Abstract
The conserved UL16 tegument protein of herpes simplex virus exhibits dynamic capsid-binding properties with a release mechanism that is triggered during initial virus attachment events. In an effort to understand the capsid-association and subsequent release of UL16, we sought to define the mechanism by which this protein is packaged into virions. The data presented here support a model for the addition of some UL16 to capsids prior to their arrival at the TGN. UL16 was found on capsids isolated from cells infected with viruses lacking UL36, UL37 or gE/gD, which are defective for budding and accumulate nonenveloped capsids in the cytoplasm. Additionally, membrane-flotation experiments showed that UL16 co-purified with cytoplasmic capsids that are not associated with membranes. Moreover, the amount of UL16 packaged into extracellular particles was severely reduced in the absence of two conserved binding partners, UL21 or UL11.
Keywords: Herpes simplex, UL16, virus assembly, capsid, HSV
Introduction
The tegument of herpesviruses is comprised of an assortment of proteins found between the viral envelope and the DNA-filled capsid. This compartment contains more than 15 different virally-encoded proteins for which many of their functions remain unknown. Much effort has been placed on elucidating the molecular interactions between various tegument proteins with the hope of understanding the mechanisms by which this complex virus is assembled (Mettenleiter et al., 2006). Tegument addition is thought to originate in the nucleus, then more tegument proteins are added to capsids in the cytoplasm following nuclear egress, and finally at trans-Golgi network (TGN)-derived membranes during maturation budding (Mettenleiter, 2006). The goal of the experiments described here was to define where the conserved tegument protein UL16 is packaged into the virus particle along this assembly pathway, and to determine which protein-protein interactions may contribute.
UL16 has been shown to be present in the nucleus and cytoplasm during an infection (Nalwanga et al., 1996); however, it has only been detected on capsids purified from the cytoplasm (Meckes, Jr. & Wills, 2007). Recently, we showed that UL16 has different capsid-binding properties in the virion compared to capsids isolated from the cytoplasm (Meckes, Jr. & Wills, 2007). Specifically, the majority of UL16 is removed from extracellular capsids following NP40 treatment, but is bound to capsids isolated from the cytoplasm of cells using similar lysis conditions (Meckes, Jr. & Wills, 2007). These data suggest that the interaction may be changed or destabilized during virus egress, possibly as a maturation mechanism to prepare the virus for entry into the next cell (Meckes, Jr. & Wills, 2008).
The actual function of UL16 for any herpesvirus remains a mystery, but it has been speculated to provide a bridging function between the capsid and the membrane during budding events through its interactions with the capsid and the membrane-bound tegument protein UL11 (Loomis et al., 2003). UL11 is a small (~15 kDa) myristylated and palmitylated protein which accumulates at TGN-derived vesicles in the absence of other viral proteins (Loomis et al., 2001). Approximately 700 molecules of UL11 are packaged into virus particles independently of its interaction with UL16 (Loomis et al., 2006). UL16 also interacts with another conserved tegument protein named UL21 that associates with capsids and microtubules (de Wind et al., 1992;Takakuwa et al., 2001;Klupp et al., 2005). These findings suggest a possible role for UL21 in capsid transport to TGN-derived vesicles. Prior to the experiments presented here, it seemed probable that UL16 was attached to capsids through an interaction with the capsid-bound UL21. Capsids containing UL21 and UL16 could then interact with membranes through the UL11 protein. Consistent with this simple model of capsid transport and budding, herpes viruses lacking UL16, UL21, or UL11 (or their homologs) have defects in virus egress compared to wild type, resulting in decreased amounts of extracellular virus produced and the accumulation of non-enveloped capsids in the cytoplasm (Baines & Roizman, 1991;Baines & Roizman, 1992;Britt et al., 2004;Kopp et al., 2004;Kopp et al., 2003;MacLean et al., 1989;MacLean et al., 1992;Schimmer & Neubauer, 2003;Silva et al., 2005;Silva et al., 2003;Baines et al., 1994;Guo et al., 2009).
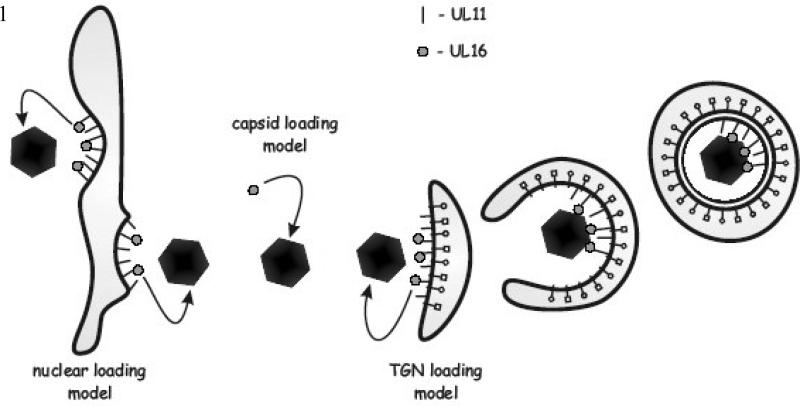
Based on the available data, there were at least three possible mechanisms of UL16 packaging into virus particles during assembly (Fig. 1). In the first, UL16 binds to nuclear capsids either prior to or during nuclear egress (nuclear-loading model). In the second model, UL16 attaches to DNA-containing C-capsids in the cytoplasm prior to their arrival at the TGN for maturation budding (capsid-loading model). Subsequent interaction of UL16 with UL11 would fasten capsids to the membrane and drive the budding process. In the third mechanism of packaging, UL16 is transported to the TGN independently of capsids, likely through an interaction with UL11 (TGN-loading model). When capsids arrive at the membrane, their binding to UL16-UL11 complexes would then promote budding. The purpose of the experiments described here was to test these loading models for UL16 packaging.
FIG. 1.
Potential UL16 packaging mechanisms. Mature DNA containing C-capsids exit the nucleus of infected cells via a budding then fusion mechanism across the nuclear envelope. It is at this step during the assembly process that UL16 could be loaded onto capsids through its direct interaction with the membrane-bound UL11 protein that is present on the nuclear envelope (nuclear loading model). Following exit from the nucleus, non-enveloped capsids transverse the cytoplasm and provide another potential location for the incorporation of UL16 (capsid loading model), possibly through an interaction with the capsid-bound UL21 protein. And finally, cytoplasmic capsids reach TGN-derived membranes where the final budding event is orchestrated. It is here that capsids acquire additional tegument proteins and their full complement of glycoproteins. Since UL11 targets to these membranes, it could bring UL16 to this location for packaging into virus particles (TGN loading model).
Results
UL16 is added to nucleocapsids prior to their arrival at the TGN
ULl6 has been shown to be associated with capsids purified from the cytoplasm, which suggested that the capsid-loading model was a potential mechanism of UL16 incorporation into virions (Meckes, Jr. & Wills, 2007). However, since these capsids include ones from cytoplasmic enveloped virions that are stripped of their membranes by the detergent-lysis conditions, it was unclear whether UL16 was added to capsids prior to maturation budding.
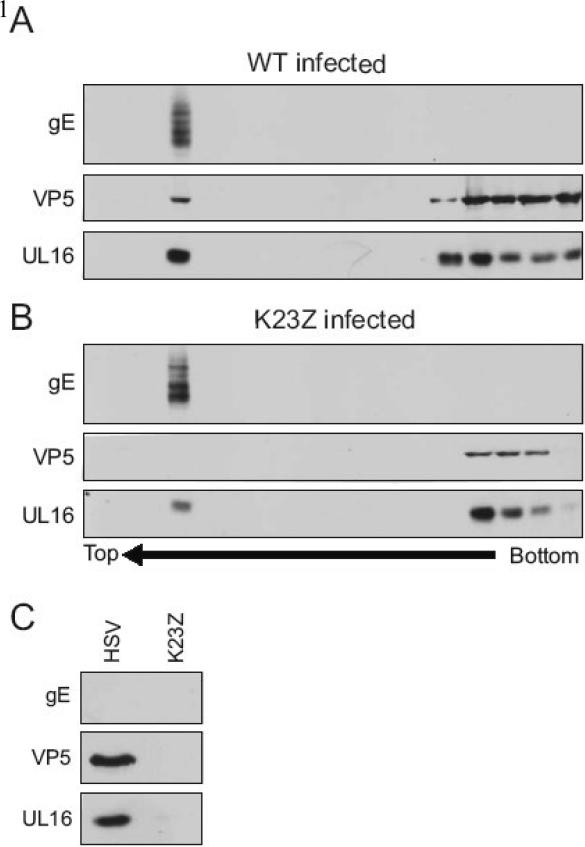
To separate enveloped from non-enveloped capsids, a membrane floatation assay was employed following osmotic lysis of infected cells (Baird et al., 2008). Briefly, the cells were lysed by dounce homogenization, and the cytoplasmic fraction was layered on the bottom of a sucrose step gradient. Then, the gradients were centrifuged for 18 h to separate membranes (and associated proteins) from the non-membrane bound proteins, which remain in the bottom fractions. In this assay, enveloped virions will float to the top of the gradient (Baird et al., 2008); and as expected, the major capsid protein (VP5), UL16, and glycoprotein E (gE) were found in this fraction (Fig. 2A). If enveloped virions were trapped in the bottom fractions, then we would expect the trans-membrane viral glycoprotein to be found there, but this was not the case (gE panel, bottom fractions). VP5 and UL16 were found in the bottom fractions of the gradient where you would expect soluble, non-membrane associated proteins to remain during centrifugation. As a control, we analyzed the cytoplasm of cells infected with a mutant virus that lacks the triplex protein VP23 (designated K23Z), which does not make capsids and therefore cannot produce enveloped particles (Desai et al., 1993). Despite the inability to detect capsids in the membrane-floating fraction (VP5 panel, top fractions), a population of UL16 was found there which implies that during an infection UL16 can associate with membranes in the absence of capsid budding events (Fig. 2B). This membrane association is likely due to an interaction with membrane-bound viral proteins (e.g., UL11), since UL16 was not detected in the floating fraction of UL16 transfected cells (data not shown). Consistently less VP5 (and UL16) was found in the bottom-most pellet fraction of K23Z compared to wild-type gradients (Fig. 2A and B, rightmost lane). This is most likely due to the inability of the K23Z mutant virus to construct capsids, which would remain in the bottom fraction in this assay. The overall reduced levels of VP5 in the K23Z gradient are likely due to degradation since the protein is not assembled into stable capsids. In contrast, the levels of gE remain the same for both the wild-type and mutant, as expected since equal amounts of infected cells were lysed and loaded on the bottom of each gradient.
FIG. 2.
UL16 is associated with non-membrane-bound capsids. Cells were infected with (A) HSV or a (B) mutant that does not make capsids (K23Z) for 20 h and mechanically lysed in a hypotonic buffer. Lysates were then loaded on the bottom of a sucrose step gradients, and centrifuged for 18 h to separate membranes (top) from non-membrane bound proteins (bottom). Fractions (800 μl) were collected from the top of the gradient, proteins in the fractions were then TCA-precipitated, separated by SDS-PAGE in 10% gels and analyzed by immunoblot with antibodies specific for gE, VP5, and UL16. (C) The bottom two non-membrane associated fractions from the gradients were incubated in NP-40 lysis buffer for 15 min. Capsids in these fractions were subsequently pelleted through a 30% (wt/vol) sucrose cushion and analyzed by immunoblot.
To analyze capsids that are not associated with membranes, we combined the two bottom-most fractions from the sucrose gradient, treated them with NP-40 lysis buffer, and re-pelleted the capsids through a 30% sucrose cushion. UL16 was readily found co-purifying with non-membrane-bound cytoplasmic capsids from cells infected with wild-type virus, but it did not pellet in the case of K23Z, for which there is an absence of capsids (Fig. 2C). It is possible that the lysis conditions used in these experiments could disrupt nuclei, releasing wild-type capsids into the cytoplasmic fraction. If this were the case, the data presented would represent an underestimate for the amount of UL16 relative to capsids since UL16 is not found on capsids purified from nuclei (Meckes, Jr. & Wills, 2007).
UL16 is present on capsids that are unable to perform secondary envelopment
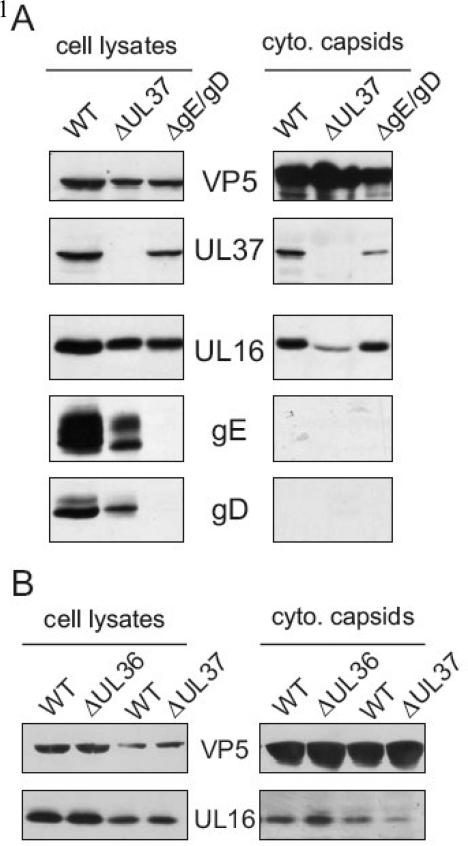
Although the membrane floatation experiments support the capsid loading model, the possibility remained that capsids may have interacted with membranes and “fallen” off during the disruption process. Therefore, to test the capsid-loading model with a different approach, cytoplasmic capsids were sucrose gradient purified from cells infected with mutant viruses that are blocked for maturation budding (Farnsworth et al., 2003;Desai, 2000;Desai et al., 2001). UL16 was readily detectible with capsids purified from the cytoplasm of cells infected with gE-/gD- and UL37-null viruses (Fig. 3A, right panels). To test for the purity of the mutant virus preparations, immunoblot analysis for gE, gD, and UL37 was performed with portions of the cell lysates (Fig. 3A, left panels) and by comparing virus titers on complementing and non-complementing cells (data not shown). Typically, virus titers were down three to five logs when grown on non-complementing Vero cells. Capsids lacking UL36, another tegument protein critical for final envelopment, also contained equivalent amounts of UL16 compared to WT capsids (Fig. 3B, right panels). Unfortunately, we were unable to detect the presence of UL36 in cell lysates from wild-type or mutant virus infected cells by immunoblotting with a rabbit polyclonal antibody (Bucks et al., 2007); however, the virus could only be propagated on UL36 expressing cells (data not shown), confirming that the defect in assembly this virus exhibited was due to the absence of the UL36 gene (Desai, 2000).
FIG. 3.
UL16 is bound to cytoplasmic capsids that are blocked in maturation budding. Cytoplasmic capsids from detergent lysed (A) WT, ΔUL37, or gE-/gD- infected cells were sedimented in 20 to 50% (wt/vol) sucrose gradients for 1 h. Fractions containing capsid bands were pulled with a needle syringe and pelleted through another 30% sucrose cushion. Purified capsids and associated proteins were separated by SDS-PAGE in 10% gels and analyzed by immunoblot with anti-VP5, anti-UL37, anti-UL16, anti-gE, and anti-gD rabbit serum. (B) ΔUL36 and ΔUL37 capsids were isolated from infected cells in parallel with WT as described above. cyto, cytoplasmic. WT, wild-type.
Unlike the capsids harvested from the gE-/gD- and UL36-null viruses, which contained roughly equivalent levels of UL16 compared to WT capsids, the amount of UL16 on ΔUL37 capsids relative to WT was somewhat reduced (42% ± 33, Fig. 3A and B). This variability may be due to the fact that the ΔUL37 virus accumulates large clusters of capsids in the cytoplasm and nucleus of infected cells, which might prevent them from acquiring normal amounts of tegument proteins. However, it does remain a possibility that some UL16 may be added to capsids through an interaction with UL37 or one of its binding partners since in the absence of UL37, less UL16 was detected on these capsids. Taken together, these data provide further evidence that at least some UL16 is added to capsids prior to their arrival at the TGN for maturation budding.
UL16 is packaged independently of its interaction with UL11 or UL21
UL16 is a direct binding partner of the conserved tegument protein UL11 (Yeh et al., 2008;Lee et al., 2008;Vittone et al., 2005;Loomis et al., 2003). During our work on the UL11-UL16 interaction in HSV, an additional 65 kDa protein consistently purified with these proteins (Loomis et al., 2003), and it has been identified as UL21 by mass spectrometry. Detailed studies have shown that UL16 directly interacts with UL21 (Harper et al., unpublished results). To ascertain the importance of UL11 and UL21 for UL16 packaging, mutant viruses lacking their individual genes where analyzed.
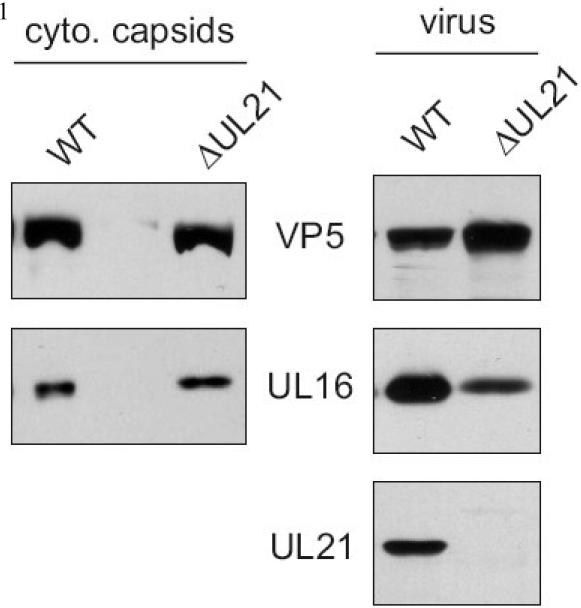
Based on published results defining UL21 as a capsid-bound protein (de Wind et al., 1992), it was anticipated that this protein might link UL16 to capsids. However, when capsids were purified from cells infected with a UL21-null virus, UL16 was detected at equivalent levels compared to wild-type capsids (Fig. 4, left panels). If there is only one mechanism of UL16 packaging (e.g., through a capsid interaction), then equal levels of UL16 might be expected in extracellular UL21-null virions, but this was not the case. Analysis of 3 independent experiments revealed a decrease of 80% (±13%) in the amount of UL16 packaged into this virus compared to wild-type virus (Fig. 4, right panels). While this does not prove that multiple incorporation pathways are used (see Discussion), it does show that UL21 is somehow critical for efficient incorporation of UL16 into assembling particles but not for cytoplasmic capsid association.
FIG. 4.
UL16 is packaged and associates with cytoplasmic capsids in the absence of UL21. Cytoplasmic capsids from detergent lysed HSV or ΔUL21 infected cells were sedimented in a 20 to 50% (wt/vol) sucrose gradient for 1 h. Regions of the gradient containing capsid bands were isolated with a syringe and then pelleted through a 30% (wt/vol) sucrose cushion. Virions present in the media were sucrose gradient purified as described. Virions and capsids with their associated proteins were separated by SDS-PAGE in 10% gels and analyzed by immunoblot with VP5 and UL16 specific antibodies. cyto, cytoplasmic.
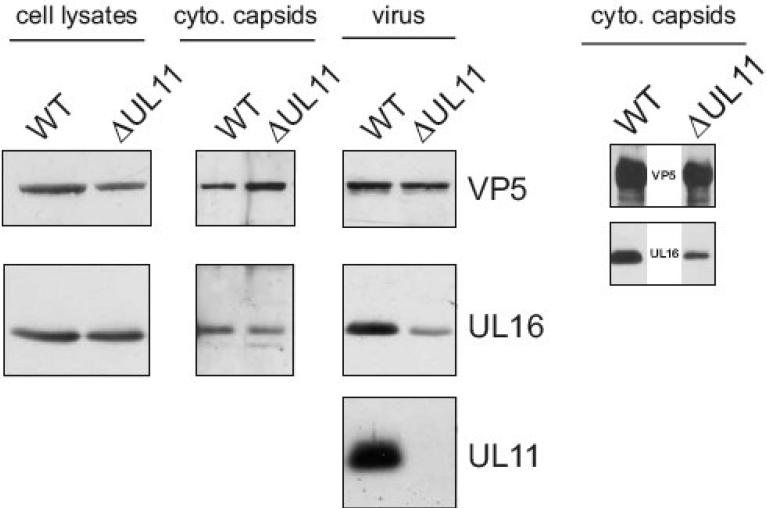
To test whether the UL11-UL16 interaction is required for the incorporation of UL16, virus particles lacking the UL11 protein were purified from the extracellular media and analyzed for the presence of UL16 (Baines & Roizman, 1992). The amount of UL16 packaged into UL11-null virions from three independent experiments was down 70% (±16%) when compared to WT virus (Fig. 5, virus panels). These results were expected based on published results with PRV, which demonstrated a decreased amount of UL16 packaged into UL11-null virions (Klupp et al., 2005). But, unlike the results obtained with the UL21-null virus (Fig. 5), a decreased level of UL16 was also detected on cytoplasmic capsids when normalized for VP5 levels and compared to WT (72% ± 7, Fig. 5, cyto. capsids panels). Since the UL16 expression level was normal in the absence of UL11 (Fig. 5, cell lysate panels), these results also demonstrate that the presence of UL16 on the wild-type cytoplasmic capsids is not an artifact of the detergent lysis conditions used throughout these studies.
FIG. 5.
An interaction with UL11 is not essential for UL16 packaging. WT or ΔUL11 extracellular virus and cytoplasmic capsids were purified in 20-50% (wt/vol) sucrose gradients and the region of the gradient containing the virions and capsids were pulled with a needle syringe and pelleted through a sucrose cushion. Cell lysates, purified capsids, and virions were then analyzed for the presence of VP5, UL16, and UL11 by immunoblot. cyto, cytoplasmic.
Discussion
The experiments described in this report substantiate the capsid-loading model, where a population of UL16 is added to capsids prior to their arrival at the TGN for maturation budding. However, two observations suggest that UL16 may be loaded onto capsids even as they are budding into, or exiting from, the nuclear membrane (Fig. 1, nuclear-loading model). First, the UL11-binding partner of UL16 has been shown to be present on nuclear membranes (Loomis et al., 2001;Baines et al., 1995), raising the possibility that UL16 molecules could be recruited there. Second, a decreased amount of UL16 was found on cytoplasmic capsids harvested from cells infected with the UL11-null mutant (this report). Whatever the timing of addition, interactions of UL16 with tegument proteins UL11 and UL21 are not absolutely required for capsid binding; however, in the absence of either protein, a severe decrease was observed in the amount of UL16 packaged into virions.
There are two entirely different—and difficult to resolve—molecular models that could explain why the packaging levels for UL16 are reduced in the absence of UL11 or UL21. In the first, low levels of UL16 are added to cytoplasmic capsids but additional molecules are added at the TGN, where complexes reside that contain UL11, UL21, and UL16 (Fig. 1, TGN-loading model). In the second model (not illustrated), all of the UL16 destined for the virion is loaded onto capsids prior to their reaching the TGN, but an absence of UL11 at the membrane or UL21 on the capsid eliminates a regulatory mechanism, causing UL16 to be prematurely destabilized and lost from capsids during budding. Thus, similar to the construction of a machine, if one component is missing during the assembly process, then other components may not be held in place properly during subsequent steps. In support of this regulatory hypothesis, we recently demonstrated that the UL16-capsid interaction is destabilized during capsid egress (Meckes, Jr. & Wills, 2007;Meckes, Jr. & Wills, 2008). Moreover, extensive studies of PRV have shown that viruses lacking UL11, UL16, and UL21 contain multiple changes in their tegument composition (Klupp et al., 2005;Michael et al., 2006;Michael et al., 2007).
Discovering what UL16 is bound to on the capsid would provide one more piece of this machine that is important for the capsid release mechanism. It is clear from the data presented here that the inner tegument proteins UL21, UL36 and UL37 are not required for UL16 to associate with capsids, although the presence of UL37 may have a minor role. Since the block in the UL36- and UL37-null viruses is at the stage of secondary envelopment, at least some UL16 must be added to capsids prior to maturation budding and subsequent steps in the assembly process may lead to the destabilization of UL16 on the capsid. These data also suggest a potential linkage through a tegument/capsid protein other than UL37 and UL36 or a protein(s) that associates with them. Thus, a yet-to-be identified protein must link UL16 to capsids. Recent data by Lee et al. may help explain why UL16 still associates with capsids even in the absence of UL36 or UL37 (Lee et al., 2008). In their study the authors identified interactions of UL16 with the major capsid protein (VP5) and a minor capsid protein (VP26) using a yeast two-hybrid assay (Lee et al., 2008). These potential interactions have not been confirmed using other methods or demonstrated in the context of an infection. However, if these interactions do indeed occur on the capsid during egress, then UL16 would bind directly to the capsid shell through an interaction with VP26 or VP5. The most direct way to identify what UL16 is bound to on the capsid would be through the use of chemical crosslinkers and mass spectrometry. Additionally, the analysis of UL16 mutants for their ability to bind and be released from capsids will likely provide valuable information into the packaging and release mechanisms.
In summary, the experiments described here provide another example of a tegument protein that is added to capsids prior to them reaching TGN vesicles (Bucks et al., 2007;Naldinho-Souto et al., 2006;Read & Patterson, 2007;Padula et al., 2009). The discovery that UL11 and UL21 likely participate in additional mechanisms of UL16 packaging further emphasizes the complex network of interactions that takes place for the proper construction of the tegument. Deciphering exactly where each tegument protein is acquired during egress and which protein interactions are necessary is critical to obtaining a better understanding of the herpesvirus assembly process.
Materials and Methods
Virus and cells
Vero cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS), 5% bovine calf serum (BCS), penicillin and streptomycin (Gibco 15140-148). The KOS strain of HSV-1 used in these studies was grown in Vero cells with DMEM supplemented with 2% FBS, 25 mM HEPES buffer, glutamine (0.3 ug/ml), penicillin, and streptomycin. The HSV-1 VP23, UL36, and UL37 deletion viruses and complementing cells C32, HS30, and BD45, respectively, were kindly provided by Prashant Desai (Johns Hopkins University). The complementing cells were grown in DMEM supplemented 1mg/mL of G418 as previously described (Desai, 2000;Desai et al., 2001). The gE-/gD- virus and complementing gD expressing cells (VD60) were a gift from David Johnson (Oregon Health and Science Center) (Farnsworth et al., 2003). The VD60 cells were maintained in DMEM lacking histadine (Hyclone) and supplemented with 10% FBS and 1.0 mM histidinol (Sigma H6647) as described previously (Farnsworth et al., 2003). HSV mutant viruses containing deletions of UL11, UL21, and UL16 were generous gifts of Joel Baines (Cornell University) (Baines & Roizman, 1992;Baines & Roizman, 1991;Baines et al., 1994).
Antibodies
UL16, UL11, UL36, gD, and VP5-specific antibodies were produced in rabbits and have been described previously (Loomis et al., 2001;Loomis et al., 2003;McNabb & Courtney, 1992;Baird et al., 2008). Rabbit polyclonal antibody raised against gE was a kind gift from Harvey Friedman (University of Pennsylvania) (Saldanha et al., 2000). Rabbit polyclonal antibody generated against the UL37 protein was generously provided by Frank Jenkins (University of Pittsburgh) (Schmitz et al., 1995).
Membrane floatation assay
Membrane-associated proteins were separated from non-membrane-bound proteins using a previously established protocol (Brignati et al., 2003;Baird et al., 2008). Briefly, five confluent 100 mm dishes of HSV infected or UL16-GFP transfected Vero cells were harvested in cold PBS at 20 h post-infection or post-transfection, respectively. Cells were pelleted at 1,000 × g for 5 min at 4°C, resuspended in hypotonic lysis buffer (10 mM Tris–HCl, pH 7.4, 0.2 mM MgCl2) with protease inhibitors, and incubated 30 min on ice. Following incubation, cells were lysed by dounce homogenization (35 strokes). Unbroken cells and nuclei were pelleted at 1,000 × g for 10 min at 4°C. The supernatants (.3 ml) were mixed with 2.7 ml of 65% wt/wt sucrose in TNE, placed in the bottom of a Beckman SW41 Ti tube, and sequentially overlayed with 45% (7 ml) and 2.5% (2 ml) wt/wt sucrose. The resulting sucrose step gradient was centrifuged at 100,000 × g for 18 h at 4°C in a Beckman SW41 rotor. 800 μl fractions were taken from the top using a piston gradient fractionator (Brandel). Trichloroacetic acid (TCA) was added to each fraction at final concentration of 13%, and the samples were incubated overnight at 4°C. The precipitated proteins were collected by centrifugation in a microcentrifuge at 18,000 × g for 30 min, washed with 100% ethanol, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (3.5% SDS, 8.5% β-mercaptoethanol, 130 mM dithiothreitol, 0.5 M urea, 290 mM Tris-HCl, pH 8.8), and boiled for 15 min at 95°C.
To analyze non-membrane bound capsids, the bottom two fractions from the sucrose step gradient were combined, and treated with NP-40 (.5% final). Capsids were then pelleted through a 30% wt/vol sucrose cushion and analyzed by SDS-PAGE and immunoblot.
Capsid analysis
Capsids were harvested from the cytoplasm of infected cells as previously described (Meckes, Jr. & Wills, 2007). Briefly, twenty 100 mm plates of confluent Vero cells were infected at a MOI of 5. At 20 to 22 h post-infection, cells were scraped into 20 ml of phosphate buffered saline (PBS), collected by centrifugation at 1,000 × g for 10 min, resuspended in 6 ml of NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl pH 8.0) containing protease inhibitors (Sigma, P8340), and incubated for 15 min on ice. The cytoplasmic fraction was separated from the nuclei by centrifugation at 1,000 × g for 10 min. Insoluble material from the cytoplasmic fractions was cleared by centrifugation at 8,000 × g for 30 min. The capsids remaining in the soluble supernatant were pelleted through a 1.7-ml 30% (wt/vol in TNE; 20 mM Tris-HCl pH 7.6, 150 mM NaCl, 1mM EDTA) sucrose cushion in a SW41 rotor at 83,500 × g for 1 h. Pellets were resuspended in 500 μls of TNE, sonicated for 2 min at moderate power, layered onto a 20 to 50% (wt/vol sucrose in TNE) continuous gradient, and centrifuged at 74,000 × g for 1 h in a SW41 rotor. The center fraction (4 mL) of the gradient containing the light scattering capsid bands was pulled with a needle syringe, diluted with 6 ml of PBS and repelleted through a 30% sucrose cushion to concentrate the capsids. All centrifugation steps were carried out at 4°C. The purified capsids were dissolved in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (62.5 mM Tris-HCl pH 6.8, 2% SDS, 5% BME, 50mM DTT, .0025% bromophenol blue, 10% glycerol), and boiled for 5 min at 95°C. Samples were separated in SDS-10% polyacrylamide gels and electrotransferred to nitrocellulose membranes. The enhanced chemiluminescence (ECL) method of immunoblot analysis was performed according to the manufacturer's instructions (Amersham). Anti-UL16 and anti-VP5 were used as the primary antibodies at dilutions of 1:3,000 and 1:7,500 (in 1% nonfat milk in TBS-T [20mM Tris pH 7.6, 135mM NaCl, .1% Tween 20]), respectively.
Analysis of deletion viruses
Due to the inefficiency of virus release with UL11- and UL21-null mutants (Baines & Roizman, 1992;Baines et al., 1994), confluent monolayers of Vero cells were infected with a low MOI (.01) and incubated until complete cytopathic effect was visualized (4-5 days). Infected cells and media were collected and centrifuged for 5 min at 3,828 × g. Virions from the extracellular supernatant were then pelleted through a 30% sucrose cushion in an SW32 rotor for 1 h at 83,500 × g. The resulting pellets were resuspended overnight at 4°C in TNE, sonicated for three 1 min pulses in an ice-water bath at moderate power, and sucrose gradient (20 to 50%) purified in a SW41 rotor for 1 h at 74,000 × g. The region of the gradient containing the virion band was pulled with a needle syringe (4 ml), diluted to 10 ml with PBS, and re-pelleted through an additional 30% sucrose cushion in a SW41 rotor. The purified virus pellets were dissolved in sample buffer and analyzed by immunoblot as described above. Using densitometry, the amount of UL16 within virions was normalized to VP5 and represented as a percentage of WT packaging (also normalized to VP5).
Acknowledgments
Special thanks to Todd Wisner (David Johnson's Laboratory) and Elizabeth Wills (Joel Baines’ Laboratory) for providing technical help on growing mutant viruses used in these studies. Additional thanks to Nicholas Baird, Jun Han, Michael Murphy and Michelle Bucks for helpful discussions and to Pooja Chadha for critical review of this manuscript. This work was supported by a NIH grant to J.W.W. (AI071286). D.G.M. was supported by a training grant from the NIH (CA60395).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baines JD, Jacob RJ, Simmerman L, Roizman B. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Koyama AH, Huang T, Roizman B. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J. Virol. 1994;68:2929–2936. doi: 10.1128/jvi.68.5.2929-2936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NL, Yeh PC, Courtney RJ, Wills JW. Sequences in the UL11 tegument protein of herpes simplex virus that control association with detergent-resistant membranes. Virology. 2008;374:315–321. doi: 10.1016/j.virol.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignati MJ, Loomis JS, Wills JW, Courtney RJ. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 2003;77:4888–4898. doi: 10.1128/JVI.77.8.4888-4898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 2004;78:539–543. doi: 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology. 2007;361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N, Wagenaar F, Pol J, Kimman T, Berns A. The pseudorabies virus homology of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J. Virol. 1992;66:7096–7103. doi: 10.1128/jvi.66.12.7096-7103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, DeLuca NA, Glorioso JC, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P, Sexton GL, McCaffery JM, Person S. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 2001;75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth A, Goldsmith K, Johnson DC. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 2003;77:8481–8494. doi: 10.1128/JVI.77.15.8481-8494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wang L, Peng L, Zhou ZH, Deng H. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J. Virol. 2009;83:10582–10595. doi: 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Bottcher S, Granzow H, Kopp M, Mettenleiter TC. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 2005;79:1510–1522. doi: 10.1128/JVI.79.3.1510-1522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M, Granzow H, Fuchs W, Klupp B, Mettenleiter TC. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 2004;78:3024–3034. doi: 10.1128/JVI.78.6.3024-3034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M, Granzow H, Fuchs W, Klupp BG, Mundt E, Karger A, Mettenleiter TC. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 2003;77:5339–5351. doi: 10.1128/JVI.77.9.5339-5351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology. 2008;378:347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Loomis JS, Bowzard JB, Courtney RJ, Wills JW. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 2001;75:12209–12219. doi: 10.1128/JVI.75.24.12209-12219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis JS, Courtney RJ, Wills JW. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 2003;77:11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis JS, Courtney RJ, Wills JW. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 2006;80:10534–10541. doi: 10.1128/JVI.01172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean CA, Clark B, McGeoch DJ. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 1989;70(Pt 12):3147–3157. doi: 10.1099/0022-1317-70-12-3147. [DOI] [PubMed] [Google Scholar]
- MacLean CA, Dolan A, Jamieson FE, McGeoch DJ. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 1992;73(Pt 3):539–547. doi: 10.1099/0022-1317-73-3-539. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Courtney RJ. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 1992;190:221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- Meckes DG, Jr., Wills JW. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 2007;81:13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes DG, Jr., Wills JW. Structural rearrangement within an enveloped virus upon binding to the host cell. J. Virol. 2008;82:10429–10435. doi: 10.1128/JVI.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet.Microbiol. 2006;113:163–169. doi: 10.1016/j.vetmic.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: a tale of two membranes. Curr.Opin.Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Michael K, Bottcher S, Klupp BG, Karger A, Mettenleiter TC. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J. Gen. Virol. 2006;87:3503–3507. doi: 10.1099/vir.0.82168-0. [DOI] [PubMed] [Google Scholar]
- Michael K, Klupp BG, Karger A, Mettenleiter TC. Efficient incorporation of tegument proteins pUL46, pUL49, and pUS3 into pseudorabies virus particles depends on the presence of pUL21. J. Virol. 2007;81:1048–1051. doi: 10.1128/JVI.01801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldinho-Souto R, Browne H, Minson T. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 2006;80:2582–2584. doi: 10.1128/JVI.80.5.2582-2584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalwanga D, Rempel S, Roizman B, Baines JD. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology. 1996;226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- Padula ME, Sydnor ML, Wilson DW. Isolation and preliminary characterization of herpes simplex virus 1 primary enveloped virions from the perinuclear space. J. Virol. 2009;83:4757–4765. doi: 10.1128/JVI.01927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read GS, Patterson M. Packaging of the virion host shutoff (Vhs) protein of herpes simplex virus: two forms of the Vhs polypeptide are associated with intranuclear B and C capsids, but only one is associated with enveloped virions. J. Virol. 2007;81:1148–1161. doi: 10.1128/JVI.01812-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha CE, Lubinski J, Martin C, Nagashunmugam T, Wang L, van Der KH, Tal-Singer R, Friedman HM. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J. Virol. 2000;74:6712–6719. doi: 10.1128/jvi.74.15.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer C, Neubauer A. The equine herpesvirus 1 UL11 gene product localizes to the trans-golgi network and is involved in cell-to-cell spread. Virology. 2003;308:23–36. doi: 10.1016/s0042-6822(02)00060-0. [DOI] [PubMed] [Google Scholar]
- Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- Silva MC, Schroer J, Shenk T. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc.Natl.Acad.Sci.U.S.A. 2005;102:2081–2086. doi: 10.1073/pnas.0409597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Yu QC, Enquist L, Shenk T. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 2003;77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakuwa H, Goshima F, Koshizuka T, Murata T, Daikoku T, Nishiyama Y. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells. 2001;6:955–966. doi: 10.1046/j.1365-2443.2001.00475.x. [DOI] [PubMed] [Google Scholar]
- Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 2005;79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh PC, Meckes DG, Jr., Wills JW. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J. Virol. 2008;82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]