Abstract
Human NHA2 is a poorly characterized Na+/H+ antiporter recently implicated in essential hypertension. We used a range of computational tools and evolutionary conservation analysis to build and validate a three-dimensional model of NHA2 based on the crystal structure of a distantly related bacterial transporter, NhaA. The model guided mutagenic evaluation of transport function, ion selectivity and pH dependence of NHA2 by phenotype screening in yeast. We describe a cluster of essential, highly conserved titratable residues located in an assembly region made of two discontinuous helices of inverted topology, each interrupted by extended chain. Whereas in NhaA, oppositely charged residues compensate for partial dipoles generated within this assembly, in NHA2, polar but uncharged residues suffice. Our findings led to a model for transport mechanism that was compared to the well known electroneutral NHE1 and electrogenic NhaA subtypes. This study establishes NHA2 as a prototype for the poorly understood, yet ubiquitous, CPA2 antiporter family recently recognized in plants and metazoans and illustrates a structure-driven approach to derive functional information on a newly discovered transporter.
Keywords: CPA, mechanism, model-structure, mutagenesis, NHA2
Introduction
Intracellular salt and pH must be tightly regulated for cell viability. Therefore, organisms from all kingdoms of life require pH-regulated cation/proton antiporters to maintain homeostasis of H+ and Na+.1; 2; 3 To date, 11 human cation/proton antiporters have been identified and assigned to specific subfamilies of the monovalent Cation/Proton Antiporter family (CPA) with orthologs throughout the biological world. The CPA1 subfamily includes many orthologs (NHE1-9) that are well characterized and believed to be electroneutral. In contrast, most CPA2 members are virtually unknown or poorly studied, with the exception of bacterial NhaA, which is an electrogenic antiporter with a stoichiometry of 2H+/Na+. More recently, phylogenetic analysis identified human NHA1 and NHA2 as members of the CPA2 subfamily.4
Based on tissue expression, genome location and phloretin sensitivity, the human transporter NHA2 was considered a candidate gene for sodium–lithium countertransport activity,5 which has been described in red blood cells,6 lymphoblasts,7 and fibroblasts,8 as a marker for essential hypertension. Pairwise alignment between bacterial NhaA and human NHA2 identified a shared pair of conserved Asp residues in the fifth transmembrane segment, implicated in electrogenic transport. Mutation of these residues to Cys abolished the ability of NHA2 to confer growth in Na+ or Li+ supplemented medium in a salt-sensitive yeast strain.5 Elevations of sodium–lithium countertransport activity are likely to correspond to physiologically relevant increases in Na+/H+ antiport activity, and are hypothesized to underlie excessive salt absorption leading to hypertension. A more mechanistic understanding of transport, however, has been hampered by the lack of a structural model. To date, there are no crystal structures of any eukaryotic member of the CPA family.
The recently determined crystal structure of NhaA from Escherichia coli is the first and only available structure of a cation/proton antiporter, revealing unique structural features that provide insight into the mechanism of antiport and its regulation.9 Of twelve transmembrane segments, TM4 and TM11 are discontinuous and interrupted by extended chains in the middle of the membrane (referred to as the TM4-TM11 assembly). Included in the unique fold of NhaA is an inverted topology repeat comprised of TMs 3-5 and TMs 10-12. Such inverted topology repeats with interrupted transmembrane helices have recently been found in structures of other ion coupled secondary transporters.10 The opposite orientation of the interrupted helices results in electrostatically-unfavorable positioning of the dipoles that face each other within the membrane. It has been proposed that the charged side chains, Asp133 (TM4) and Lys300 (TM10), located in the same region, compensate for these dipoles. Further, it has been suggested that the delicate, electrostatic balance of the TM4-TM11 assembly may facilitate conformational changes, associated with the transport mechanism. A pair of conserved and essential residues in TM5, Asp163 and Asp164, are located in close proximity to the unwound regions of TM4 and TM11, and implicated as binding sites for H+ and Na+.9; 11 Other remarkable structural features of NhaA include two funnels, facing the cytoplasm and periplasm, respectively.
Homology modeling is a useful computational approach for producing reliable structural data on membrane proteins for which structure determination is still a challenge.12 Landau and co-workers recently generated a model-structure of human NHE1 using NhaA as template, which was supported by existing mutagenesis data, inhibitor binding13 and recent NMR studies.14; 15 Because of the lack of existing biochemical data on NHA2 and the extremely low sequence similarity between mammalian and bacterial CPA proteins, it was critical to use a complex modeling technique that combined several computational tools to produce high-quality pairwise alignment between the two proteins. In this study, we optimized the pairwise alignment between human NHA2 and NhaA by combining fold recognition, profile-to-profile alignment and hydrophobicity analysis. The new alignment was used to produce a 3D model of NHA2 that was supported by evolutionary conservation analysis. We refer to it as ‘model-structure’, rather than ‘homology model’, because of the low sequence similarity between NHA2 and the NhaA template. Most significantly, the model guided the design of mutations that revealed new key residues for function. Together, the experimental and structural data presented in this study identified novel structural attributes of NHA2 likely to contribute to a mechanism of antiport distinct from the previously characterized NhaA and NHE1 type transporters. In future studies, the model can be used to assess sequence variants in the human population that may be associated with hypertension and other diseases.
Results and Discussion
Construction of a Model-Structure for Human NHA2
Both human NHA2 and E. coli NhaA have been identified as members of the CPA2 subfamily4 and are also part of the same phylogenetic clan according to the Pfam database,16 suggesting that they are evolutionarily-related and may share the same fold. Indeed, two fold recognition algorithms, FFAS0317 and INUB,18 identified NhaA as a structural template for NHA2. The low sequence similarity between NHA2 and NhaA (<15% sequence identity) required the use of a composite modeling approach in order to optimize the alignment between the target and template sequences. Similar to the attempt to model the NHE1 structure,13 our modeling procedure also employed pairwise alignments computed by the FFAS0317 and the HMAP19 algorithms (Fig. 1a). Additionally, we integrated results of other methods to aid the selection of the TM helices boundaries (Figs. 1a and 1b). This included a hybrid fold recognition method, INUB, 18 TM and secondary structure prediction algorithms,20; 21; 22 hydrophobicity analysis and the previously presented pairwise alignment between NHA2 and NhaA.5
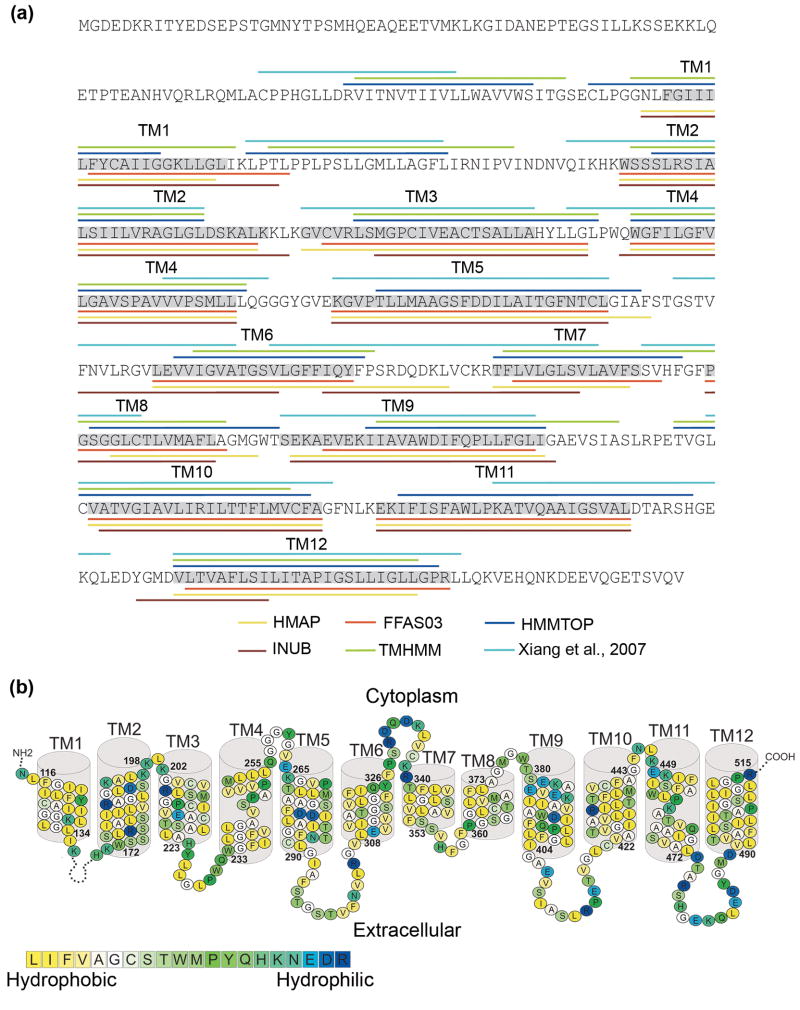
Fig. 1. Building the NHA2 model-structure.
(a) The location of the helices of NHA2 according to secondary structure predictions (HMMTOP and TMHMM), profile-to-profile alignment (HMAP), fold recognition (FFAS03 and INUB) and pairwise alignment with NhaA5 are marked in different colors according to the legends. The boundaries of the TM helices that were used for the modeling here are highlighted in grey, and the TM helix numbers are marked above. (b) The suggested TM topology of NHA2. Residues are colored according to the hydrophobicity scale of Kessel and Ben-Tal,30 using the color bar, with blue-through-yellow indicating hydrophilic-through-hydrophobic. The long loop connecting TM1 and TM2 was omitted for clarity. Overall, the helices are hydrophobic, but they do feature polar and titratable residues, as anticipated for a transporter.
(i) Initial TM Helices Assignment
The majority of the computational methods, especially the fold recognition and profile-to-profile alignment algorithms, predicted a similar helix assignment, albeit with minor deviations in the exact boundaries (Fig. 1a). To establish the initial boundaries of the TM helices for most of the segments, we utilized the helix assignment produced using the HMAP pairwise alignment.19 This decision was based on the following observations. First, a recent study found that HMAP is more accurate than sequence-to-sequence, sequence-to-profile and MSAs in cases of weak sequence similarity between membrane proteins,12 although the study did not compare this method to fold recognition or other profile-to-profile alignment methods. Second, the HMAP result presented at least a partial match to the results of all other computational tools (Fig. 1a). Third, examination of a hydrophobicity analysis of NHA2 and its close homologues revealed that the HMAP TM predictions are in most cases very similar to the preserved hydrophobic segments within the NHA2 family (data available upon request).
Deviations from the HMAP predictions are described here: For TMs 3, 5 and 6, the HMAP prediction placed gaps one residue before the end of the helix. We manually removed the gaps and assigned these helices according to the gap-less TM segments. The same procedure was performed for TM12, in which the gaps occurred in the middle of the predicted helix. In addition, we assigned TM8 according to the identical prediction of FFAS03 and INUB since the region detected for TM8 by these methods better corresponded to the MSA and hydrophobicity analysis than the HMAP assignment.
(ii) 3D Model Building and Modification
We then matched the TM segments according to the initial prediction to their corresponding TM helices in NhaA and used this alignment to construct an initial 3D model of NHA2. On examining the hydrophobicity of the preliminary model, the selected boundaries of most of the TM segments maximized the exposure of hydrophobic residues to the lipids while maintaining most of the polar and charged residues in the protein core (Supplementary Figs. S1a and S1b), as they should. In the case of TM1, however, our initial assignment placed two highly hydrophilic residues towards the lipids. We thus shifted the boundaries of TM1 so it would begin one residue N-terminally to the initial assignment. Furthermore, the evaluation process (described below) revealed that the initial assignment of TM9 placed its evolutionarily conserved face towards the membrane. We therefore shifted the original assignment of TM9 by a single residue towards the N-terminal so it would better correspond to hydrophobicity and conservation (Figs. 1b and 2a, Supplementary Fig. S1).
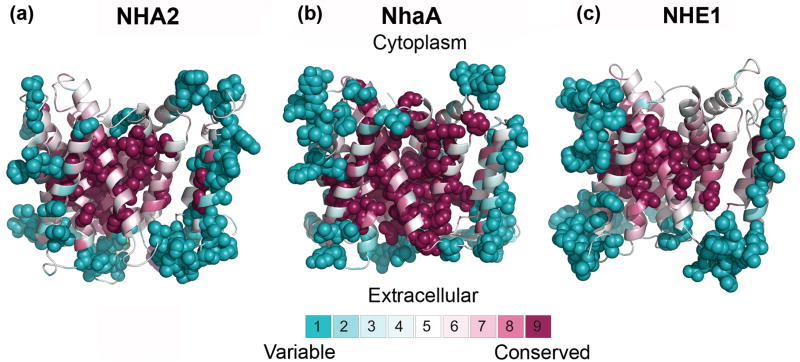
Fig. 2. Evolutionary conservation.
The evolutionary conservation profiles of NHA2, NhaA and NHE1, calculated via the ConSurf web-server (http://consurf.tau.ac.il),28 are shown in panels (a),(b) and (c), respectively. The intra-cellular side is facing up in all panels. The structure of NhaA9 and models of NHA2 and NHE113 are colored according to their conservation-grades using the color-coding bar, with turquoise-through-maroon indicating variable-through-conserved. Positions are colored yellow in cases in which there were insufficient data to determine the conservation scores with high statistical significance. The most variable and most conserved positions of each transporter are shown as spheres. The three proteins exhibit a highly conserved core while loops and lipid-facing residues are variable, as they should.
The final pairwise alignment of NHA2 and NhaA, after the slight modifications in TM1 and TM9, was utilized for building the model-structure (Supplementary Figs. S1 and S2). The model showed the TM4-TM11 assembly of NHA2, embedded between TM1, TM3, TM5, TM10 and TM12 and the cytoplasmic and external funnels shaped by the cytoplasmic parts of TM2, TM4, TM5, and TM9 and the external regions of TM2, TM8, and TM11, respectively. The model-structure possessed the expected physicochemical properties of a typical transporter, with hydrophobic residues facing the membrane, while most of the polar residues clustered in the core or on extra-membrane loops (Supplementary Figs. S1a and S1b). It should be noted that exact overlap between the current helix boundaries and the boundaries deduced from the previous pairwise alignment5 occurred only in four of the twelve TM segments (i.e. TM3, TM5, TM9 and TM12) (Fig. 1a). We note the work of Forrest and co-workers, who demonstrated the increased accuracy of structure-based profile alignments over pairwise alignments for modeling of structures of TM proteins, especially in sequence pairs sharing low sequence identity.12
In silico Validation of the NHA2 Model
There are no previous experimental data that can be used to examine the validity of the model-structure of this newly-discovered transporter.5 Structures of membrane proteins, however, have been shown to display distinct common features, especially in regard to the distribution of positively charged residues at the ends of the TM helices and the extramembrane loops,23 and the pattern of evolutionary conservation.24 We evaluated the compatibility of the NHA2 model with these generic characteristics.
(i) NHA2 Follows the ‘Positive-Inside’ Rule
The ‘positive-inside’ rule describes the empirical observation that intracellular regions of membrane proteins are enriched with positively-charged residues (lysine and arginine) relative to extracellular regions.25; 26 This distribution has been readily demonstrated in NhaA and NHE1.13 Likewise, the model-structure of NHA2 has 15 lysines and arginines in the cytoplasmic side versus 8 in the extracellular face of the membrane (Supplementary Fig. S3a).
(ii) Evolutionary Conservation Supports the NHA2 Model-Structure
As previously exemplified for the NHE1 model-structure,13 evolutionary conservation can be utilized to assess the validity of structures and model-structures.27 Proteins are usually subjected to evolutionary pressure in regions of structural or functional importance. For ion transporters, this consists of regions that take part in ion binding or translocation, but also of inter-helical interfaces, crucial for stabilizing the architecture of the helix bundle. Therefore, the expectation is that the protein core would be conserved whereas residues that face the lipids would be variable.27 We projected evolutionary conservation scores, calculated using the ConSurf server (http://consurf.tau.ac.il/)28 on the model-structure of NHA2. The model-structure is compatible with the conservation pattern in that, overall, the variable residues face the lipids or are located in extramembrane regions, whereas the protein core is highly conserved (Fig. 2a, Supplementary Figs. S1c and S1d). The NHA2 conservation distribution is similar to that exhibited for the structure of NhaA and the model of NHE113 (Figs. 2b and 2c). Moreover, the NHA2 and NHE1 models along with the NhaA crystal structure9 all show high conservation of the TM4-TM11 assembly and its flanking helices, each containing a cluster of at least four extremely conserved charged residues (Fig. 3). It is noteworthy that the conservation scores were calculated individually for NhaA, NHE1 and NHA2 by using three distinct sets of homologues. The similarity detected between the conservation patterns of the 3D structures of these distantly-related transporters reflects their presumed structural resemblance, strengthening the notion that NhaA is indeed a suitable template for modeling the structures of human proteins belonging to the CPA superfamily.
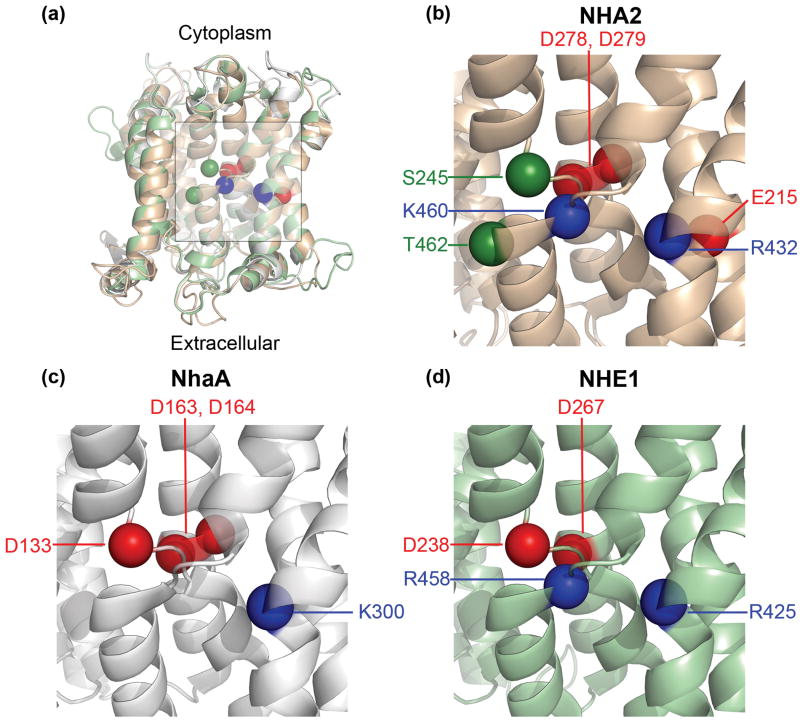
Fig. 3. Titratable residues in the TM4-TM11 assembly region.
In all panels, TM12 was omitted for clarity and the intra-cellular side is at the top. Locations of Cα atoms of highly conserved titratable residues in the TM4-TM11 assembly region are shown as spheres. Red spheres denote aspartates and glutamates, while blue spheres indicate lysines or arginines. Ser245 and Thr462 of NHA2 are shown as green spheres. (a) The model-structure of NHA2 (light brown) is aligned to the crystal structure of NhaA (grey)9 and the model of NHE1 (green).13 The center of the TM4-TM11 assembly and flanking region is marked by a square. (b)-(d) Focus on the marked region of NHA2, NhaA and NHE1, respectively. While the assembly region of all three transporters includes conserved charged residues, each displays distinct features, possibly corresponding to functional divergence. Please refer to the main text for details.
Additionally, our analysis revealed that three non-synonymous SNPs detected in the sequence of NHA2 all map to variable residues on extracellular loops, supporting a neutral effect for substitutions occurring in these positions (see Supplementary Data, Supplementary Fig. S3b).
Experimental Validation of the NHA2 Model
We undertook a detailed validation of the model-structure based on directed mutagenesis. Previously, we described functional complementation of the salt sensitive phenotype of a yeast host by heterologous expression of NHA2.5 Wild type human NHA2 rescued the dose-dependent Na+- and Li+-growth sensitivity of the yeast strain AB11c lacking endogenous cation pumps and antiporters (ena1-4Δnhx1Δnha1Δ29) (Fig. 4, Supplementary Fig. S6), but could not rescue the K+-sensitive phenotype, consistent with ion selective transport of Na+ and Li+ but not K+. Insight into proton transport and regulation was derived from the pH-dependence of phenotype complementation which occurred between a pH range of 3-4.5 (Fig. 5, and Supplementary Fig. S7). We note that this acidic pH optimum of the NHA2 activity (Fig. 5) probably reflects the H+ motive force set up by the plasma membrane H-ATPase in the yeast cell, and other pH dependency ranges could emerge under different physiological conditions in human cells. Nevertheless, we still interpreted shifts from the pH optimum as potential changes in the pH dependence of NHA2. Our experimental approach allowed relatively large numbers of newly designed mutants to be rapidly and quantitatively screened for functional changes. Mutants were evaluated for protein folding defects by monitoring expression levels by Western blot analysis (Supplementary Fig. S4) and plasma membrane localization by fluorescence imaging of GFP-NHA2 (Supplementary Fig. S5).
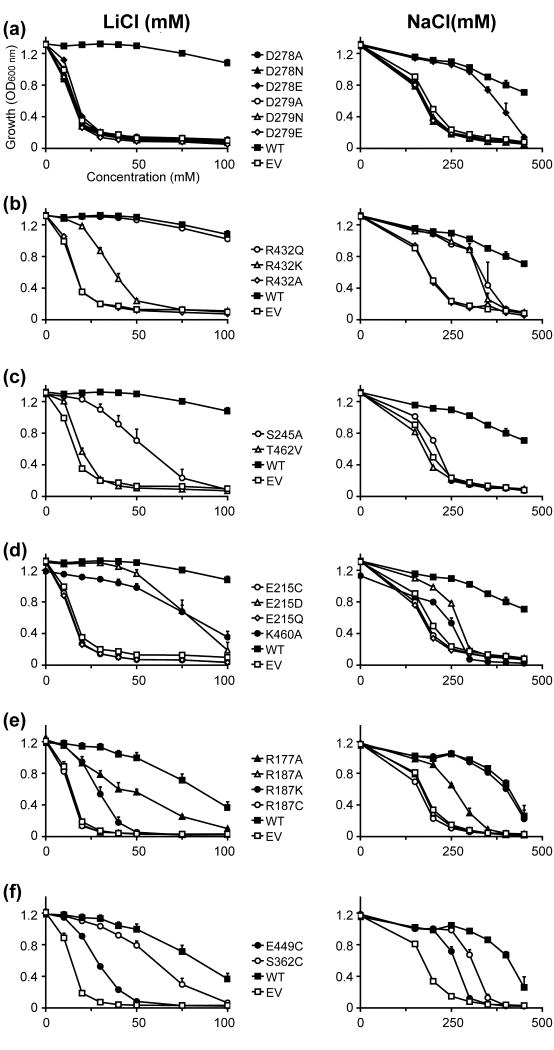
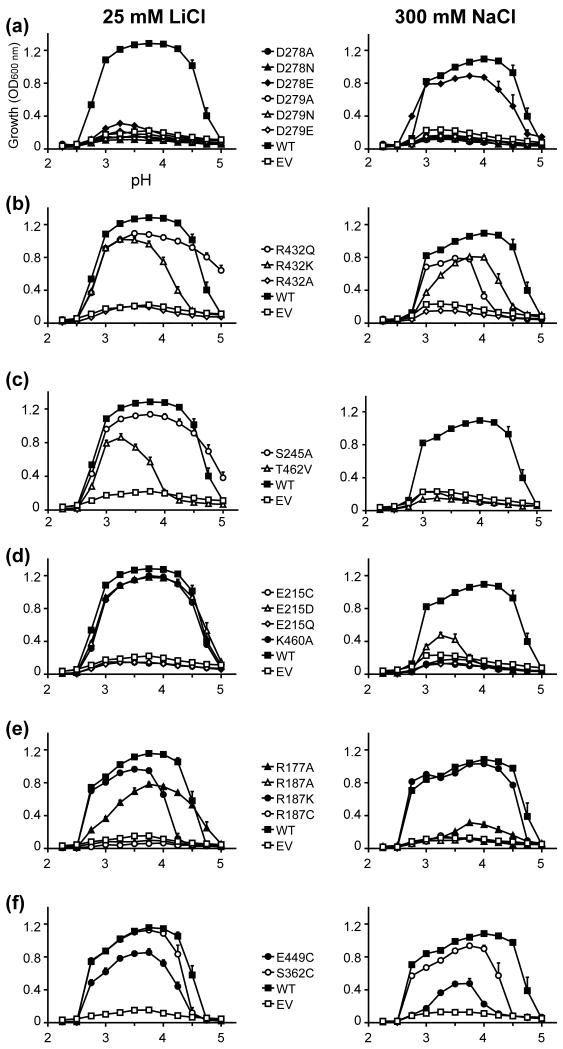
Fig. 4. Functional analysis of NHA2 mutants predicted by homology modeling: dosage response.
The salt-sensitive yeast strain AB11C was transformed with His-tagged NHA2 (WT), NHA2 mutant as indicated, or empty vector (EV). Yeasts were grown in APG medium supplemented with LiCl (Left panels), or NaCl (Right panels), and growth was determined by optical density of the culture at 600 nm (OD600) after 24 hrs (LiCl) or 48 hrs (NaCl) at 30°C. Data are the average of triplicate determinations
Fig. 5. Functional analysis of NHA2 mutants predicted by homology modeling: pH profile.
Same as Fig. 4 except that LiCl concentrations was fixed at 25 mM and NaCl concentration at 300 mM. Media pH was adjusted by using phosphoric acid.
(i) Structure Guided Design of Mutations
The insertion of charged residues into the membrane bilayer is energetically unfavorable, and the common view is that they do reside in the membrane only to conduct physiological functions.30; 31; 32 Indeed, several such charged and conserved residues within the core of bacterial NhaA and human NHE1 have been assigned specific roles in ion binding and structural stabilization of the TM4-TM11 assembly based on structural and mutagenesis data (Figs. 3a, 3c and 3d). 9; 13; 33; 34; 35; 36 The model-structure of NHA2 accommodates eight charged residues in the core of the membrane domain, five of which are highly conserved positions located in or near the TM4-TM11 assembly (Figs. 3b and 6). Many other charged residues are located in the extramembrane regions. In order to obtain structural and functional insight on NHA2, we undertook site-directed mutagenesis of the highly conserved charged residues in the core and several peripheral titratable residues, including both conservative and nonconservative replacements. While our general aim was to investigate charged residues, we also performed substitutions in few other residues of interest, chosen based on the comparison of NHA2 with previous data available for NhaA and NHE1.
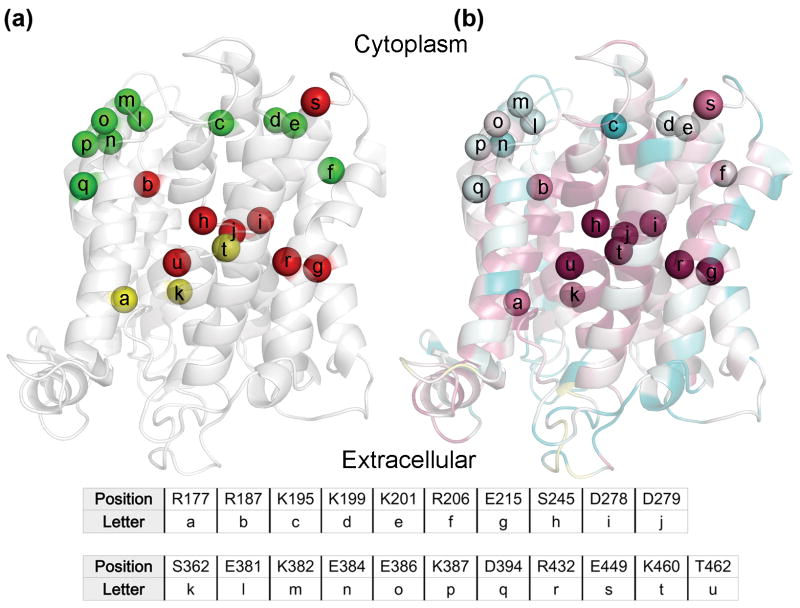
Fig. 6. Mutagenesis results in light of the evolutionary conservation analysis.
In panels (a) and (b), the NHA2 model is shown as transparent ribbons, with its cytoplasmic-facing side up. Cα atoms of mutated positions are shown as spheres, and are marked by lower-case letters with their corresponding residues shown in the table. (a) Residues mutated in this study are colored by the resulting phenotype of the mutations, with red, green and yellow corresponding to non-functional, functional and partially functional. Positions were colored red even if a single mutant failed to rescue the salt-sensitive yeast in the presence of either Li+ or Na+, whereas residues were marked as partially-functional if mutations mildly lowered yeast growth in comparison to WT. (b) The model-structure is colored by conservation, as in Fig. 2a. All positions which were sensitive-to-mutation are evolutionary conserved (grades 8 and 9), while all non-sensitive positions receive significantly lower conservation scores, as anticipated.
(ii) Two Conserved and Essential Aspartates in TM5 of NHA2
In our previous study, we identified two sequential Asp residues in TM5 of NHA2, Asp278 and Asp279, which are homologous to Asp163 and Asp164 of NhaA (Figs. 3b, 3c and 6b, Supplementary Fig. S2), referred to as the ‘DD motif’. Mutagenesis demonstrated that the NHA2 double mutant DD→CC failed to rescue the salt-sensitive phenotype of the host yeast strain,5 consistent with a role of these aspartates in cation (Na+, Li+ or H+) binding.37 Herein, we expanded investigation of this essential motif by individually replacing each asparatate with glutamate, asparagine or alanine (Figs. 4a and 5a). All the mutants except for D278E failed to restore growth of the salt-sensitive strain under increasing concentrations of both Na+ and Li+, supporting a specific requirement for an acidic residue at these locations. Western-blot and fluorescence microscopy confirmed that both the expression level and localization of the NHA2 mutants were comparable to that of WT NHA2 (Supplementary Figs. S4a and S5), confirming that D278 and D279 play a functional role, probably in transport.
Notably, the ND motif in the CPA1 family, corresponding to Asn and Asp in positions 266 and 267 of NHE1, has also been implicated in ion binding and translocation.13; 35; 38 We explored the possible relation between the predicted ion binding sites of NHE1 and NHA2 by generating the D278N mutant in NHA2. The mutant, in which the original DD motif was substituted to an ND motif, failed the functional complementation test. Because similar results were obtained in bacterial NhaA,33 it appears that contrary to CPA1, CPA2 exchangers with an ND motif are unable to perform transport.
As with other substitutions in the DD motif, the D278E mutant failed to confer Li+ tolerance (Figs. 4a and 5a). However, the yeast strain expressing this NHA2 mutant showed Na+ tolerant growth comparable to the WT protein, suggesting a limited retention of function by Glu in place of Asp (Figs. 4a and 5a). Interestingly, a similar partial retention of function was observed in the D164E mutant of NhaA, with both Na+ and Li+,36 and the corresponding D267E mutant in NHE1 was also deemed active.38 This suggests that a small change in placement of the carboxyl group within the DD motif may be accommodated in all three structures.
(iii) Compensation of Helix Dipoles in the TM4-TM11 Assembly
In the NhaA structure, both TM4 and TM11 are interrupted by an extended chain,9 flanked by partial charges formed at the N- and C- termini of the respective small helices connecting the extended chains. These partial charges in the middle of the membrane were proposed to be compensated by the highly conserved Asp133 of TM4 and Lys300 of TM10 (Fig. 3c).9; 37 The model of NHE1 suggested a similar compensation for these helix dipoles, with D238 and R425 corresponding to D133 and K300, respectively (Figs. 3a, 3c and 3d).13 However, the model-structure of NHA2 suggested some deviations from this pattern: while the highly conserved Arg432 of TM10 (Figs. 3b and 6b) corresponds to Lys300 in NhaA and Arg425 in NHE1 (Figs. 3c and 3d, respectively), there was no conserved negatively charged residue in the position equivalent to D133 or D238 of NhaA and NHE1, respectively. Mutant R432A failed to rescue salt-tolerant growth in the host yeast strain and R432K retained partial function with both Na+ and Li+ (Figs. 4b and 5b), consistent with the proposed mechanism of dipole compensation. Surprisingly, mutant R432Q retained significant levels of function equal to, or greater than, mutant R432K, indicating that a polar residue with a partial positive dipole may suffice for stabilization of the two negative dipoles of TM4 and TM11.
Following this, we speculated that the partial-positive charge of the TM4-TM11 helix dipoles could be compensated through partial charges of hydroxyl side-chains of two highly conserved residues, Ser245 and Thr462, positioned at the N-termini of the small helices connecting the extended chains of TM4 and TM11, respectively (Figs. 3b and 6b). Indeed, the S245A and T462V mutants failed to rescue the salt-sensitive phenotype in the presence of Na+ (Figs. 4c and 5c), while substitution of these residues with cysteine retained activity (Supplementary Fig. S6c). Together, these data suggest that the partial dipoles of the TM4-TM11 assembly, both positive and negative, can be compensated via polar residues possessing the opposite partial charges.
(iv) Novel Features of the TM4-TM11 Assembly
A unique feature of the region in the vicinity of the TM4-TM11 assembly of NHA2 is the highly conserved Glu215 in TM3 (Figs. 3b and 6b). Neither NhaA nor NHE1 possess a charged residue in the same position (Figs. 3c and 3d, respectively). We therefore tested the effect of a series of substitutions at this site, ranging from profound to mild. Whereas substitutions lacking a carboxylic group, E215A, E215C and E215Q, failed to rescue the salt-sensitive phenotype of the host strain, partial function was retained in the E215D mutant (Figs. 4d and 5d). We note that Li+ tolerance was more prominent than Na+ tolerance in this mutant. We can conclude that structural, phylogenetic and experimental data all point to the significance of Glu215. Whether Glu215 is directly involved in ion or proton binding or contributes to function in another manner (for instance, participates in conformational changes during the transport cycle or in pH regulation) remains to be elucidated.
Another highly conserved and positively charged residue, Lys460 in TM11 (Fig 3b), has an equivalent in CPA1 family members (Fig. 3d), but not in bacterial NhaA (Fig. 3c). Thus, the corresponding residue in NhaA is Gly338, which oddly enough, is implicated in pH regulation,39 whereas NHE1 possesses the conserved and positively charged Arg458 in the equivalent position. However, replacement of this residue in NHE1 failed to express,40 so the functional effect of this mutant could not be assessed. We show that in NHA2, the mutant K460A exhibited a Li+ selective phenotype reminiscent of the effect of E215D in the vicinity of the TM4-TM11 assembly (Figs. 4d and 5d).
(v) Three Unique Charged Residues in the Core Outside the Assembly Region
The membrane domain of NHA2 incorporates three other titratable residues outside the TM4-TM11 assembly: Arg177 and Arg187 in TM2, and Asp394 in TM9 (Fig. 6b). Given the highly conserved nature of Arg187 within the protein core, it was not surprising that mutant R187A failed to rescue salt-sensitive growth of the host strain. Nevertheless, maintaining a positive charge in this position (R187K) allowed partial function (Figs. 4e and 5e). While these results suggest that a positive charge in this position is needed for NHA2, NhaA and NHE1 do not possess a similar charge in this position.
Mutation to alanine of the conserved Arg177 allowed only partial compensation (Figs. 4e and 5e). Since Asp394 is not conserved, we hypothesized that it does not have a significant role for function or structure stabilization (Fig. 6b). Verifying this notion, mutants D394E and D394C were deemed functional (Supplementary Figs. S6a and S7a),
(vi) Ser362 and Glu449 Line the Gateways to the Transport Funnels
Next, we selected two other residues of interest for mutagenesis, Ser362 and Glu449, which are both evolutionarily conserved (Fig. 6b). Ser362 is the equivalent position of Ser351 of NHE1, which was predicted to take part in the ion transport mechanism of NHE1.13 Indeed, Reddy et al.14 recently showed that Ser351 was seemingly a pore-lining residue. Interestingly, the S362C variant of NHA2 did allow complementation almost as WT (Figs. 4f and 5f), maybe owing to the physicochemical similarity between Ser and Cys.
On the other hand, Glu449 was located at the cytoplasmic side of TM11, facing the putative entrance gate for the TM4-TM11 assembly (Fig. 6b). The E449C mutant significantly reduced activity, with a more profound effect for Na+ than for Li+ (Figs. 4f and 5f). Since NhaA and NHE1 do not encompass a similar conserved and charged residue in this region, this was also considered as a novel feature of NHA2.
(vii) Mutations in Non-Conserved Residues Support Model-Structure
Integrating structural data with phylogenetic analysis, we identified and verified the functionality of several NHA2 residues. While this provided some primary validation for our model-structure, we also initiated a different validation approach. We focused on relatively variable segments which are enriched with charged residues: the cytoplasmic ends of TM2, TM3 and TM9, along with the loop between TM2 and TM3 (Fig. 6b). We anticipated that mutations in charged but variable residues in these regions, which are exposed to the cytoplasm or residing on the membrane boundary, would not impair function. Reassuringly, substitutions performed in 10 randomly-selected residues in these regions still allowed growth of the salt-sensitive strain in the presence of Na+ or Li+ (Supplementary Figs. S6a, S6b, S6d, S7a, S7b and S7d).
On the Mechanism
(i) Ion Selectivity and the Translocation Pathway
Studies have shown that different CPA transporters exhibit selectivity for a distinct range of cations.4 Mutations that alter ion selectivity of a transporter are of special interest because they help identify the residues that most likely contribute to the ion translocation pathway. Several NHA2 mutants located at the TM4-TM11 assembly and its flanking helices displayed asymmetric phenotypes with Na+ and Li+, including S245A, R432Q, K460A, T462V along with D278E and E215D (Figs. 4, 5 and 6) consistent with a role for these residues in ion discrimination, binding and translocation.
Although neither Na+ nor Li+ were shown in the crystal structure of NhaA determined at pH 4, structure-based calculations have suggested that the non hydrated ions reach the binding site.9 Based on differences in non-hydrated ionic radii (0.68 Å for Li+ and 0.95 Å for Na+) and the previous suggestion that Li+ requires fewer coordinating residues than Na+,41 we can speculate that the ion binding sites overlap only partially. We observed that mutant D278E strongly complemented Na+ phenotypes but grew poorly in Li+ supplemented medium, whereas mutant E215D displayed the opposite phenotype, with preferred selectivity for Li+ over Na+ (Figs. 4a and 5b). Considering that the single mutants D278E and E215D were reversed in terms of both side-chain size and complementation phenotype, we generated a double mutant D278E/E215D to test whether this may restore activity with both ions. Interestingly, the double mutant restored the deficient activity of the E215D single mutant in the presence of Na+, although it did not change the observed D278E Li+ phenotype (Supplementary Figs. S6e and S7e). This functional interaction supports the proximity of these two residues in the model-structure (Fig. 3B) and suggests that the unique residue E215, along with D278, is indeed located along the ion translocation pathway. The specificity of this interaction was validated by another double mutant, D278K/K460D which failed to restore activity (Supplementary Figs. S6f and S7f). This result is also in line with the NHA2 model, in which D278 and K460 are both part of the assembly region, but are not in close proximity like D278 and E215 (Fig. 3b).
(ii) Proposed Transport Mechanism
Generally speaking, the NHA2 model predicted that the external and cytoplasmic funnels, along with the TM4-TM11 assembly and nearby regions, are enriched with highly conserved polar residues (Supplementary Fig. S1). Consistent with the previous suggestions for NHE113 and NhaA,9; 11 we propose that these regions, especially the specific residues discussed above, take part in ion translocation in NHA2. More specific ion transport mechanisms have been proposed for NhaA and NHE1.9; 11; 13 We therefore examined whether these mechanisms can provide an initial understanding of the mechanism of NHA2. Based on the crystal structure it has been suggested that the irregularities in the α-helical structure in the TM4-11 assembly, exposes main–chain hydrogen bonding partners and creates a delicately balanced electrostatic environment essential for ion-binding and antiport.9 MD simulation explained electrogenic transport by suggesting that Asp163 binds only H+ while Asp164 binds both Na+ and H+, and proposed that there are two translocation pathways (Fig. 7a).11 Mutation in Glu252 of TM2 increased Km for Na+, implicating this residue in ion binding.42 In the model of the mechanism of NHE1, one Na+ ion is exchanged for one H+ via Asp267, while Glu262 and Ser351 were suggested to play a role in proton attraction and cation binding, respectively (Fig. 7b).13
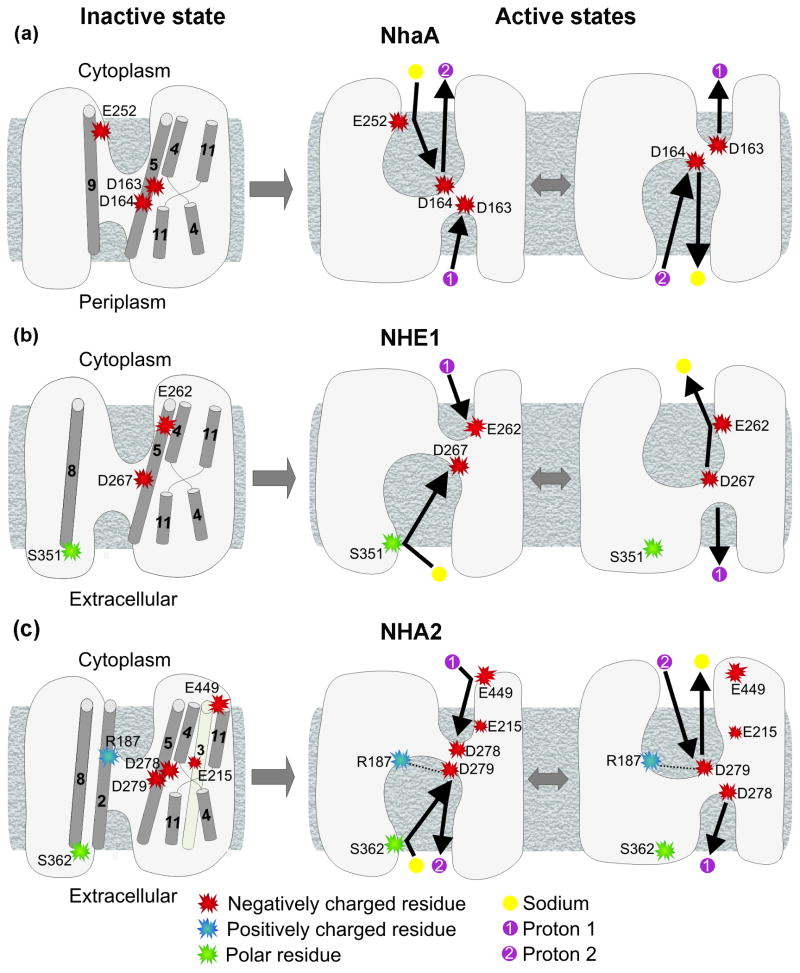
Fig. 7. Comparison of the suggested transport mechanisms of NhaA, NHE1 and NHA2.
The intracellular side is facing upwards. The schemes on the left depict the inactive conformations, which correspond to the crystal structure of NhaA and models of NHE1 and NHA2 (panels (a), (b) and (c), respectively). Indeed, pH-dependant inactive and active states were identified for NhaA and NHE, 37; 43, and here we showed that the activity of NHA2 also depends on pH conditions (Fig. 5). Residues proposed to take part in transport and their corresponding helices are illustrated, along with the TM4-TM11 assembly. The right hand side represents the putative active conformations and the suggested transport cycles, with the residues of interest, sodium ions and protons according to the legend below. The number of transported protons matches the stoichiometry of NhaA and NHE1, and putative electrogenicity of NHA2. Since the physiological direction of transport for NHA2 is yet to be determined, its transport direction in panel (c) is for illustration purposes only. While Glu262 serves as a proton attractor in the cytoplasmic side of NHE1 (panel (b),13) and Glu252 participates in sodium binding in NhaA (panel (a),42), Glu449 may possess a similar role for NHA2 (panel (c)). The two Asp of TM5 in NhaA (panel (a)) and NHA2 (panel (c)) along with the single one of NHE1 (panel (b)) are the proposed binding sites for protons and cations in the protein core. Uniquely for NHA2, a salt bridge between Arg187 and Asp279 (panel (c)) may be formed in the active state. In the extracellular side, Ser352 and Ser362 of TM8 of NHE1 and NHA2, respectively, perhaps participate in cation binding (panels (b) and (c)). Additionally, essential Glu215 is probably situated in the ion translocation pathway of NHA2, and its precise functional role is yet to be discovered.
Fig. 7 summarizes the comparison between the mechanisms of NhaA, NHE1 and NHA2. In the case of NHA2, Asp278 and Asp279 are likely to possess the same role proposed for Asp163 and Asp164 of NhaA (Figs. 7a and 7c). Considering the predicted structural location of Ser362 at the extracellular end of TM8 (Fig. 6), Ser362 might participate in ion attraction or initial binding, just like its equivalent residue in NHE1, Ser351 (Figs. 7b and 7c). This may be possible if TM8 indeed rotates in the active state relative to its conformation in the inactive state, as suggested for NHE1.13 Although there was no corresponding residue for Glu262 of NHE1 nor for Glu252 of NhaA in NHA2, we did find Glu449, a negatively charged and conserved position at the cytoplamsic end of TM11 (Fig. 6b), facing the entrance of the TM4-TM11 assembly. In line with the necessity of the negative charge at this position, we suggest that Glu449 serves as a proton attractor that can aid the transport process but is not crucial for function (Fig. 7c). Relying on the model-structure, we also speculate that a slight rotation of TM5 could result in a salt bridge between the conserved and essential Arg187 of TM2 and Asp279 of TM5 (Fig. 7c). Such an interaction may stabilize the active conformation during transport and explain the importance of Arg187 for function. Alternatively, Asp279 may replace its interaction with Arg187 for Na+, in the same manner in which H+ is replaced by Na+ in the mechanism proposed by Arkin et al. for NhaA (Figs. 7a and 7c). If indeed the positively charged side chain of Arg187 interacts with Asp279 instead of a proton, one might consider the possibility that NHA2 is electroneutral rather than electrogenic. Last, highly conserved and essential Glu215 of TM3, positioned near the TM4-TM11 assembly, is predicted to take part in the function mechanism, in manner that remains to be discovered.
Concluding Remarks
Helices and specific residues which are part of the proposed NHA2 antiport mechanism are marked on the model-structure presented in Fig. 8. Despite an overall similarity in architecture with other CPA family members, our studies revealed striking differences and unique features not seen in other CPA subtypes (Figs. 3 and 7). First, our results suggest that partial dipoles of the TM4-TM11 assembly of NHA2 can be compensated by polar side-chains (Asn, Ser, Thr, Cys), in the absence of oppositely charged residues seen in both NhaA and NHE1 (Fig. 3). Second, we describe a uniquely conserved negative charge, E215, in the vicinity of the TM4-TM11 assembly and the essential charged residue D278 in TM5. Another unique and conserved charge, R187, is located in TM2 where it has the potential to stabilize the oppositely charged D279 in TM5 (Fig. 8). Whereas the DD motifs of TM5 in NHA2 and NhaA are clearly functional equivalents and characteristic of CPA2 family members, other residues in NHA2 are potentially equivalent to functionally important residues seen in the CPA1 family prototype, NHE1, including S362 and K460, proposed to line the translocation pathways.
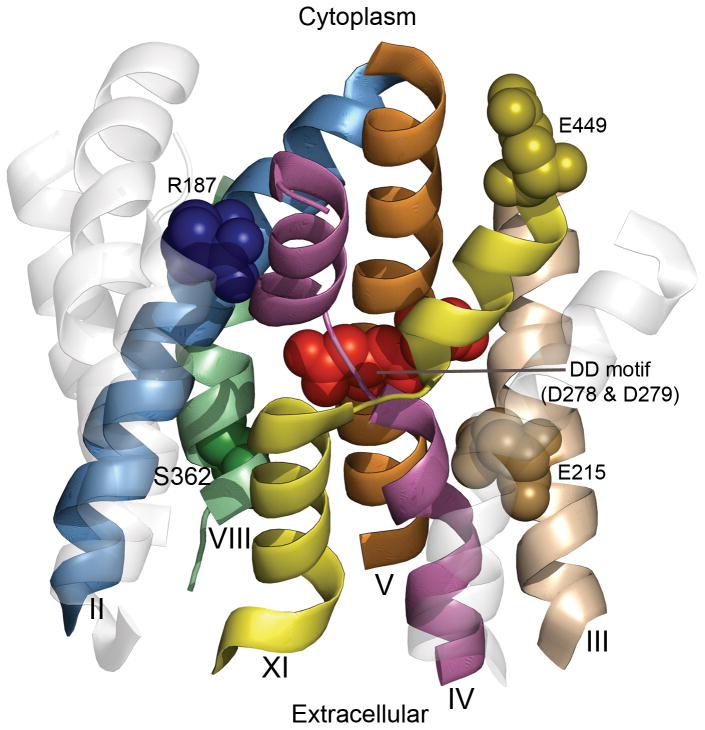
Fig. 8. Functional residues mapped on the structural architecture of NHA2.
The cytoplasmic side is upwards. The model-structure of NHA2 is shown as transparent ribbons, whereas helices implicated in the function mechanism are marked. Helices 9 and 12, along with the extramembrane loops, were omitted for clarity. Specific residues suggested to participate in transport, illustrated in Fig. 7C, are shown as spheres and highlighted.
In this study we applied an innovative reverse genetic approach to study the mechanism and function of a recently identified ion transporter. Traditionally, investigation of membrane proteins begins with elaborate biochemical data that ultimately validate structural information, often the culmination of years of trial and effort. Here, the model-structure of NHA2 allowed us to obtain early molecular level insight into structure-function relationship of a novel human protein implicated as a marker in essential hypertension (Figs. 3b, 6, 7 and 8). Our findings establish NHA2 as a prototype for the poorly understood, yet ubiquitous CPA2 family in plants and metazoans.
Materials and Methods
Identifying the Fold
We initiated a PSI-BLAST search44 of three iterations against the UniProt database45 using the NHA2 sequence, but did not succeed in finding NhaA as a sequence homologue. The FFAS03 fold recognition server17 identified the structure of NhaA (PDB ID 1ZCD) as the best template with a significant score of -36.1. According to the FFAS03 benchmark, scores lower than -9.5 exhibit less than 3% of false-positives.17 The INUB hybrid fold recognition method also identified NhaA as a suitable template.18
Sequence Data and Evolutionary Conservation Analysis
Studies have shown that evolutionary conservation analysis is a useful tool for prediction of TM protein structures, as conserved residues often reside in the protein core, whereas variable positions face the membrane or are part of extramembrane loops.24; 46; 47; 48; 49 When not employed for model-building, the conservation profile can also be used for model evaluation.27 To this end, we conducted a BLAST search44 against the UniProt database45 to collect homologous sequences to NHA2. We discarded hits shorter than 60% of the original sequence and identical sequences. This resulted in a multiple sequence alignment (MSA) of 55 sequences that we computed using the MUSCLE algorithm.50 We then utilized the MSA to calculate conservation scores via the ConSurf webserver (http://consurf.bioinfo.tau.ac.il/, 28) with Bayesian inference.51 Scores were projected on the model-structure of NHA2. We also calculated conservation scores using several additional MSAs produced for NHA2 (described in the Supplementary Text), which were similar to the results computed using the 55 homologues collection. The MSAs and conservation scores for NhaA and NHE1 were taken from a previous study.13
Prediction of NHA2 TM Helices and Pairwise Alignments between NHA2 and NhaA
The low similarity between NhaA and NHA2 makes it difficult to construct an accurate pairwise alignment using simple means, but requires a more complex procedure. We estimated the secondary structure of NHA2 using PSIPRED22 and utilized HMMTOP20 and TMHMM21 to predict TM helices. In addition, we colored the MSA produced for NHA2 according to a hydrophobicity scale30 to identify segments conserved as hydrophobic throughout the NHA2 family. We computed pairwise alignments via the FFAS03 server,17 the HMAP profile-to-profile method19 and the INUB server.18 An additional pairwise alignment was taken from a preceding study.5
Building a 3-Dimensional Model by Homology Modeling
We used the homology modeling program NEST52 with default parameters, and the pairwise alignment between NHA2 and NhaA (Supplementary Fig. 2), for modeling the TM domain of NHA2 (residues 114-516) according to the template structure NhaA (PDB ID 1ZCD). The model's accuracy is directly correlated to the sequence identity with the template.12 Because of the high sequence divergence between NhaA and NHA2 we refer to the model as “model-structure” to emphasize that it might be less accurate than typical homology models.
Site-Directed Mutagenesis, Yeast Strains and Growth Media
Subcloning of Human NHA2 with N-terminal His9 or GFP epitope tag into plasmid pSM1052 were described previously.5 Mutations in NHA2 were carried out using QuikChange (Stratagene) method and confirmed by sequencing.
Saccharomyces cerevisiae strains AB11c or B31W lacking endogenous Na+ pumps and antiporters29 were used as host for heterologous expression of NHA2 and its mutants. Yeast cultures were grown at 30°C in APG, a synthetic minimal medium with minimal salt.53 Salt-sensitive growth was monitored by inoculating 0.2 ml of APG medium in a 96-well microplate with 4 μl of a saturated seed culture. After incubation at 30°C for 20–72 h, cultures were gently resuspended, and the OD600 was recorded on a FLUOStar Optima plate reader (BMG Labtechnologies).
SDS/PAGE, Biochemical Techniques and Fluorescence Microscopy
Total yeast lysates were prepared by breaking the cells with 27 mM NaOH, 1% (v/v) 2-mercaptoethanol54 from equal numbers of cells (4 OD600 units). Total samples were treated with 6.4% trichloroacetic acid (TCA) and the pellets were washed once with acetone. Samples were resuspended in100 ul of SDS lysis buffer (1xPBS, 1% [w/v] SDS, 3 mM EDTA, 5 mM EGTA) with 1× protease inhibitors. Samples (50 μg protein) were subjected to SDS/PAGE and Western blotting. NHA2 was detected on a Western blot by anti-NHA2 antibody (1:2,000 dilution), as described in the figure legends. Actin as loading control was detected using mouse monoclonal antibody to beta actin (1:2,000, mAbcam 8224). IRDye 700DX conjugated anti-rabbit IgG (1:20,000, Rockland, code: 611-130-122) or IRDye 800CW conjugated anti-mouse secondary antibody (1:20,000; Rockland, code: 610-131-121) were used to visualize protein bands on the Odyssey Fluorescence Imaging System (Li-Cor Biosciences, Lincoln, NE). Live yeast cells harboring GFP-HsNHA2 and its mutants were grown overnight and then visualized and imaged directly under a Zeiss Axiophot fluorescence microscope.
Proper expression and plasma membrane localization of all examined mutants was indeed confirmed (Supplementary Figs. S4 and S5).
Supplementary Material
Acknowledgments
This work was supported by grant 611/07 from the Israel Science Foundation to N.B-T, a grant from the USA-Israel Binational Science Foundation (BSF, Grant 501/03-16.2) to EP and R.R, a grant from the National Institutes of Health (NIDDK 054214) to R.R., and EDICT (EU FP 7, European Drug Initiative on Channels and Transporters) to EP. MS was supported by the Edmond J. Safra Bioinformatics program at Tel-Aviv University.
Abbreviation
- TM
transmembrane
- CPA
cation/proton antiporter
- MSA
multiple sequence alignment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orlowski J, Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 2.Padan E, Tzubery T, Herz K, Kozachkov L, Rimon A, Galili L. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter. Biochim Biophys Acta. 2004;1658:2–13. doi: 10.1016/j.bbabio.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Counillon L, Pouyssegur J. The expanding family of eucaryotic Na(+)/H(+) exchangers. J Biol Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 5.Xiang M, Feng M, Muend S, Rao R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc Natl Acad Sci U S A. 2007;104:18677–18681. doi: 10.1073/pnas.0707120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canessa M, Adragna N, Solomon HS, Connolly TM, Tosteson DC. Increased sodium-lithium countertransport in red cells of patients with essential hypertension. N Engl J Med. 1980;302:772–776. doi: 10.1056/NEJM198004033021403. [DOI] [PubMed] [Google Scholar]
- 7.Schork NJ, Gardner JP, Zhang L, Fallin D, Thiel B, Jakubowski H, Aviv A. Genomic association/linkage of sodium lithium countertransport in CEPH pedigrees. Hypertension. 2002;40:619–628. doi: 10.1161/01.hyp.0000037131.41957.a8. [DOI] [PubMed] [Google Scholar]
- 8.Zerbini G, Podesta F, Meregalli G, Deferrari G, Pontremoli R. Fibroblast Na+-Li+ countertransport rate is elevated in essential hypertension. J Hypertens. 2001;19:1263–1269. doi: 10.1097/00004872-200107000-00011. [DOI] [PubMed] [Google Scholar]
- 9.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 10.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459:347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arkin IT, Xu H, Jensen MO, Arbely E, Bennett ER, Bowers KJ, Chow E, Dror RO, Eastwood MP, Flitman-Tene R, Gregersen BA, Klepeis JL, Kolossvary I, Shan Y, Shaw DE. Mechanism of Na+/H+ antiporting. Science. 2007;317:799–803. doi: 10.1126/science.1142824. [DOI] [PubMed] [Google Scholar]
- 12.Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landau M, Herz K, Padan E, Ben-Tal N. Model structure of the Na+/H+ exchanger 1 (NHE1): functional and clinical implications. J Biol Chem. 2007;282:37854–37863. doi: 10.1074/jbc.M705460200. [DOI] [PubMed] [Google Scholar]
- 14.Reddy T, Ding J, Li X, Sykes BD, Rainey JK, Fliegel L. Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger. J Biol Chem. 2008;283:22018–22030. doi: 10.1074/jbc.M803447200. [DOI] [PubMed] [Google Scholar]
- 15.Lee BL, Li X, Liu Y, Sykes BD, Fliegel L. Structural and Functional Analysis of Transmembrane XI of the NHE1 Isoform of the Na+/H+ Exchanger. J Biol Chem. 2009;284:11546–11556. doi: 10.1074/jbc.M809201200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer D. Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac Symp Biocomput. 2000:119–130. [PubMed] [Google Scholar]
- 19.Tang CL, Xie L, Koh IY, Posy S, Alexov E, Honig B. On the role of structural information in remote homology detection and sequence alignment: new methods using hybrid sequence profiles. J Mol Biol. 2003;334:1043–1062. doi: 10.1016/j.jmb.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 20.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 21.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 22.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 23.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 24.Fleishman SJ, Unger VM, Ben-Tal N. Transmembrane protein structures without X-rays. Trends Biochem Sci. 2006;31:106–113. doi: 10.1016/j.tibs.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Heijne GV. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleishman SJ, Ben-Tal N. Progress in structure prediction of alpha-helical membrane proteins. Curr Opin Struct Biol. 2006;16:496–504. doi: 10.1016/j.sbi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maresova L, Sychrova H. Physiological characterization of Saccharomyces cerevisiae kha1 deletion mutants. Mol Microbiol. 2005;55:588–600. doi: 10.1111/j.1365-2958.2004.04410.x. [DOI] [PubMed] [Google Scholar]
- 30.Kessel A, Ben-Tal N. Free energy determinants of peptide association with lipid bilayers. Current Topics in Membranes. 2002;52:205–253. [Google Scholar]
- 31.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 32.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 33.Inoue H, Noumi T, Tsuchiya T, Kanazawa H. Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 1995;363:264–268. doi: 10.1016/0014-5793(95)00331-3. [DOI] [PubMed] [Google Scholar]
- 34.Galili L, Rothman A, Kozachkov L, Rimon A, Padan E. Trans Membrane Domain IV Is Involved in Ion Transport Activity and pH Regulation of the NhaA-Na+/H+ Antiporter of Escherichia coliℷ€. Biochemistry. 2001;41:609–617. doi: 10.1021/bi011655v. [DOI] [PubMed] [Google Scholar]
- 35.Ding J, Rainey JK, Xu C, Sykes BD, Fliegel L. Structural and functional characterization of transmembrane segment VII of the Na+/H+ exchanger isoform 1. J Biol Chem. 2006;281:29817–29829. doi: 10.1074/jbc.M606152200. [DOI] [PubMed] [Google Scholar]
- 36.Olkhova E, Kozachkov L, Padan E, Michel H. Combined computational and biochemical study reveals the importance of electrostatic interactions between the “pH sensor” and the cation binding site of the sodium/proton antiporter NhaA of Escherichia coli. Proteins: Structure, Function, and Bioinformatics. 2009;76:548–559. doi: 10.1002/prot.22368. [DOI] [PubMed] [Google Scholar]
- 37.Padan E. The enlightening encounter between structure and function in the NhaA Na(+)-H(+) antiporter. Trends Biochem Sci. 2008;33:435–443. doi: 10.1016/j.tibs.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Murtazina R, Booth BJ, Bullis BL, Singh DN, Fliegel L. Functional analysis of polar amino-acid residues in membrane associated regions of the NHE1 isoform of the mammalian Na+/H+ exchanger. Eur J Biochem. 2001;268:4674–4685. doi: 10.1046/j.1432-1327.2001.02391.x. [DOI] [PubMed] [Google Scholar]
- 39.Rimon A, Gerchman Y, Kariv Z, Padan E. A point mutation (G338S) and its suppressor mutations affect both the pH response of the NhaA-Na+/H+ antiporter as well as the growth phenotype of Escherichia coli. J Biol Chem. 1998;273:26470–26476. doi: 10.1074/jbc.273.41.26470. [DOI] [PubMed] [Google Scholar]
- 40.Wakabayashi S, Pang T, Su X, Shigekawa M. A novel topology model of the human Na(+)/H(+) exchanger isoform 1. J Biol Chem. 2000;275:7942–7949. doi: 10.1074/jbc.275.11.7942. [DOI] [PubMed] [Google Scholar]
- 41.Kaim G, Wehrle F, Gerike U, Dimroth P. Molecular Basis for the Coupling Ion Selectivity of F1F0 ATP Synthases: Probing the Liganding Groups for Na+ and Li+ in the c Subunit of the ATP Synthase from Propionigenium modestum. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 42.Tzubery T, Rimon A, Padan E. Mutation E252C Increases Drastically the Km Value for Na+ and Causes an Alkaline Shift of the pH Dependence of NhaA Na+/H+ Antiporter of Escherichia coli. Journal of Biological Chemistry. 2004;279:3265–3272. doi: 10.1074/jbc.M309021200. [DOI] [PubMed] [Google Scholar]
- 43.Slepkov ER, Rainey JK, Sykes BD, Fliegel L. Structural and functional analysis of the Na+/H+ exchanger. Biochem J. 2007;401:623–633. doi: 10.1042/BJ20061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adamian L, Liang J. Prediction of transmembrane helix orientation in polytopic membrane proteins. BMC Struct Biol. 2006;6:13. doi: 10.1186/1472-6807-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donnelly D, Overington JP, Ruffle SV, Nugent JH, Blundell TL. Modeling alpha-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 1993;2:55–70. doi: 10.1002/pro.5560020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleishman SJ, Harrington S, Friesner RA, Honig B, Ben-Tal N. An automatic method for predicting transmembrane protein structures using cryo-EM and evolutionary data. Biophys J. 2004;87:3448–3459. doi: 10.1529/biophysj.104.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens TJ, Arkin IT. Substitution rates in alpha-helical transmembrane proteins. Protein Sci. 2001;10:2507–2517. doi: 10.1110/ps.10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayrose I, Mitchell A, Pupko T. Site-specific evolutionary rate inference: taking phylogenetic uncertainty into account. J Mol Evol. 2005;60:345–353. doi: 10.1007/s00239-004-0183-8. [DOI] [PubMed] [Google Scholar]
- 52.Petrey D, Xiang Z, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh IY, Alexov E, Honig B. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53:430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Navarro A, Ramos J. Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol. 1984;159:940–945. doi: 10.1128/jb.159.3.940-945.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.