagents to vials, subjecting them to repeated cycles of vacuum and argon, and adding anaerobic water using a syringe. Protein was added to a 1-cm path length, 300 μL, black-walled cuvette and made anaerobic by the same method followed by scanning with stirring at 9 °C. Data were corrected for baseline shifts arising from cuvette repositioning to have a uniform absorbance at 900 nm by using the software IGOR.
2.7. Enzyme assays
Oxygen consumption assays were performed in a YSI Inc. model 5300 Clark-style biological oxygen monitor coupled to a circulating water bath to maintain a temperature of 33 °C. TLP5 or TLP7 was assayed at 1 μM in 25 mM imidazole, 100 mM KCl, and 5 mM MgCl2 using variations of the standard conditions: 50 μM FeII, 500 μM αKG, 400 μM ascorbate, and 500 μM of the potential substrates.
Succinate production assays utilized 1 μM enzyme that was incubated with 10 μM FeII, 10 μM ascorbate, and 100 μM αKG (and sometimes MgCl2) along with potential substrates in 25 mM Tris HCl, pH 8.0, at either 28 °C (PF conditions) or 37 °C (BF conditions) for 30 min. Aliquots (300 μL) were taken at various time points, quenched with 5 μL of 6 M sulfuric acid, centrifuged at 13,800 g for 5 min, passed through a 0.45 μm spin filter, and 200 μL was analyzed by Aminex HPX-87H (BioRad) ion-exchange chromatography (Waters, Corp.). Organic acids were eluted with 0.013 M H2SO4 as the mobile phase and detected using a refractive index detector (Waters, Corp.) set at 35 °C and a sensitivity level of 16. Standards of αKG and succinate eluted at 19 and 26 min, respectively, and succinate was detectable to a lower limit of 1 nmol.
Purified TLP5 and TLP7 (both at 10 μM), and trypanosome whole cell protein extracts from both the PF and BF life stages (from ~6 × 106 and ~5 × 106 cells, respectively) were incubated with thymine (250 μM) in 1 mM HEPES, pH 7.4, at 30 °C for 1 h with added αKG (500 μM) and ferrous ammonium sulfate (100 μM). Reactions were stopped by addition of hot ethanol, incubated at 60 °C for 5 min, and dried in a speed vac for 2h. Lyophilized pellets were resuspended in 100 μL anhydrous acetonitrile and silylated via treatment with 100 μL N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide + 1% tert-butyldimethylchlorosilane (Pierce) for 30 min at 135 °C. The solutions were analyzed by gas chromatography-mass spectrometry (GC-MS) using electrospray ionization on a 30 m DB-5MS, I.D. 0.25 column for analysis as previously described (Neidigh et al., 2009).
3. Results and discussion
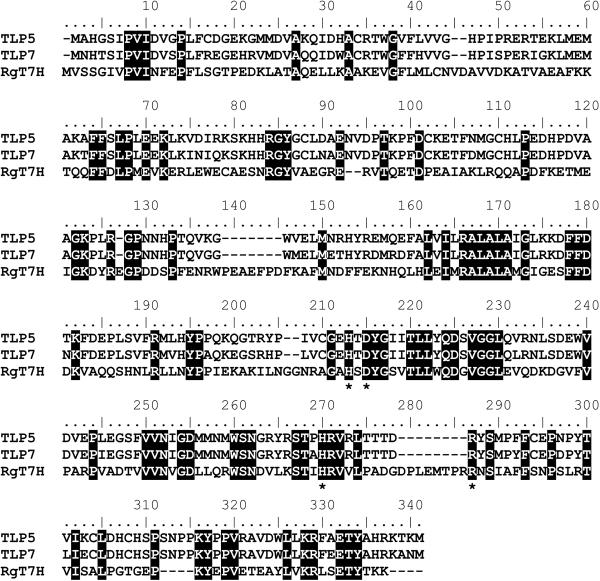
The T. b. brucei gene products TLP5 and TLP7 are approximately 30% identical to the fungal T7H sequence and are 85% identical to each other at the amino acid level (Fig. 1). All residues suspected to be critical to the active site are conserved, including those likely to coordinate the metal (His 200, Asp 202, and His 257 for both TLP5 and TLP7 numbering) and a residue that could stabilize binding of the αKG cosubstrate (Arg 266). The TLP5 and TLP7 sequences contain several short gaps (of 2, 7, 2, and 8 amino acid residues) compared to the fungal sequence, but the significance of these gaps, if any, cannot be ascertained because the structure of T7H is unknown.
Fig 1.
Sequence comparison of TLP5, TLP7, and T7H from R. glutinis. Identities are indicated by black shading. Putative metal- or αKG-binding residues are highlighted by asterisks.
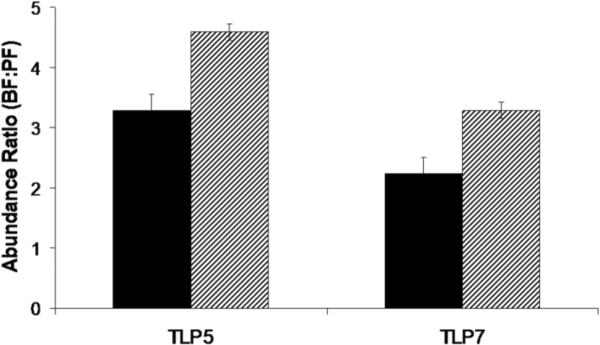
Quantitative real-time PCR methods were used to examine the steady-state expression levels of the two genes in both the BF and PF life stages of T. b. brucei. Due to the high sequence identity of TLP5 and TLP7 (87% at the DNA level), none of the available programs for primer selection in trypanosomes could be used. Therefore, gene sequences were manually analyzed and primers carefully chosen in order to be unique to each gene. Triplicate runs with mRNA containing TLP5 and TLP7 from T. b. brucei strain 427 (MiTat 1) (BF and PF), 427 clone 221 (BF), and 427 cell line 29–13 (PF) yielded similar results consistent with both genes being up-regulated in the BF parasite over the insect-form by approximately 4 and 2.5 fold, respectively (Fig. 2). This seemingly small effect is considered significant as only about 2% of genes in trypanosomes have been found to be regulated at least 2 fold at the level of mRNA abundance (Brems et al., 2005).
Fig. 2.
Quantitative real-time PCR analysis of TLP5 and TLP7 expression in BF- and PF-stages of T. b. brucei. The solid bars represent the fold increase of BF over PF in 427 (MITat-1) for the two genes. The hatched bars compare the fold increase of the BF 427 (221) over the PF 29–13. All data are normalized to tubulin internal controls that were included in each run.
TLP5 and TLP7 were PCR amplified out of T. b. brucei 29–13 genomic DNA and subcloned into pET28b for expression as His-tagged proteins in E. coli BL21 Gold (DE3). Sequencing of plasmids isolated from individual colonies revealed a number of differences from the published T. b. brucei TREU 927 sequence, most involving amino acid changes in non-catalytic areas of the protein. Of potential interest were a frame shift mutation in TLP5 and a nonsense mutation in TLP7 detected in multiple clones from independent PCR amplifications suggesting that one allele may be non-functional in this strain. This phenomenon may be due to an issue inherent to the trypanosomes being studied. The 29–13 cell line used in this study was developed from strain 427 which has been in laboratory culture for over 30 years and is now incapable of differentiating to the BF. Loss of the ability to differentiate back into infective blood forms may reflect the mutation of genes essential for function during this stage without conferring negative selection in culture.
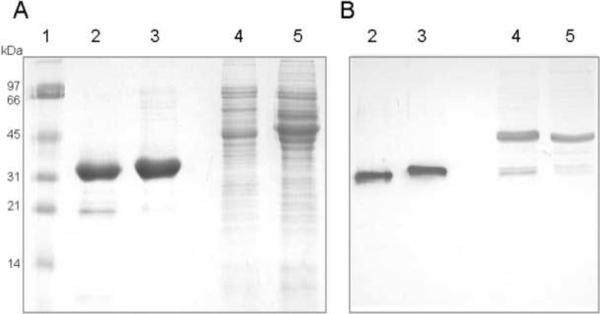
E. coli BL21 (DE3) cells containing the pET28b TLP5 and TLP7 derivatives were used as the source of purified TLP5 and TLP7, typically yielding 5 mg of protein per gram of wet cellular weight. The proteins were >95% homogeneous (Fig. 3A, lanes 2 and 3) and of the size predicted by the sequences (37.5 kDa when accounting for the His tag), with slight differences in electrophoretic mobility observed. The proteins behaved as monomers when examined by Blue Native gels (Invitrogen) or Sephadex 75 gel filtration chromatography (data not shown). Both proteins precipitated when exchanged into a low salt buffer after purification, but exchange back into the column binding buffer provided enhanced stability.
Fig 3.
Electrophoretic analysis of purified TLP5, TLP7, and extracts of PF and BF cells. Samples were subjected to SDS-PAGE and examined by (A) Coomassie staining or (B) Western blot analysis with anti-TLP5 antibodies. Lanes: 1, molecular weight ladder; 2, TLP5; 3, TLP7; 4, extracts of PF cells; 5, extracts of BF cells.
Polyclonal anti-TLP5 antibody was used to perform Western blots of purified recombinant TLP5 and TLP7 along with cell extracts of both PF and BF trypanosomes (Fig. 3B). The antibody detected both proteins, as expected from their close similarity. A major cross-reactive species in both BF and PF electrophoresed more slowly than the TLP5/TLP7 doublet; this species may be an unrelated contaminant or it might arise from post-translational modification of these proteins (leading to an apparent ~10-kDa increase in size) in the trypanosome, but lacking in the E. coli expression system. Future work could include efforts to identify this band and determine what, if any, modifications are added to the protein in vivo. In contrast to the RT-PCR results that indicate higher transcript levels in the BF cells, the immunological analysis suggests that TLP5/7 is more abundant in PF than BF cells; these results are compatible with the protein(s) being turned over more rapidly in the BF life stage or being translated at a greater rate in the PF life stage. Of primary importance, these results indicate that both proteins appear to be present in both life stages.
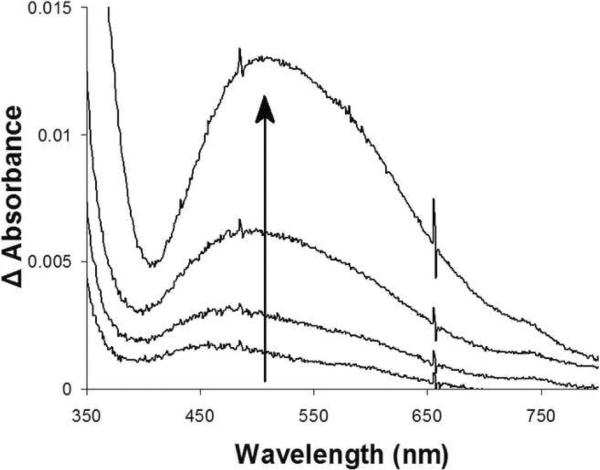
To confirm proper folding of TLP5 and TLP7, the purified proteins were examined for their ability to form a characteristic chromophore seen in other members of the FeII/αKG-dependent dioxygenases and associated with binding of the iron cofactor and the αKG cosubstrate (Ryle et al., 1999). The proteins were monitored by UV-visible spectroscopy under anaerobic conditions, using low temperature to maintain stability, while titrating in αKG and FeII. As shown in Figure 4, anaerobic TLP5 generated the diagnostic metal-to-ligand charge-transfer transition at 505 nm when both metal and cofactor were present. The extinction coefficient for this feature was approximately 40 mol−1 cm−1, indicating that the chromophore was incompletely formed compared with previously described intensities (Ryle et al., 1999), but addition of more metal ions led to protein precipitation. TLP7 also exhibited the diagnostic transition (data not shown), confirming correct folding of the protein, but it was less stable than TLP5 and precipitation during the experiment precluded estimation of the extinction coefficient.
Fig. 4.
Spectroscopic evidence for binding of FeII and αKG by TLP5. The anaerobic UV/visible spectrum of TLP5 (250 μM in binding buffer) was examined for the sample as isolated, after adding 1 mM αKG, and while titrating in FeII with stirring at 9 °C. The (FeII-αKG-protein minus αKG-protein) difference spectra shown correspond to the addition of 0.5, 1, 2, and 3 equivalents of metal ions.
Despite extensive efforts, no in vitro thymine hydroxylase activity was detected for purified recombinant TLP5 or TLP7 when assayed by using an oxygen electrode to monitor oxygen consumption, HPLC to assess succinate production, or GC-MS to measure thymine hydroxylation. We cannot exclude the possibility that a post-translational modification, not occurring in E. coli, is required for activity. It is also conceivable that the His-tag hinders activity in the recombinant proteins or that, despite formation of the FeII/αKG chromophore indicating appropriate folding of the active site domains, the full length proteins do not fold properly. On the other hand, these results are compatible with the proteins utilizing alternative substrates. Other members of the FeII/αKG dioxygenase family recognize a wide array of substrates ranging from small molecules to proteins and DNA (Simmons et al., 2008). Several additional small molecules were tested as putative substrates including other bases, nucleosides, and amino acids, but again no activity was detected. With regard to the possible use of DNA as a substrate, trypanosomes are known to contain the hyper-modified base J in their nuclear DNA (Borst and Sabatini, 2008, Gommers-Ampt et al., 1993), which requires hydroxylation of an unactivated methyl group of thymidine in DNA to create hydroxymethyl-deoxyuracil (Gommers-Ampt et al., 1993, Ulbert et al., 2002) that is subsequently glucosylated. Thus, we examined whether TLP5 or TLP7 reacted with polynucleotides or thymidine; however no such activity was observed. Furthermore, neither protein bound to linear DNA fragments under any conditions examined (incubation with or without 50 μM FeII, 150 μM αKG, and 100 μM MgCl2 at 37 °C for 30 min) according to gel band shift studies. Thus, TLP5 and TLP7 are unlikely to possess DNA modifying activity, consistent with the recent proposals that J-binding protein 1 (JBP1), identified on the basis of its ability to bind to J-containing chromatin, and JBP2, related to JBP1, are critically involved in base J synthesis (Cliffe et al., 2009, Cross et al., 1999, DiPaolo et al., 2005, Vainio et al., 2009, Yu et al., 2007).
Extracts of PF and BF cells were directly assayed for thymine hydroxylase activity by using GC-MS. No significant hydroxylase activity could be detected (i.e., less than 0.5% product formed from 250 μM thymine) after a 1 h incubation.
In conclusion, we've identified two genes encoding T7H-like proteins in T. b. brucei, demonstrated that the genes are expressed at greater levels in the BF over the PF stage, shown that both proteins are present in each life stage (possibly with greater abundance in the PF life stage), and expressed the genes in E. coli. We purified the two recombinant proteins, demonstrated that they form an FeII/αKG chromophore (indicating proper folding), and tested for catalytic activity and DNA binding. Our results provide no indication of the recombinant proteins being able to hydroxylate free thymine or thymidine within DNA or to bind DNA. Furthermore, no hydroxylase activity was detected in extracts from trypanosomes of either life stage. The roles of these proteins in T. b. brucei remain unknown, but our evidence provides strong arguments against their participation in pyrimidine salvage.
Acknowledgments
We thank Piotr Grzyska, William Kettleman, and Greg Moyerbrailean for assistance with several of these studies and Dr. Laurie Read for providing extracts of bloodstream form cells. This work was supported by NIH grants AI45835 (to D.J.K.) and GM063584 (to R.P.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. Lancet. 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 3.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UCM, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth T-J, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, MacLeod A, Mooney PJ, Moule S, Martin DMA, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream M-A, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CMR, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 4.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annual Review of Microbiology. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- 5.Brems S, Guilbride DL, Gundlesdodjir-Planck D, Busold C, Luu VD, Schanne M, Hoheisel J, Clayton C. The transcriptomes of Trypanosoma brucei Lister 427 and TREU927 bloodstream and procyclic trypomastigotes. Molecular and Biochemical Parasitology. 2005;139:163–172. doi: 10.1016/j.molbiopara.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Research. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, Borst P. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO Journal. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Molecular Cell. 2005;17:441–451. doi: 10.1016/j.molcel.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Gommers-Ampt JH, Teixeira AJ, van de Werken G, van Dijk WJ, Borst P. The identification of hydroxymethyluracil in DNA of Trypanosoma brucei. Nucleic Acids Research. 1993;21:2039–2043. doi: 10.1093/nar/21.9.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. β-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 11.Gudin S, Quashie NB, Candlish D, Al-Salabi MI, Jarvis SM, Ranford-Cartwright LC, de Koning HP. Trypanosoma brucei: A survey of pyrimidine transport activities. Experimental Parasitology. 2006;114:118–125. doi: 10.1016/j.exppara.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Holme E, Lindstedt G, Lindstedt S, Tofft M. 7-Hydroxylation of thymine in a Neurospora strain coupled to oxidative decarboxylation of 2-ketoglutarate. Biochimica et Biophysica Acta. 1970;212:50–57. doi: 10.1016/0005-2744(70)90177-4. [DOI] [PubMed] [Google Scholar]
- 13.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Neidigh JW, Darwanto A, Williams AA, Wall NR, Sowers LC. Cloning and characterization of Rhodotorula glutinus thymine hydroxylase. Chemical Research in Toxicology. 2009;22:885–893. doi: 10.1021/tx8004482. [DOI] [PubMed] [Google Scholar]
- 16.Randolph LB, Krug CE, Marr JJ. Purine and pyrimidine metabolism. In: Marr JJ, Muller M, editors. Biochemistry and Molecular Biology of Parasites. Academic Press; 1995. pp. 89–114. [Google Scholar]
- 17.Ryle MJ, Padmakumar R, Hausinger RP. Stopped-flow kinetic analysis of Escherichia coli taurine/α-ketoglutarate dioxygenase: interactions with α-ketoglutarate, taurine, and oxygen. Biochemistry. 1999;38:15278–15286. doi: 10.1021/bi9912746. [DOI] [PubMed] [Google Scholar]
- 18.Shaffer PM, Arst HN. Regulation of pyrimidine salvage in Aspergillus nidulans: a role for the major regulatory gene areA mediating nitrogen metabolite repression. Molecular and General Genetics. 1984;198:139–145. doi: 10.1007/BF00328713. [DOI] [PubMed] [Google Scholar]
- 19.Simmons JM, Müller TA, Hausinger RP. FeII/α-ketoglutarate hydroxylases involved in nucleobase, nucleoside, nucleotide, and chromatin metabolism. Dalton Transactions. 2008;38:5132–5142. doi: 10.1039/b803512a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiley JA, Kundracik M, Landfried DA, Barnes VR, Sr., Axhemi AA. Genes of the thymidine salvage pathway: thymine-7-hydroxylase from a Rhodotorula glutinis cDNA library and iso-orotate decarboxylase from Neurospora crassa. Biochimica et Biophysica Acta. 2005;1723:256–264. doi: 10.1016/j.bbagen.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Ulbert S, Cross M, Boorstein RJ, Teebor GW, Borst P. Expression of the human DNA glycosylase hSMUG1 in Trypanosoma brucei causes DNA damage and interferes with J biosynthesis. Nucleic Acids Research. 2002;30:3919–3926. doi: 10.1093/nar/gkf533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vainio S, Genest PA, ter Riet B, van Luenen H, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base J. Molecular and Biochemical Parasitology. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 23.van den Born E, Omelchenko MV, Bekkelund A, Leihne V, Koonin EV, Dolja VV, Falnes PO. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Research. 2008;36:5451–5456. doi: 10.1093/nar/gkn519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HG, Rigden DJ, Sabatini R, Borst P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Research. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]