Abstract
Microarray expression profiling has become a valuable tool in the evaluation of the genetic consequences of metabolic disease. Although 3′-biased gene expression microarray platforms were the first generation to have widespread availability, newer platforms are gradually emerging that have more up-to-date content and/or higher cost efficiency. Deciphering the relative strengths and weaknesses of these various platforms for metabolic pathway level analyses can be daunting. We sought to determine the practical strengths and weaknesses of four leading commercially-available expression array platforms relative to biologic investigations, as well as assess the feasibility of cross-platform data integration for purposes of biochemical pathway analyses.
METHODS
Liver RNA from B6.Alb/cre,Pdss2loxP/loxP mice having primary Coenzyme Q deficiency was extracted either at baseline or following treatment with an antioxidant/antihyperlipidemic agent, probucol. Target RNA samples were prepared and hybridized to Affymetrix 430 2.0, Affymetrix Gene 1.0 ST, Affymetrix Exon 1.0 ST, and Illumina Mouse WG-6 expression arrays. Probes on all platforms were re-mapped to coding sequences in the current version of the mouse genome. Data processing and statistical analysis were performed by R/Bioconductor functions, and pathway analyses were carried out by KEGG Atlas and GSEA.
RESULTS
Expression measurements were generally consistent across platforms. However, intensive probe-level comparison suggested that differences in probe locations were a major source of inter-platform variance. In addition, genes expressed at low or intermediate levels had lower inter-platform reproducibility than highly expressed genes. All platforms showed similar patterns of differential expression between sample groups, with steroid biosynthesis consistently identified as the most down-regulated metabolic pathway by probucol treatment.
CONCLUSIONS
This work offers a timely guide for metabolic disease investigators to enable informed end-user decisions regarding choice of expression microarray platform best-suited to specific research project goals. Successful cross-platform integration of biochemical pathway expression data is also demonstrated, especially for well-annotated and highly-expressed genes. However, integration of gene-level expression data is limited by individual platform probe design and the expression level of target genes. Cross-platform analyses of biochemical pathway data will require additional data processing and novel computational bioinformatics tools to address unique statistical challenges.
INTRODUCTION
Microarray expression profiling offers a useful means of surveying the global genetic consequences of complex diseases, such as inborn errors of metabolism. 3′-biased gene expression microarrays were the first generation to gain widespread use, with data from more than 12,000 Affymetrix (Mo4302) GeneChips alone deposited by August 2009 in the NCBI Gene Expression Omnibus (GEO) microarray public repository (www.ncbi.nlm.nih.gov/geo). Newer platforms are gradually emerging that offer more comprehensive coverage, updated content, alternative technical designs, and/or greater cost efficiency for the study of new biological specimens. While the original 3′-biased arrays utilize probes localized near the distal end of each transcript, increasing probe coverage is provided on newer Affymetrix platforms. Affymetrix Gene arrays provide even probe coverage along the entire 5′ to 3′ transcript, whereas Exon arrays select multiple probes from every probe selection region such as exons and UTRs [1]. Compared to Affymetrix platforms that interrogate each transcript with multiple 25-mer probes, the Illumina platform measures each transcript with multiple copies of a single 50-mer probe sequence. The MicroArray Quality Control (MAQC) project offers a large-scale bioinformatics effort to evaluate the technical variability of seven expression platforms along parameters such as data reproducibility and signal variation between test sites [2]. Smaller scale bioinformatics studies also tend to provide technical comparisons of global array performance, irrespective of investigator-driven aims. However, there is a lack of comparative microarray studies that are either directed at scientists rather than bioinformaticians or focus on well-defined biological questions.
Integration of microarray data obtained from multiple independent studies either for data mining or validation purposes is increasingly possible owing to growing public accessibility to expression microarray data [3–6]. However, compatibility between older and newer expression microarray platforms has yet to be fully explored. A recent study suggested that although Affymetrix and Illumina platforms do generate highly comparable data, inter-platform correlation is significantly impacted by expression level and probe location [7]. Comparative analysis of gene expression accuracy and precision in cancer cell lines between the Affymetrix GeneChip, Amersham Codelink, and Agilent Technologies two-color platforms concluded that one-color platforms provided greater precision, while Affymetrix GeneChip provided slightly higher sensitivity for differential gene expression [8]. Probe design by different teams at different times is a critical issue which may prohibit direct comparison of data generated on different microarray platforms, even when probes were originally assigned to the same genes. Furthermore, the relative virtues and weaknesses of various platforms have not been reported for purposes of cluster analyses, such as gene grouping by biochemical pathway involvement. Indeed, determination of cross-platform data integration compatibility relevant to biochemical pathways necessitates careful dissection of individual platform performance at both gene and probe levels.
To address these needs, we compared the functional consequences of inter-platform differences in array data assessed by gene clustering analyses using a well-studied mouse model of therapy for primary coenzyme Q deficiency [9]. This work provides timely descriptions of the practical strengths and weaknesses of four commercially-available expression arrays with particular relevance to planning studies involving pathway-level analyses. Indeed, the four expression platforms differed substantially in terms of content, annotation, and software support. The data demonstrated significant inter-platform agreement in terms of differential gene expression between treatment groups, and show that pathway-level expression data can be integrated across different platforms. However, significant limitations to direct integration of gene-level expression data from different platforms existed based largely on probe design and the expression level of target genes.
METHODS
Mouse treatment groups
Liver-conditional knockout mice for Pdss2, a coenzyme Q biosynthetic pathway gene, were generated as previously described [9]. Briefly, the mutation was targeted to hepatocytes by utilizing mice homozygous for the floxed gene (B6.Pdss2loxP/loxP) crossed with partners that expressed cre under the control of an albumin/cre promoter (B6.Cg-Tg(Alb-cre)21 Mgn/J (Alb/cre)) obtained from The Jackson Lab. Mice with the Pdss2kd/kd missense mutation on the B6 background have been previously reported [10,11]. Two B6.Alb/cre, Pdss2loxP/loxP mice (one male and one female) were fed standard mouse chow supplemented with probucol (1% wt/vol) from weaning beginning on day of life 44. Untreated controls consisted of two B6.Alb/cre, Pdss2loxP/loxP mice (one male and one female) fed standard mouse chow. Animals were sacrificed and liver specimens flash frozen for RNA extraction at 140 to 169 days old. All procedures were approved by the Institutional Animal Care and Use Committee of both the University of Pennsylvania and The Children s Hospital of Philadelphia.
RNA isolation and microarray hybridization
Total RNA from each mouse was isolated by Trizol extraction, purified, and combined into a single aliquot from 100 mg flash-frozen liver specimens collected at the time of sacrifice, as previously described [9]. RNA isolated from each of the four mice, described above, was individually hybridized on each of four platforms: Affymetrix 430 2.0, Affymetrix Gene 1.0 ST, Affymetrix Exon 1.0 ST, and Illumina Mouse WG-6 v2.0. Each male sample was hybridized to two Illumina arrays as technical replicates. All RNA labeling and hybridization was performed in the CHOP Nucleic Acid and Protein Core Facility according to protocols specified by the manufacturers.
Microarray data processing
Probe sequences of all platforms were downloaded from Affymetrix and Illumina websites, parsed into FASTA format, and sent to AffyProbeMiner, a web-based tool that re-maps probes to the updated version of mouse coding sequences based on RefSeq and GenBank databases. A perfect match to the reference sequences was required for a probe to be used. Probes matching the same transcript were grouped as a probe set. Raw data from scanned microarray chips were retrieved using Affymetrix Expression Console (as CEL files) and Illumina BeadStudio (as tab-delimited files) software. The raw probe-level data were processed using the RMA method [12] to obtain probe set-level expression measurements. Details of data processing are provided as Supplemental File 1. Microarray data as well as the library and annotation files generated by this study are publicly available in a MIAME-compliant database (GEO accession GSE18677) (www.ncbi.nlm.nih.gov/geo).
Statistical analysis of microarray data
All statistical analyses were performed within R/Bioconductor statistical environment (www.bioconductor.org). The four microarray samples permitted analysis of two possible group comparisons, gender and treatment. Gender difference was used to evaluate platform similarity at the gene level, as some genes are expected to have differential expression between males and females (eg. y chromosome genes). Treatment differences were used to evaluate platform similarity at the gene set level, which was more relevant to the underlying biological question. Differential gene expression between untreated and probucol-treated samples was first calculated as the average of the male and female pairs. The values were then adjusted by penalizing genes with low expression or high variance between males and females (see details in Supplemental File 1). Gene lists ranked by adjusted group difference were input into two functional annotation tools for pathway analysis. KEGG Atlas (www.genome.jp/kegg/atlas.html) was used to visualize differential expression of metabolic genes on a well-organized global metabolism map. GSEA [13] was used to draw statistical conclusions about the modification of metabolic pathways by probucol treatment. Gene-to-pathway mapping information was downloaded from the KEGG web site.
RESULTS and DISCUSSION
Content analysis of four expression microarray platforms
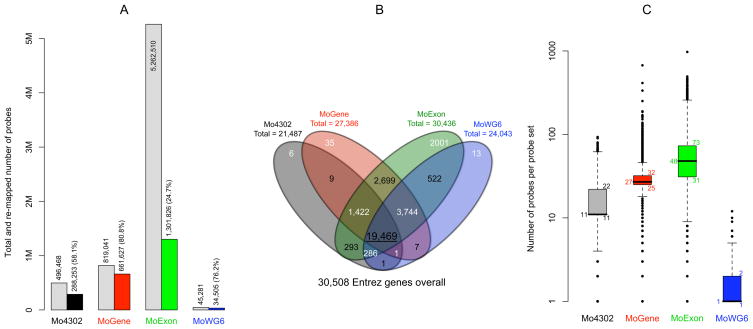
Specific microarray platforms compared in this study were Affymetrix Mouse Genome 430 2.0 (Mo4302), Mouse Gene 1.0 ST (MoGene), and Mouse Exon 1.0 ST (MoExon), as well as Illumina Mouse WG-6 v2.0 (MoWG6). Previous studies have noticed that a considerable portion of Affymetrix probes became obsolete as the quality of genome sequences improved [14] and suggested that microarray probes should be re-annotated over time [15–17]. We therefore applied a probe re-mapping tool, AffyProbeMiner [18], to map all probes to the most recent version of consensus coding sequences in RefSeq and GenBank databases. Probes mapped to the same transcript were grouped into probe sets to permit comparison at the level of transcripts or genes. Only probes perfectly mapped to one or multiple locations within coding sequences were subsequently studied. Less than 60% of Mo4302 probes mapped perfectly (Figure 1A), which was not unexpected since the quality of mouse reference sequences has substantially improved since the platform was introduced more than five years ago. Substantial improvement was seen with the MoGene platform, which had 80.8% of its probes perfectly mapped to the mouse reference genome. However, re-mapping filtered out more than 75% of probes on the MoExon platform, since most of its probes were selected from regions of incomplete and predicted transcripts. Nevertheless, because of the overall greater number of probes, more perfectly matched probes remained on the MoExon platform than on any other platform after filtering. 76.2% of Illumina MoWG6 probes were perfectly mapped, although they were twice as long as Affymetrix probes (50mer vs. 25mer). These results reflect the high probe quality on Illumina platforms, since their typical interrogation of each transcript by only one probe requires more stringent probe selection.
Figure 1. Content of four expression microarray platforms.
A) For each platform, total number of unique probes and probe number perfectly re-mapped to the current version of the mouse coding sequence are indicated by the first and second bars, respectively. B) When re-grouped probe sets were mapped to NCBI Entre genes, 35,508 unique genes were included on one or multiple platforms. Over 60% of unique genes were commonly shared by all four platforms. C) Re-mapped probes were grouped intro probe sets which were uneven in size. Boxplots numbers from bottom upward indicate 1st, 2nd, and 3rd quartiles of the number of probes in 12,783 probes sets common to all platforms.
Since the only array studied capable of detecting alternative splicing was MoExon, the four platforms were compared at the level of genes rather than transcripts. To this end, probe sets were further mapped to Entrez genes. MoExon was the most comprehensive platform, missing only 72 genes covered by other platforms (Figure 1B). Most of the genes unique to MoExon were predicted and not yet functionally annotated. The Mo4302 3′ array missed 3,744 genes covered by the other three platforms, including mainly predicted genes and approximately 1,000 olfactory receptor genes. The 1,422 genes included on all three Affymetrix platforms but not on the Illumina platform were largely also predicted genes. In contrast, 80% of the 19,469 genes common to all four platforms were functionally annotated. Notably, probe sets and genes did not have a one-to-one relationship. Some probe sets mapped to multiple genes due to sequence similarity, and some genes were mapped by multiple probe sets due to alternative splicing. To avoid ambiguity, 12,783 Entrez genes that were exclusively mapped by exactly one probe set on all platforms were used for downstream platform comparisons.
Transcripts were originally mapped by probe sets containing eleven probes on Mo4302 and usually by just one probe sequence on MoWG6. In contrast, MoGene and MoExon originally used variable numbers of probes to interrogate each transcript. Re-mapping resulted in probe sets being comprised of uneven probe numbers (Figure 1C). However, not all probe sets were affected by the re-mapping procedure. For example, 49% of the Mo4302 probe sets and 62% of the MoWG6 probe sets still included eleven and one probe, respectively.
Comparative analysis of gene expression measurements on four microarray platforms
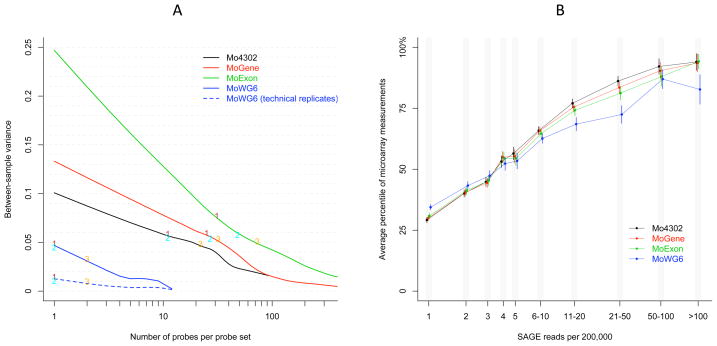
Microarray platforms are expected to generate repeatable expression measurements to distinguish subtle difference in RNA abundance. We evaluated the repeatability and sensitivity of expression measurements on all four microarray platforms. Among the three Affymetrix platforms, MoGene showed the least variability and MoExon showed the highest variability in probe set size. Average probe set size was largest in MoExon (57.9 probes per probe set) and smallest in MoWG6 (1.56 probes per probe set). Analysis of between-sample variance of 12,783 common genes demonstrated an inverse relationship of variance and probe set size on all platforms (Figure 2A). The four platforms distinctly differed in variance of small probe sets (10 or less probes). Probe replication and longer probe size likely accounted for MoWG6 probe sets having the lowest variance. Increasing variance from Mo4302 to MoGene to MoExon may be attributable to reduced feature (spot) size, which is a necessary consequence of higher chip density. The majority of genes were not substantially affected, however, since variance among median-sized (1st to 3rd quartiles) probe sets were at similar levels both within and across platforms. Biological variation may have accounted for most of the between-sample variance in this data set since the mouse samples studied belonged to different gender and treatment groups, although neither biological or experimental factors satisfactorily explained the dependence of variance on probe set size. To further verify this point, identical hybridization material from two pairs of technical replicates was applied to two MoWG6 arrays. Overall variance between technical replicates was substantially reduced in this data set but remained negatively correlated to probe set size.
Figure 2. Repeatability and sensitivity of gene expression measurements across four expression microarray platforms.
A) The between-sample variance of probe sets was plotted versus the probe size. The lines were smoothed by lowess [23] function implemented in R. The blue dashed line was generated based on two pairs of technical replicates measured on the same Illumina Mouse-6 BeadChip. The labels indicate the 1st, 2nd, and 3rd quartiles of probe set size, corresponding to the labeled numbers in Figure 1C. B) Individual platform sensitivity to varying mRNA abundance was evaluated with a SAGE library of male mouse liver as reference (mLSAGE_Liver_normal_P84_MMD_SM100, SAGE Genie). Given a range of SAGE read frequencies, microarray probe set average measurement in two male mice and 95% confidence intervals are represented by nodes and vertical bars, respectively. Node positions were slightly shifted along the X-axis to improve visibility of individual error bars.
Microarray measurements are expected to be solely based on RNA abundance. Perfect correlation is not practically achievable due to technical variables such as length, GC content, and probe cross-hybridization. Nevertheless, a properly designed platform should make measurements that are generally consistent with RNA abundance. This parameter was evaluated by comparing microarray measurements to the frequency of short reads using a mouse liver serial analysis of gene expression (SAGE) library as a reference. The two technologies were moderately correlated, with Spearman correlation coefficients equal to 0.66 (Mo4302), 0.64 (MoGene), 0.60 (MoExon), and 0.48 (MoWG6). Approximately 70% of the genes in the SAGE library were mapped by five or less reads per 200,000 bases, indicating that most genes were expressed at medium to low levels. The average measurements made by all four platforms differed significantly among genes mapped by only one or two SAGE reads [p values equal to 5.1e-31 (Mo4302), 6.7e-27 (MoGene), 1.8e-23 (MoExon), and 6.5e-19 (MoWG6)] (Figure 2B).
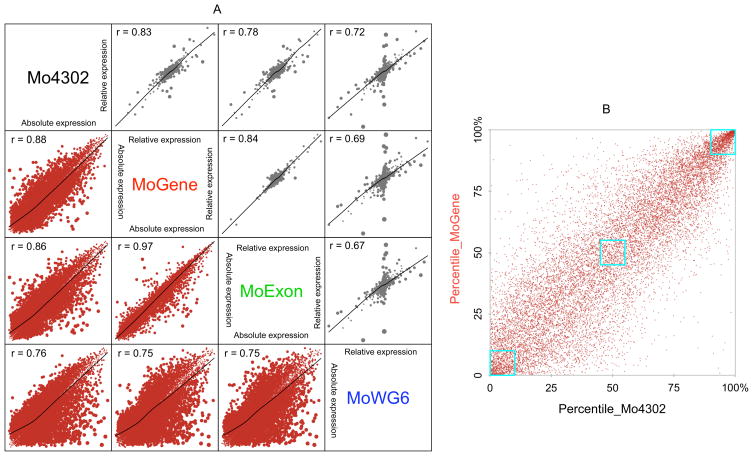
Individual platform reliability in determining absolute expression measurements and relative expression changes between groups was assessed by directly comparing each platform pair (Figure 3A). Comparison between male and female samples demonstrated the lowest pair-wise correlation between MoWG6 and any of the three Affymetrix platforms, which was most likely attributable to differing technologies. MoGene and MoExon displayed the best agreement (r = 0.97), which was not unexpected since these two platforms share many common probes. Most genes were randomly distributed without differential expression between gender groups, with cross-platform correlations higher among absolute measurements than relative expression changes. Although outlier gene expression existed between all platform pairs, all platforms largely agreed on most differentially expressed genes. Interestingly, pair-wise correlations of absolute measurements depended on gene expression levels. Highly expressed genes showed the best agreement when expression measurements were rescaled to percentiles (Figure 3B), likely owing to near-saturation of their signals on all platforms. Conversely, measurements from genes with medium expression levels varied dramatically between platforms. Although non-expressed genes should have measurements close to background on all platforms, disagreement for many was observed between platforms. Since highly expressed and housekeeping genes are rarely the focus of microarray studies, this observation suggests that absolute expression levels measured by different platforms are not directly comparable.
Figure 3. Inter-platform correlation.
A) The absolute expression measurement (brown) and relative expression changes based on gender (grey) were compared between each pair of expression array platforms. The absolute measurement is the average of four samples and the relative change is the difference between two male samples and two female samples. The size of the dots is proportional to platform difference. Data have been processed by quantile-quantile normalization before plotting, so both axes are in the same scale. B) The platform-platform correlation between Affymetrix 3′ array (Mo4302) and gene (MoGene) platforms of absolute expression measurements is dependent on gene expression levels. Average expression measurements were transformed to percentiles. Three blue boxes correspond to low (0 to 10th percentile), intermediate (45th to 55th percentile), and high (90th to 100th percentile) transcript abundance as measured by both platforms. Transcript overlap within these three boxes in sequential descending expression order was 48.4%, 24.7%, and 73.2%, respectively.
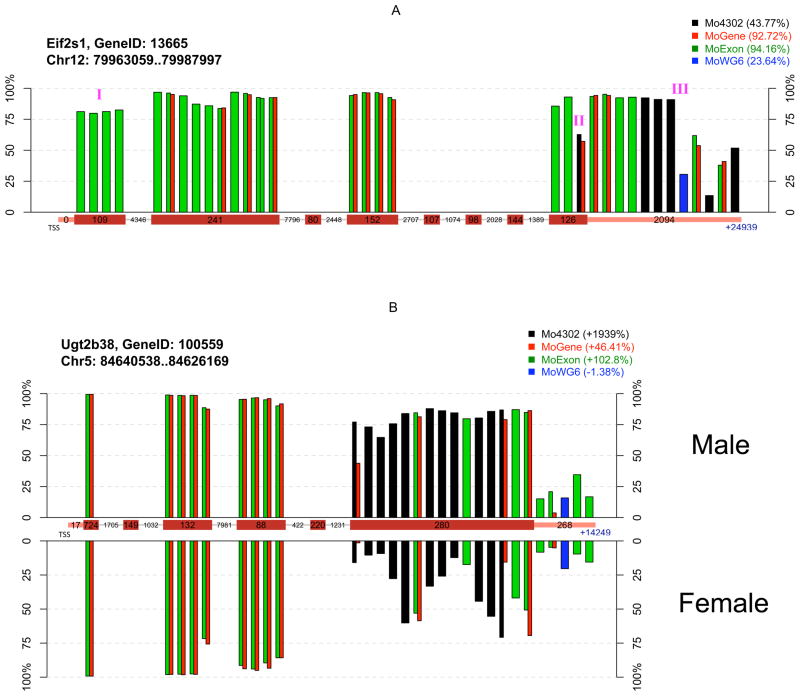
The potential basis for platform inconsistencies was explored by scrutinizing a few genes at the probe level between different sample groups (Figure 4). Eif2s1 encodes a key translation initiation factor having two expressed mRNAs, which differ by one having an extended 3′ UTR that regulates its mRNA stability [19]. Its average gene-level expression among four mouse samples was high according to MoGene and MoExon, moderate according to Mo4302, and low according to MoWG6. Analysis of probe-level measurements along the complete Eif2s1 transcript permitted several remarkable observations (Figure 4A). First, probes mapped to the same location provided very similar expression measurements even when assessed on different platforms. Second, probes located in the same exon usually showed greater similarity to each other than to probes located in different exons. Lastly, probe distribution was uneven across the transcript and differed greatly between platforms. Eif2s1 expression measurements were overall high but dropped significantly near the end of the 3′ UTR, probably due to alternative transcription. Consequently, gene-level measurements were highly dependent on probe location. Since most MoGene and MoExon Eif2s1 probes were located in exons, they showed high gene-level expression. However, the single MoWG6 probe for this gene was located in the 3′ UTR, which had low expression and missed the majority of the transcript. Detailed analysis was performed for a second gene, Ugt2b38, which encodes an enzyme whose substrate includes steroid hormones [20]. Ugt2b38 probe-level expression analysis in samples grouped by gender suggested that its last exon was the only differentially expressed region of this gene (Figure 4B). Since all Mo4302 probes were located in that last exon, this platform reported dramatic differential gene-level expression of Ugt2b38 by gender. MoGene and MoExon probes in the last exon of Ugt2b38 were diluted by additional probes in other gene regions, although its differential expression was still apparent at the gene level. Exon level analysis of MoExon data would identify the differential expression of the last exon. Since the sole MoWG6 probe interrogating Ugt2b38 expression was located in its 3′ UTR, it failed to detect gender-based expression differences. These two focused gene examples suggest probe location is an important factor underlying inter-platform variation. Additional gene-specific analyses are provided in Supplemental File 2.
Figure 4. Interplatform comparison of probe-level expression measurements between sample groups.
Each bar represents individual probe(s) color-coded by platform at unique chromosomal locations, with bar height indicating probe-level measurement percentile. Probes common to multiple platforms are indicated by adjacent bars. Probes are ordered relative to their distance from the transcriptional start site (TSS), and packed into their corresponding gene segment indicated on the central x-axis (exon, intron, or 3′ UTR). Box width is not proportional to segment size, but numbers within each box indicate actual base number per segment. A) Absolute measurements at the probe set-level are highly variable among platforms due to differences in probe location and number. Legend indicates probe set-level average percentiles of four mouse samples on each platform. (I) These probes are located in a non-coding exon. (II) This probe is located at the exon-3′ UTR junction, with 14 bases in the last exon and 11 bases in the 3′ UTR. The measurements from this probe are considerably lower than those from nearby probes, suggesting an excision event. (III) The gap between these two probes is about 800 bases long, which separates the 3′ UTR into two parts. The first part of the UTR is highly conserved across species according to the UCSC genome browser, but the second part is not. B) Probe location and number impacts the relative change at the probe-set level between sample groups. Probe-level percentiles were plotted separately by sample gender. Legend indicates probe set-level change of two male samples relative to two female samples. Eif2s1 and Ug2b38 are examples of genes having highly different results by platforms. Their analysis is not suggested to indicate overall platform performance, as expression patterns may substantially differ under different experimental conditions. Additional probe-level plots are available as supplementary materials.
Metabolic pathway analysis in probucol-treated vs control B6.Alb/cre, Pdss2loxP/loxP mice
Downstream analysis of microarray data commonly involves grouping genes into common categories, such as Gene Ontology terms or metabolic pathways, to coherently assess altered gene expression patterns at a systems level. This perspective allows researchers to efficiently link their experiments to known information to gain an overall biologic gestalt. Many tools have been developed to support such analyses, including Expression Analysis Systematic Explore (EASE, www.david.abcc.ncifcrf.gov/ease) and Gene Set Enrichment Analysis (GSEA, www.broad.mit.edu/gsea). Inter-platform comparison can further benefit from gene set-based analyses. As long as overall expression patterns across gene clusters are consistent, it will be possible to draw the same biologic conclusion from different platforms regardless of particular distinctions in single genes.
To this end, we compared expression alterations in Kyoto Encylcopedia of Genes and Genomes (KEGG) defined metabolic pathways following probucol treatment on four different expression platforms. Probucol is an antihyperlipidemic and antioxidant drug known to increase the rate of LDL catabolism and inhibit cholesterol synthesis [21,22]. Consequently, we postulated that probucol treatment may alter intermediary lipid metabolism and other inter-connecting biochemical pathways. Visualization of global differential expression among metabolic pathway genes was performed by mapping their expression in KEGG Atlas. Focused analysis of the lipid metabolism section of the Atlas demonstrated that all four platforms conveyed similar expression patterns at the pathway level despite variation at the level of individual genes (Figure 5). Indeed, all platforms reported significant concordant down-regulation of two metabolic pathways, “biosynthesis of steroids” and “fatty acid biosynthesis”.
Figure 5. Interplatform comparison of pathway-level transcriptional response of lipid metabolism to Probucol treatment.
Kegg Atlas maps permit visualization of a subset of pathway-level expression changes in sample groups related to lipid metabolism (www.kegg.jp/kegg/atlas). All four expression pathways indicated Probucol therapy results in significant down-regulation of fatty acid biosynthesis. The complete metabolic pathway maps are available as supplementary materials. Map elements were colored red or green if the corresponding genes were up- or down-regulated by Probucol, respectively. The magnitude of change was coded in color scale. Unavailable elements were labeled gray.
Quantitative pathway-level analyses were performed by applying GSEA to KEGG-pathway defined genes that were pre-ranked based on differential expression between samples grouped by probucol treatment. GSEA outputs of metabolic pathway analyses are provided in Supplemental File 3. “Biosynthesis of steroids” was consistently identified as the most significantly altered pathway by all four platforms, consistent with KEGG Atlas findings. However, different platforms yielded inconsistent, or even opposite, results for several marginally-altered pathways. For example, “drug metabolism – cytochrome P450” was identified as one of the most up-regulated pathways by MoGene, only marginally up-regulated according to Mo4302, and down-regulated according to MoExon and MoWG6. A closer look at expression of individual genes in this pathway (Figure 6) suggested that three genes (Cyp3a16, Cyp3a41, and Cyp3a44) underlie the discrepancy. Indeed, expression of these three genes was highly up-regulated only according to MoGene, which decisively affected the GSEA statistical result by being included as leading edge elements [13]. These same genes also resulted in inter-platform inconsistencies among other pathways, including “drug metabolism – other enzymes” and “linoleic acid metabolism”. Furthermore, while the overall expression of the “biosynthesis of steroids” pathway and several of its component genes were commonly upregulated (Figure 6), a subset of genes in this pathway was significantly down-regulated on all four platforms. Thus, while pathway-level analysis does yield an overall initial biologic gestalt, closer analysis of component pathway genes is necessary to achieve an accurate understanding of the underlying biology.
Figure 6. Inter-platform comparative heatmap of gene-level expression alterations in two metabolic pathways up-regulated by Probucol therapy.

Consistent upregulation of steroid biosynthesis is seen across all four expression platforms. However, greater variability in overall pathway expression alterations is seen in cytochrome P450 metabolism, with a subset of pathway genes similarly down-regulated on all four platforms despite overall pathway upregulation. Arrows indicate individual genes driving inter-platform discrepancies in pathway-level expression. Colors and scale indicate the direction and magnitude of expression change of individual genes after treatment, where red was up-regulated, green was down-regulated, black was unchanged, and white was unavailable.
DISCUSSION
We demonstrate overarching similarities and individual distinctions in data generated from the same samples applied to four expression platforms purchased from two commercial vendors (Table 1). Instead of using artificial samples characterized solely by known amounts of RNA, we selected samples from a traditional biomedical experiment whose aim was to assess drug effects in a disease model. Hence, the “truth” about the samples was unknown and the primary data analysis strategy involved assessment of expression consistency at the levels of probes, genes, and pathways across platforms. Since the samples analyzed were representative of a real-life scientific scenario, this study serves as a useful paradigm for other microarray experiments aimed at assessing global transcriptional responses of inborn errors of metabolism and their potential therapies.
Table 1.
Comparison of platform features and performance relative to Mo4302.
| MoGene | MoExon | MoWG6 | |
|---|---|---|---|
| Probe length | = | = | + |
| Probe location | + | +++ | = |
| Number of unique probes | + | +++ | — |
| Number of unique genes | + | +++ | + |
| Repeatability of measurements | = | − | +++ |
| Sensitivity to low expression | = | = | − |
| Detection of alternative splicing | + | +++ | − |
| Performance of pathway analysis | = | = | = |
| Support from third party software* | − | — | − |
All comparisons are made relative to Mo4302 as a reference platform. =, similar to Mo4302; −−−/+, worsened/improved; −−−/+++, much worsened/improved;
evaluation of support from third party software is made based on currently available software and methods for data processing/normalization, functional annotation, statistical analysis, and pathway analysis.
Understanding individual expression platform strengths and limitations was enhanced through comparison of platform data at three levels: probe, probe set/gene, and gene set/pathway. Probe level comparison suggested that probe location relative to individual genes greatly affected expression measurement. While probes located in different gene regions often resulted in dissimilar measurements regardless of originating platform, probes from different platforms that localized to the same gene region often yielded similar expression data. Therefore, the distribution of probes within genes is a major factor affecting gene level measurements, especially when genes have alternative transcript forms (Figure 4). Gene-set or pathway-level analyses can partially avoid such bias, as long as most genes in the sets are not profoundly affected by technical variation. Regardless, analysis of probe-level data among focused genes or gene sets may still be necessary when inter-platform inconsistencies are observed. In addition, while pathway-level analysis permits reduction of data complexity to provide an overall biologic gestalt, subsequent gene-level analysis of component members of key pathways may be necessary to gain complete biologic understanding since this may be more complicated than evident from the overall pathway expression profile (Figure 6).
Although high inter-platform variance was observed among individual gene expression from four different expression platforms, pathway-level analyses of differential gene expression were generally compatible. Indeed, different platforms agreed on the absolute expression level of highly-expressed genes, but often made discordant measurements of intermediately- and minimally-expressed genes. Hence, the absolute expression level of most genes could not be directly compared across platforms. Probe set size was another key factor underlying differences at the individual gene level. Larger probe sets tended to have smaller between-sample variance (Figure 2A) and presumably smaller between-platform variance. This observation suggests that larger probe sets will be favored by statistical methods that account for data variance, such as Student s t test, when evaluating differential gene expression between sample groups. A consequence might be the production of differentially-expressed gene lists that are similar in unrelated experimental scenarios. Such distinctions in gene performance will be amplified by projects involving multiple data sets, which could lead to substantial systematic bias. Although trimming large probe sets is a possible solution to the issue of their smaller between-sample variance, several new problems will arise. For example, random trimming will reduce overall data set quality, as a substantial portion of the data will be removed. If trimming leaves only the “best” probes, new bias will be introduced. It is preferable to correct this bias by adjusting common statistical methods, such as Student s t test, to include probe set size as a factor. Thus, novel statistical methods are needed for data adjustment to avoid systematic bias in integrated analyses of microarray data sets.
Ultimately, platform selection for microarray experiments must be based on multiple factors including budget, cost efficiency, inclusion of previously acquired expression data, and local support in terms of expertise and laboratory equipment. However, conclusions drawn from this study will assist researchers in making a more informed end-user decision about platform properties that are best-suited to their research project goals (Table 1). For example, since Illumina platforms utilize only one probe sequence for each transcript, they are not flexible to assess alternative mRNA splicing. Indeed, many genes expressed in the study samples according to three Affymetrix platforms were not detected on MoWG6 (Figure 3). MoExon was clearly the most comprehensive platform (Figure 1), permitting expression measurement at the individual exon level not possible on other platforms. However, the tradeoff of compressing over five million probes onto the MoExon platform was reduced data repeatability and greater variability in probe set size (Figure 2). Although Mo4302 had more than 40% outdated probes (Figure 1A) and only covered approximately 70% of the genes measured by MoExon (Figure 1B), analysis results generated from the older Mo4302 platform were compatible to those from more updated and comprehensive platforms (Figure 5 and Figure 6). Lack of functional categorization for most of the genes not covered by Mo4302 implies limited practical benefit of higher transcript comprehensiveness provided by newer platforms pending substantial improvement in biologic knowledge about these novel genes. Indeed, we continue to use Mo4302 for analyses related to this mouse model because we already have substantial data generated from this platform, its genes are functionally well-characterized, and the best software support for pathway analysis, such as GSEA, is currently available on this platform.
This study demonstrated the feasibility of achieving consistent study results at the biochemical pathway level from data obtained on different expression platforms. However, the integrated analysis of data generated from platforms that exploit different technologies, such as the Affymetrix and Illumina platforms studied here, is not generally encouraged. Integration of data from different platforms using the same technology, such as the three Affmetrix platforms studied here, involves fewer technical inconsistencies and is possible to achieve. For example, data inconsistencies attributable to probe location might be avoided by including data generated only from Exon or Gene array probes that are located near the probes of 3′-biased platform. Further investigation will be essential to optimize data processing to remap and filter probes, as well as development of novel computational bioinformatics tools and analysis strategies to address statistical challenges specific to cross-platform studies.
Supplementary Material
Acknowledgments
This work was supported by grants R01-DK55852 (DLG) and K08-DK073545 (MJF) from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We are grateful to Min Peng, Ph.D., Erzsebet Polyak, Ph.D., Meera Rao, and Stephen Dingley, B.A. for their assistance with RNA isolation and specimen handling; and Juan Carlos Perin, M.S. for his generous support with computational resources.
ABBREVIATIONS
- MAQC
MicroArray Quality Control (MAQC)
- GEO
gene expression omnibus
- KEGG
Kyoto Encylopedia of Genes and Genomes
- GSEA
gene set enrichment analysis
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
ZZ, EFR, and MJF conceived of the study. DLG coordinated animal breeding, handling, and treatment. MJF coordinated molecular genetic specimen preparation and handling. EFR performed microarray hybridizations. ZZ performed bioinformatics and statistical analyses. ZZ and MJF drafted the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xing Y, Kapur K, Wong WH. Probe selection and expression index computation of Affymetrix Exon Arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Chen D, Fenstermacher DA. Integrated analysis of independent gene expression microarray datasets improves the predictability of breast cancer outcome. BMC Genomics. 2007;8:331. doi: 10.1186/1471-2164-8-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McHale CM, Zhang L, Lan Q, Li G, Hubbard AE, Forrest MS, Vermeulen R, Chen J, Shen M, Rappaport SM, Yin S, Smith MT, Rothman N. Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics. 2009;93:343–349. doi: 10.1016/j.ygeno.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Reynies A, Geromin D, Cayuela JM, Petel F, Dessen P, Sigaux F, Rickman DS. Comparison of the latest commercial short and long oligonucleotide microarray technologies. BMC Genomics. 2006;7:51. doi: 10.1186/1471-2164-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng M, Falk MJ, Haase VH, King R, Polyak E, Selak M, Yudkoff M, Hancock WW, Meade R, Saiki R, Lunceford AL, Clarke CF, Gasser DL. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4:e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock WW, Tsai TL, Madaio MP, Gasser DL. Cutting Edge: Multiple autoimmune pathways in kd/kd mice. J Immunol. 2003;171:2778–2781. doi: 10.4049/jimmunol.171.6.2778. [DOI] [PubMed] [Google Scholar]
- 11.Peng M, Jarett L, Meade R, Madaio MP, Hancock WW, George AL, Jr, Neilson EG, Gasser DL. Mutant prenyltransferase-like mitochondrial protein (PLMP) and mitochondrial abnormalities in kd/kd mice. Kidney Int. 2004;66:20–28. doi: 10.1111/j.1523-1755.2004.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Iratxeta C, Andrade MA. Inconsistencies over time in 5% of NetAffx probe-to-gene annotations. BMC Bioinformatics. 2005;6:183. doi: 10.1186/1471-2105-6-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6:107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Zeeberg BR, Qu G, Koru AG, Ferrucci A, Kahn A, Ryan MC, Nuhanovic A, Munson PJ, Reinhold WC, Kane DW, Weinstein JN. AffyProbeMiner: a web resource for computing or retrieving accurately redefined Affymetrix probe sets. Bioinformatics. 2007;23:2385–2390. doi: 10.1093/bioinformatics/btm360. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto S, Chiorini JA, Urcelay E, Safer B. Regulation of gene expression for translation initiation factor eIF-2 alpha: importance of the 3′ untranslated region. Biochem J. 1996;315(Pt 3):791–798. doi: 10.1042/bj3150791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckley DB, Klaassen CD. Mechanism of gender-divergent UDP-glucuronosyltransferase mRNA expression in mouse liver and kidney. Drug Metab Dispos. 2009;37:834–840. doi: 10.1124/dmd.108.024224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckley MM, Goa KL, Price AH, Brogden RN. Probucol. A reappraisal of its pharmacological properties and therapeutic use in hypercholesterolaemia. Drugs. 1989;37:761–800. doi: 10.2165/00003495-198937060-00002. [DOI] [PubMed] [Google Scholar]
- 22.Kuzuya M, Kuzuya F. Probucol as an antioxidant and antiatherogenic drug. Free Radic Biol Med. 1993;14:67–77. doi: 10.1016/0891-5849(93)90510-2. [DOI] [PubMed] [Google Scholar]
- 23.Cleveland WS. LOWESS: A program for smoothing scatterplots by robust locally weighted regression. The American Statistician. 1981;35:54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.