Abstract
Both appropriate DNA methylation and histone modifications play a crucial role in the maintenance of normal cell function and cellular identity. In cancerous cells these “epigenetic belts” become massively perturbed, leading to significant changes in expression profiles which confer advantage to the development of a malignant phenotype. DNA (cytosine-5)-methyltransferase 1 (Dnmt1), Dnmt3a and Dnmt3b are the enzymes responsible for setting up and maintaining DNA methylation patterns in eukaryotic cells. Intriguingly, DNMTs were found to be overexpressed in cancerous cells, which is believed to partly explain the hypermethylation phenomenon commonly observed in tumors. However, several lines of evidence indicate that further layers of gene regulation are critical coordinators of DNMT expression, catalytic activity and target specificity. Splice variants of DNMT transcripts have been detected which seem to modulate methyltransferase activity. Also, the DNMT mRNA 3′UTR as well as the coding sequence harbors multiple binding sites for trans-acting factors guiding post-transcriptional regulation and transcript stabilization. Moreover, microRNAs targeting DNMT transcripts have recently been discovered in normal cells, yet expression of these microRNAs was found to be diminished in breast cancer tissues. In this review we summarize the current knowledge on mechanisms which potentially lead to the establishment of a DNA hypermethylome in cancer cells.
Keywords: Breast cancer, DNA methylation, Dnmt3b, HuR, MicroRNA, TARBP2
Introduction
The molecular mechanisms underlying the development and progression of breast cancer are far from being understood. It is evident that the initiation of breast cancer as well as its transition towards distinct breast cancer subtypes is triggered by the accumulation of pathologically altered gene function. Like in other cancers, the increasing number of deregulated genes subsequently affects virtually any important cellular network, such as cell cycle control, apoptosis, DNA repair, detoxification, inflammation, cell adhesion or migration. According to the somatic mutation theory cancer has long been considered as a genetic disorder of fatal acquisition of multiple mutations in key genes, which coordinate these functional networks. Such mutations can result either in inactivation of tumor suppressor genes (e.g. TP53, BRCA1) or activation of proto-oncogenes (e.g. MYC), both of which contributes to the malignant state of a transformed cell.
During the past decade, the somatic mutation theory of cancer has been revolutionized for it became evident that epigenetic malfunction plays a role as equally important as genetics in cancer development. The concept of epigenetics describes mitotically stable states and changes of gene activity that do not involve alterations of the primary DNA sequence, thus provide a second layer of information above the pure genomic blueprint [1]. Epigenetic mechanisms coordinate crucial biological processes, like X-chromosome inactivation, genomic imprinting, position effect variegation, reprogramming of genomes during differentiation and development, or RNA interference leading to posttranscriptional gene silencing. It is not surprising that defects in the dynamics of these key functions were found to be associated with many human disorders, including breast cancer (for review see [2]). In recent years, two epigenetic mechanisms have emerged as the most critical players of transcriptional regulation: The methylation of DNA and chemical histone tail modifications. DNA methylation refers to the covalent post-replicative addition of a methyl group (–CH3) onto the 5-carbon of the cytosine ring within CpG dinucleotides. This enzymatic reaction is conferred by DNA methyltransferases (DNMT), which catalyze the transfer from the methyl group donor S-adenosyl methionine. Typically, such CpG dinucleotides are enriched in gene promoters or the first exon where they cluster to form a so-called CpG island. Approximately 60% of protein-coding mammalian genes harbor CpG islands in their promoter region. These are normally unmethylated in transcriptionally active genes like housekeeping genes, whereas developmental and tissue-specific genes mostly appear to be methylated and silenced in differentiated tissues (e.g. reviewed in [3, 4]). In cancer, however, numerous genes which are unmethylated in the non-malignant tissue become aberrantly methylated in the tumor. Since the first discovery of a hypermethylated gene in cancer, the retinoblastoma tumor suppressor (RB1) [5], many tumor suppressor genes have been identified being hypermethylated in tumorous tissues as compared to their normal counterparts, e.g. VHL, CDKN2A, or BRCA1 [6–8]. Although our knowledge on epigenetically inactivated genes in cancer is constantly increasing, the basic mechanisms underlying aberrant DNA methylation as well as the selection of genes that become methylated are only rudimentary understood, and shall be reviewed further on.
The second key player in chromatin conformation and transcriptional regulation are histone modifications. Histone proteins constitute the nucleosome around which DNA is tightly packaged. Their N-terminal tails reach out of the nucleosomal core and harbor numerous spots for protein modifications, such as acetylation, methylation, phosphorylation, sumoylation, ubiquitination or ADP ribosylation (reviewed in [9, 10]). Both the type of modification and the affected amino acid residue determine the tightness of the DNA-histone interaction, leading to either an open chromatin state allowing active transcription (e.g. acetylation of lysine) or to a compact chromatin state associated with transcriptional repression (e.g. deactetylation of the same residue). Altered histone modifications in breast cancer will be reviewed in a further article of this issue, but one essential relationship ought to be mentioned: DNA methylation and histone modifications interact with each other in the regulation of gene expression. It is generally believed that DNA methylation is the initiating event that marks certain genomic sites for the establishment of a transcriptionally inactive chromatin state [11, 12]. DNA methylation, however, may also depend on prior methylation of histone 3 at lysine 9 (H3K9) [13], and is followed by binding of methyl-CpG binding domain proteins (MBDs) which contribute to gene repression by the recruitment of histone deacetylases (HDACs) to the nucleosome [14, 15]. Also, for certain genes it has been shown that the initial recruitment of DNMTs to target sequences is mediated by Enhancer of zeste homologue 2 (EZH2) as a part of the repressive polycomb group (PcG) of proteins [16, 17], increasing the complexity of relations between various epigenetic repression systems.
In recent years, the discovery of a class of small non-coding RNAs, so called microRNAs (miRNAs), has gained much attention in oncological research. MiRNAs are regulatory RNAs 20–30 nucleotides in length, that perfectly match the 3′ untranslated regions (3′UTR) of target mRNAs, resulting in its degradation or inhibition of mRNA translation (reviewed in [18]). It is the function of the target mRNA that determines a miRNA acting either tumor suppressive (if directed against proto-oncogene transcripts) or oncogenic (if directed against tumor suppressor gene transcripts). Prominent members of miRNAs include the let-7 family (containing at least 11 homologous miRNAs), whose depletion in breast, lung and colon cancer causes enhanced tumorigenicity [19–21]. Another example is miR-21, whose overexpression in breast cancer confers increased invasion capacities and promotes tumor metastasis to the lung [22, 23]. The number of genes known to be regulated by miRNAs is growing rapidly. The latest release of the Sanger miRNA registry currently annotates more than 800 human miRNAs (http://microrna.sanger.ac.uk; release 13.0), yet many more miRNAs are expected to be identified in the future. It is not surprising that miRNAs, just like protein-coding genes, have to be tightly regulated in order to contribute to a distinct transcriptome of a normal cell. In cancer, however, miRNAs were found to be massively deregulated. Recent genome-wide approaches revealed that miRNAs are globally downregulated in breast cancer [22]. Signatures of deregulated miRNAs were shown to be useful in subtyping mammary carcinomas [24, 25], or determining their aggressiveness, e.g. in node-negative estrogen receptor-positive tumors [26]. Like protein-coding genes, DNA sequences encoding miRNAs were found to be a target of aberrant DNA methylation [27], explaining in part how miRNAs may be upregulated (through DNA hypomethylation) or downregulated (through DNA hypermethylation) in cancer. Besides DNA methylation, a failure of post-transcriptional regulation may also lead to impaired miRNA biogenesis, as has been shown for the miRNA maturation responsible endoribonuclease Dicer, which is commonly expressed at lower levels e.g. in progressive breast cancer [28]. The production of mature miRNAs underlies a complex process of subsequent modifications of the primary transcript, termed pri-miRNA. The primary transcript contains a stem-loop structure representing the active miRNA species. This stem-loop is liberated by the nuclear ribonuclease III Drosha, and then termed premiRNA. After export to the cytoplasm the precursor miRNA is further processed by the ribonuclease Dicer, resulting in the mature miRNA. Finally, this miRNA is loaded into the RNA induced silencing complex (RISC) where it exhibits translational repression of its target mRNA [29–31]. In tumourous cells, discrepancies between the levels of primary transcript, precursor and mature miRNA have been reported, strongly arguing for defects in the maturation pathways of miRNAs on various levels, such as Drosha or Dicer processing [32–34]. In the second part of this article the current knowledge on defects in miRNA processing shall be highlighted.
Mechanisms of Altered DNA Methylation
DNA methylation patterns differ largely between tumor tissues and corresponding normal tissues. A paradoxon commonly observed in carcinomas is that despite of the regional hypermethylation of tumor suppressor genes, the global 5-methylcytosine content is drastically decreased in the bulk of the tumor genome. Less frequent than regional DNA hypermethylation, also regional DNA hypomethylation occurs in cancer, resulting in the activation of potential oncogenes [35]. The existence of a specific enzyme conferring active demethylation of methylated DNA is still unclear. However, enzymes conferring methylation of DNA have been well characterized.
DNA Methyltransferases
The C-terminal catalytic domain of DNMTs transfers methyl groups onto cytosine residues within the DNA, thus methyltransferases represent the crucial enzyme class responsible for hypermethylation of tumor suppressor genes. In mammals, five members of the DNMT protein family have been discovered (Dnmt1, Dnmt2, Dnmt3a, Dnmt3b, and Dnmt3L), of which only three were shown to possess catalytic methyltransferase activity (Dnmt1, Dnmt3a, and Dnmt3b). Dnmt1 exhibits a strong preference for hemi-methylated over unmethylated DNA, and its particular targeting of replication foci, as shown by co-localization with the proliferating cell nuclear antigen (PCNA), is thought to allow copying of the parental DNA methylation pattern onto the newly synthesized DNA daughter strand [36]. Therefore, Dnmt1 is regarded as a maintenance methyltransferase. The Dnmt3 family consists of two catalytic members, Dnmt3a and Dnmt3b, both of which exhibit increased methyltransferase activity towards unmethylated over hemi-methylated DNA, which is why they were termed de novo methyltransferases. DNMT3A -/- knock-out mice appear to be normally developed, but die shortly after birth. Homozygous inactivation of DNMT3B leads to embryonic lethality due to multiple developmental disorders including growth impairment and rostral neural tube defects [37]. In humans, a specific mutation in the DNMT3B gene is responsible for a syndrome referred to as ICF (Immunodeficiency, Centromere instability and Facial abnormalities), which is characterized by global hypomethylation of centromeric DNA repeat sequences, chromatin decondensation and genomic instability in tissues of affected patients [38–41]. The remaining members of the DNMT family, Dnmt2 and Dnmt3L, lack cytosine methyltransferase activity, although Dnmt3L was shown to be capable of stimulating de novo DNA methylation mediated by Dnmt3a [42, 43].
DNA Hypomethylation in Breast Cancer
Among solid tumor types, global DNA hypomethylation in most evident in breast cancer with up to 50% of cases showing reduced 5-methylcytosine content when compared with normal tissue counterparts [44, 45]. Hypomethylation in breast carcinomas mainly affects repetitive DNA sequences and pericentromeric satellite DNA, which are normally heavily methylated in non-malignant cells [44, 46]. For instance, long interspersed nuclear elements (LINEs) represent retrotransposons that are methylated in all mammalian cell types. Cancer-related hypomethylation of these transposable elements induces transcriptional reactivation, thus they can relocate and integrate into other sites of the genome, leading to insertional mutagenesis and contributing to genomic instability [47]. Hypomethylation of the Sat2 and Satα repeats frequently occurs in certain cancers, such as ovarian and breast cancer [48, 49]. In the latter, Sat2 hypomethylation was shown to affect 50% and SatR-1 hypomethylation 86% of breast tumors [49, 50]. In contrast to ovarian cancer, where increased satellite DNA hypomethylation is associated with tumor progression [48], satellite DNA hypomethylation in breast cancer is involved in early tumor development [49].
Though being a relatively rare event, DNA hypomethylation can also affect individual genes. In breast cancer, this is the case for the melanoma associated cancer/testis antigens MAGE, which are methylated and silenced in adult tissues, but hypomethylated and expressed in several tumors and breast cancer cells [51]. Other hypomethylated genes in breast tumors include the gene encoding the plasminogen activator uPA (PLAU), the breast cancer specific protein 1/synuclein-γ gene (SNCG), and more recently reported, the multidrug resistance 1 gene (MDR1) [52–54].
The underlying mechanisms leading to DNA hypomethylation in cancer have not yet been clearly elucidated. There is no reduction of DNA methyltransferase activity in cancer cells [55]. Despite, in animal models knockdown or deficiencies in the activities of DNMTs lead to genome-wide DNA hypomethylation and chromosomal instability [56]. Reports from hepatocellular carcinoma and leukaemia suggested that enzymatically inactive DNMT splice variants compete with enzymatically active forms for the same binding site of pericentromeric satellite DNA, thereby inhibiting DNA methylation [57, 58]. However, this association could not be confirmed in other tumor types [59], so further investigations are needed.
De Novo Gene Methylation is a Non-random Process
As stated before, many unmethylated tumor suppressor genes in normal tissues acquire hypermethylation during tumor development, so the key question here is: does aberrant hypermethylation of genes in cancer follow a random process that is accompanied by clonal selection of those cells which gained growth advantages, or do DNMTs specifically recognize target genes, which implies that the repertoire of potential targets for inactivation is already encoded by intrinsic or extrinsic factors?
Currently, research results seem to support the latter hypothesis. First to mention, it was shown that distinct tumor types may harbor methylation in several common genes, but also in numerous different genes. This finding lead to the first hypermethylation profiles of human cancer. The analysis of 12 tumor suppressor genes in 600 primary tumors representing 15 tumor entities revealed tumor-characteristic methylation changes in all genes, displaying a unique profile of promoter methylation for each cancer type, in which some gene changes were shared and others were cancer type-specific [60]. As an example, the genes encoding secreted frizzled-related proteins (SFRP) 1 and 2, both encoding inhibitors of the Wnt signaling pathway, were found to be high-frequently methylated in virtually any human tumor type, including breast and colon cancer [61–64]. Furthermore, SFRP methylation occurs already in early tumor stages, suggesting that epigenetic inactivation of SFRP genes may be a common hallmark of human neoplasia following non-random but targeted gene selection. Second, specific nucleotide sequence patterns within gene promoters were identified that are more prone to hypermethylation than other sequences, which tend to be methylation-resistant. Feltus and co-workers have demonstrated in an ectopic Dnmt1 overexpression model that CpG islands differ in their susceptibility to de novo methylation, suggesting the existence of cis-acting intrinsic factors that facilitate methylation of specific target sequences [65]. Indeed, in a further study Feltus and colleagues were able to identify 13 short DNA sequence patterns between 11 bp and 37 bp in length, which were able to distinguish between methylation-prone and methylation-resistant promoters with 87% accuracy [66]. Interestingly, only the methylation-prone DNA sequences were closely associated with CpG islands, whereas methylation-resistant sequences were randomly distributed along the analyzed chromosome, further supporting the idea of an intrinsic hypermethylation code in the DNA sequence of affected gene promoters. However, this study used a model of Dnmt1 overexpression, which exerts only limited de novo methylation capacity. It remains to be shown whether Dnmt3a or Dnmt3b overexpression models may also identify similar methylation-prone DNA sequences in the mammalian genome.
At least in part, another finding supports the hypothesis that aberrant de novo methylation is a non-random process. It has been recently demonstrated that chromatin repressive proteins, such as PcG, can mark certain target genes for hypermethylation by DNMTs. In an approach to identify the functional relation between DNMTs and EZH2, Vire and colleagues showed that the presence of EZH2 is tightly associated with the presence of Dnmt1, Dnmt3a and Dnmt3b proteins at gene promoters, e.g. those of the MYT1 or KCNA1 gene [16]. They found that after siRNA-mediated depletion of EZH2 expression DNMTs were no more bound to the respective promoters, while depletion of DNMT expression did not prevent EZH2 from endowing target sequences with the repression mark H3K27. In another study, Schlesinger and co-workers investigated whether the H3K27 mark is specifically associated with de novo methylated CpG islands in the colon tumor cell line Caco-2. Supporting the previous idea, all analyzed genes found to be hypermethylated in the cell line were also found being enriched for H3K27 [17]. Interestingly, the acquisition of the H3K27 mark turned out to occur not only in the tumor cells themselves, but already existed in normal control tissues, as determined by chromatin immunoprecipitation (ChIP) analysis using an antibody against trimethylated H3K27. Since unmethylated control genes in normal tissues, such as blood lymphocytes, embryonic stem cells and fibroblasts were lacking this histone mark, it is conceivable that a subset of target genes may first become “primed” by the H3K27 mark through EZH2 in normal tissues, which then could represent a favored substrate for hypermethylation during cancer development. Among the identified PcG target genes were the previously mentioned SFRP1 and SFRP2 gene, suggesting that EZH2-mediated de novo methylation may be affecting in particular developmental regulator genes that are occupied by polycomb repressor complexes in embryonic stem cells [67, 68], increasing their susceptibility to non-random methylation-mediated silencing in cancer.
DNMTs are Differentially Expressed in Human Cancer
DNMTs are ubiquitously expressed at distinct levels in normal human tissues [69]. In cancer, they are overexpressed in various tumor types, e.g. in leukemia, colorectal cancer, prostate cancer, ovarian cancer, endometrial cancer and breast cancer [55, 70–75]. Surprisingly, the mean levels of Dnmt1, Dnmt3a and Dnmt3b overexpression turned out to be quite similar among different tumor types. However, these levels were not strikingly high, ranging from 1.8 to 2.9-fold in breast cancer up to 4-fold in colon cancer. In breast cancer, approximately 30% of patients revealed overexpression of Dnmt3b in the tumor tissue as compared to normal breast tissue. Taken only these overexpressing tumors into account, the Dnmt3b expression change was 82-fold, thus being significantly higher [55]. Interestingly, Dnmt1 and Dnmt3a were overexpressed in only 5% and 3% of breast carcinomas, exhibiting also a lower expression change of 17- and 14-fold in the affected tumors, respectively. These results have two implications: First, it appears that Dnmt3b plays the predominant role over Dnmt3a and Dnmt1 in breast tumorigenesis. This is consistent with a recent study in breast cancer cell lines, which demonstrated a strong correlation between total DNMT activity and overexpression of Dnmt3b, but not with the expression of Dnmt3a or Dnmt1 [76]. Second, it is noteworthy that due to the lack of a specific antibody against Dnmt3b almost any expression analysis so far has been performed on the RNA level. Only recently, a study on colon cancer employing 765 primary colorectal carcinomas revealed that Dnmt3b protein overexpression affects 15% of cases [77], in consistency with the relatively low frequency of overexpression reported in other tumor types. The fact that only a subgroup of tumors is affected by Dnmt3b overexpression raises the question whether this can be regarded as a universal feature of tumorous cells. In general, it is possible that further regulators of DNMT activity may play an important role in the dysfunction of the DNA methylation machinery. For instance, DNMT target sequence specificity may be impaired by genetic or regulatory factors, such as DNMT-associated proteins or protein complexes in which DNMTs reside. Furthermore, DNMT3B is the only DNA methyltransferase whose mRNA is expressed as several alternative splice variants. Although DNMT3B1 and DNMT3B3 are the most abundantly expressed transcripts, only DNMT3B1 and DNMT3B2 were shown to be catalytically active, while the remaining splice variants do not possess methyltransferase activity due to the lack of a C-terminal catalytic domain [69, 78]. Despite this, Ostler et al. identified over 20 novel DNMT3B transcripts from various cancer cell lines that are aberrantly spliced at the 5′-end and lacked the C-terminal catalytic domain. Surprisingly, forced expression of one of the variant transcripts (DNMT3B7) significantly changed the DNA methylation pattern in kidney HEK293 cells [79]. Since no catalytic domain could be responsible for the DNA methylation changes, it was proposed that the truncated Dnmt3b7 protein could interfere with DNA methylation processes by binding of Dnmt3b interaction partners, or that the truncated version of DNMT3b affects the activity of catalytic DNMTs by directly binding to the DNA. Adding more complexity to this, the catalytically inactive Dnmt3L directly binds Dnmt3b and positively stimulates its methylation activity [80]. Taking into account the large number of physiological and aberrant splice variants of the DNMT3B transcript, it can only be speculated that the precise mechanisms of Dnmt3b action and the biological function of the many diverse transcripts still have to be identified.
Despite the low-frequent overexpression of DNMTs in tumors, a direct evidence for Dnmt3b involvement in cancer has been previously described. Soejima and colleagues demonstrated that Dnmt3b contributes to the oncogenic phenotype in a lung cancer model. In their study, Dnmt3b was able to promote oncogenic transformation induced by SV40 T antigen in bronchial epithelial cells, whereas antisense suppression of Dnmt3b prevented tumor growth in soft agar assays [81]. In a study on colon tumorigenesis the impact of a conditional inactivation of DNMT3B in APC Min/+ mice was investigated. Although loss of Dnmt3b expression had no impact on microadenoma formation, it significantly decreased the formation of macroscopic colonic adenomas, suggesting a role of Dnmt3b in the transition from one to the other stage [82]. In breast cancer, elevated expression of Dnmt3b was shown to be significantly associated with higher histological grade, absence of estrogen receptor-α and presence of the proliferation marker Ki67, pointing to a potential involvement of Dnmt3b in breast tumor progression and aggressiveness [55]. In the same study, an association of high Dnmt3b expression and reduced relapse-free patient survival was detected, although it was only significant in a subgroup of patients receiving adjuvant hormone therapy, while in patients receiving adjuvant chemotherapy no difference could be detected.
A question of particular interest here is: Is a more malignant phenotype, which seems to be associated with higher Dnmt3b expression, indeed related to increased hypermethylation of genes in these tumors? The answer to this is still not clear. While clear associations have been demonstrated in some in vivo studies, in other studies this relationship could not be confirmed. For instance, in colorectal tumors no significant correlation could be found between the level of Dnmt3b overexpression and the methylation status of the four indicator genes adenomatous polyposis coli (APC), estrogen receptor α (ESR1), cyclin-dependent kinase inhibitor 2A (CDKN2A), and mutL homolog 1 (MLH1) [72]. Similar results have been described in hepatocarcinoma, lung cancer and gastric carcinoma, although different indicator genes were used [83–85].
Dnmt3b is Post-Transcriptionally Regulated
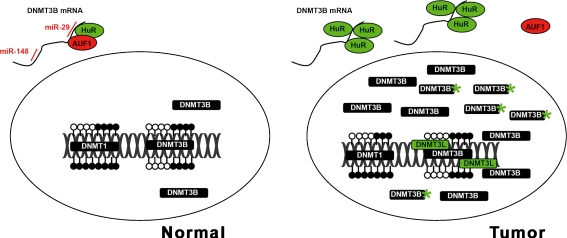
The reasons for the observed overexpression of Dnmt3b in some cancers are largely unknown. On the level of gene transcription alterations of the primary DNA sequence, such as gene amplification or activating mutations, are as reasonable to consider as potential trans-acting mechanisms, e.g. increased expression of transcription factors. Indeed, several polymorphisms were detected in the DNMT3B gene promoter which increases the risk of developing lung cancer, colorectal cancer, prostate cancer, and breast cancer [86–90]. In lung cancer the observed C➔T transition led to enhanced DNMT3B promoter activity, providing one explanation for elevated DNMT3B transcript levels in this tumor entity [86]. Despite, there is increasing evidence that Dnmt3b expression may be more relevantly regulated on the post-transcriptional level. The 3′UTRs of the DNMT3A and DNMT3B mRNA contain complementary sites for the recently detected miR-29 family. Fabbri and colleagues could show that miR-29a, miR-29b and miR-29c bind to the predicted target sequence in lung cancer cell lines, leading to a decrease of Dnmt3b protein levels in the cytoplasm of transfected cells [91]. Important to note, expression of the miR-29 family is commonly downregulated in lung cancer [92, 93]. Enforced expression of miR-29 in lung cancer cell lines restored normal patterns of DNA methylation accompanied by a decrease of tumorigenicity of these cells, as shown by in vitro and in vivo mouse xenograft models [91]. Knowing that miRNA profiles are globally downregulated in breast cancer [22], there is reason to argue that miRNAs targeting DNMT3B mRNA may also be affected by downregulation, resulting in increased transcript levels of the gene (Fig. 1).
Fig. 1.
Schematic representation of factors that potentially influence abundance of the de novo methyltransferase Dnmt3b in tumorous cells. In normal cells (left), physiological DNMT3B transcript levels are maintained by inhibitory miR-29 (a–c), miR-148 and translational inhibitor AUF1, which co-resides with the stabilization factor HuR in the 3′UTR of the mRNA. In tumorous cells (right), miR-29 and miR-148 are frequently abolished. Elevated levels of HuR may occupy the 3′UTR, thus preventing the binding of translational inhibitors. In the nucleus, Dnmt3b activity is further stimulated by binding of the catalytically inactive Dnmt3L as well as products from DNMT3B splice variants and aberrantly truncated Dnmt3b proteins (asterisks). Green color indicates stimulation/activation; red color indicates inhibition. Open circles, unmethylated DNA; filled circles, methylated DNA. For references see text.
Another study recently uncovered a further mechanism of post-transcriptional regulation of Dnmt3b expression. The human family of embryonic-lethal, abnormal vision (ELAV)-like proteins contains four members, of which only Hu-antigen R (HuR) is expressed ubiquitously in many cell types [94]. HuR protein possesses three RNA-recognition motifs by which it binds target mRNAs bearing AU- and U-rich sequences, leading to enhanced stability and decreased decay of the transcript [95, 96]. HuR has become recognized as a pivotal regulator of gene expression, whose function was found to be impaired in many tumor entities. In ovarian carcinoma, HuR was reported as being overexpressed together with one of its target genes, cyclooxegenase-2 (COX2). Moreover, high expression of HuR was shown to be a prognostic factor in patient survival [97]. So far, most studies on HuR expression and function focussed on breast and colon cancer. In breast cancer, a global change in HuR-bound mRNAs is implicated in the evolution of a more tumorigenic phenotype and tumor progression [98]. HuR is overexpressed in 29–39% of breast carcinomas, which correlates with increased Cox-2 expression and adverse survival of the patient [99–101]. Moreover, cytoplasmic HuR accumulation could be identified as a significant factor of tamoxifen resistance in estrogen receptor positive breast cancer cell lines [102]. HuR overexpression mediated decreased tamoxifen sensitivity in these cells, while decreasing the levels of HuR increased tamoxifen sensitivity. Moreover, high HuR expression levels were associated with tamoxifen responsiveness and advanced tumor grade also in primary breast carcinomas, arguing for a role of HuR target mRNAs in the acquisition of resistance to anti-hormonal therapy. Further evidence of a role of HuR in tumor biology was provided from studies in colon cancer. There, HuR was described as being increasingly expressed in consecutive stages of the adenoma-carcinoma sequence, whereas HuR expression in normal mucosa was low and infrequent. [103]. High HuR expression in colon cancer, similar to breast cancer, was an independent predictor of adverse patient survival [104], largely attributable to its tumorigenic phenotype which was increased in mouse xenograft models after enforced overexpression, and decreased after HuR-based antisense approaches [105].
Interestingly, besides many important proto-oncogenes, like FOS, MYC, COX2, CCND1 or CTNNB1 [105–108], DNMT3B mRNA was identified as a putative HuR target in an en masse approach, in which one HuR binding motif was computationally detected in the DNMT3B 3′UTR [104]. Later on, HuR binding to DNMT3B mRNA and its function was further characterized in colorectal carcinoma RKO cells. In immunoprecipitation assays, Lopez de Silanes and colleagues showed for the first time that DNMT3B mRNA co-precipitates with HuR protein under conditions that preserved the HuR-mRNA complex [109]. After siRNA-mediated silencing of HuR expression, DNMT3B mRNA levels were reduced by 25%. In order to test whether this reduction was due to changes in mRNA stability and to exclude a possible change on the level of transcription, the authors inhibited de novo transcription in RKO cells by means of actinomycin D, and then monitored DNMT3B mRNA levels over time. Indeed, cells in which HuR expression has been silenced by siRNA showed a substantial decrease of DNMT3B transcript halftime as compared to control cells. Therefore, HuR binding to its target was responsible for DNMT3B mRNA stability in cancerous cells. Furthermore, when HuR levels were lowered in RKO cells this was associated with a decrease of the global DNA methylation content as well as with a decrease of methylation in two indicator DNA regions, the pericentromeric Sat2 and the subtelomeric D4Z4 sequence. These regions were shown to be specifically hypomethylated when a certain mutation in the DNMT3B gene occurs, which is associated with the rare autosomal ICF syndrome [41]. In summary, it was proven that the association of HuR with DNMT3B mRNA possibly influences its expression on the level of transcript stabilization, which effects global DNA methylation content as well as Dnmt3b-specific methylation sites (Fig. 1).
Another interesting point in terms of post-transcriptional regulation of DNMT3B mRNA is that of its occupation with trans-acting regulatory factors in the 3′UTR. HuR was shown to bind at least three different regions within the 3′UTR of DNMT3B mRNA, consistent with studies on other HuR targets like CYCS (cytochrome c) and DUSP1 (MAP kinase phosphatase 1), which contain three and two different binding regions for HuR, respectively [96, 109, 110]. The reason for this is currently unknown, but it is assumed that the presence of several HuR binding sites in the 3′UTR increases the accessibility of the target sequence even in the presence of other competing RNA-binding factors. For instance, the three HuR binding motifs in the DUSP1 mRNA 3′UTR were shown to be bound by the translational inhibitor TIAR [96]. TIAR and a further translational inhibitor termed AUF1 were found to bind to the DNMT3B 3′UTR, too, [109, 111], although it remains to be determined if the HuR recognition motifs were the actual site of binding. However, upon induction of cellular stress, for instance by treatment with the drug cisplatin, HuR and AUF1 dissociate from the DNMT3B transcript, whereas the binding of TIAR to the 3′UTR was found to be even tighter [109]. It is possible that such changes in binding patterns at regulatory regions of mRNA as a response to external stimuli is a key to fast and efficient adaption of gene expression. In germ cell development, the absence of Dnmt3b protein despite the presence of its transcripts is a strong indicator of a regulatory level downstream from gene transcription [112].
Further complexity in post-transcriptional regulation of DNMT3B expression adds from the finding that members of the miR-29 family of miRNAs were shown to bind to the 3′UTR in lung cancer cells, most probably competing with other regulatory factors for binding mRNA within this region [91]. Since the miR-29 family was reported to be downregulated in lung cancer, the loss of a translational inhibitor may ease the accessibility of the 3′UTR for other stabilizing factors. Recently, a further miRNA was detected to target the DNMT3B transcript. Duursma et al. reported that, in contrast to the current knowledge on miRNA biology, miR-148 does not target the 3′UTR of the DNMT3B mRNA but its coding sequence (CDS), leading do decreased expression level of the DNMT3B transcript [113]. The binding of miRNAs to the CDS is a common mechanism in plants [114], but was functionally not detected in animals yet. So far, targeting of CDS by miRNAs in animals has only been hypothesized based on bioinformatics approaches. Interestingly, the CDS region that is targeted by miR-148 is evolutionary conserved and present in the splice variants DNMT3B1, DNMT3B2, and DNMT3B4, but not in the most abundantly expressed transcript of DNMT3B3, indicating that the DNMT3B3 variant is resistant to regulation by miR-148. In two tumor entities miR-148 expression was recently reported to be downregulated. In primary gastric cancers miR-148 as well as miR128b and miR-129 were downregulated as compared to normal gastric tissue [115]. Also in primary breast carcinoma samples Lehmann and colleagues reported the downregulation of a panel of miRNAs, including miR-148 [116]. Furthermore, these authors found that the DNA sequence encoding miR-148 is a target of hypermethylation, leading to the observed expression loss of this miRNA species. This finding adds an interesting aspect to the link between DNMT expression and aberrant DNA hypermethylation, because it may be possible that elevated Dnmt3b expression in cancer may be, at least in part, initiated, maintained or increased by hypermethylation of genes encoding Dnmt3b-repressive factors. Recapitulating the idea that various regulatory factors compete in binding to target sequences of the DNMT3B gene, even minor shifts in the dynamics of these factors together with a altered pattern of various splice variants, e.g. through hypermethylation of potential repressor genes, represents the most probable explanation to date for the increased expression and activity of methyltransferases observed in human cancer.
MicroRNAs and Breast Cancer
MiRNAs have been shown to play a critical role in the regulation of a wide range of biological and pathological processes. Recent large-scale profiling approaches have revealed that miRNAs are globally downregulated in several cancer types, including breast cancer (for overview see Table 1). The first study describing genome-wide profiling of miRNAs in breast cancer identified 29 differentially expressed candidates, of which 15 predictive miRNAs were able to distinguish between breast cancer and normal breast tissue [22]. In addition, several correlations between downregulation of certain miRNAs and clinicopathological features, such as ER/PgR positivity, tumor size, lymph node status and the expression of p53 were found. In particular the expression loss of members of the let-7 family was associated with clinical features, such as PgR status (let-7c), a positive lymph node status (let-7f-1, let-7a-3 and let-7a-2), or a high proliferation index (let-7c and let-7d). Moreover, panels of miRNAs in breast carcinomas characteristic for the Her2/neu or ER status of the analyzed tumor have been detected [117]. There is now increasing evidence that signatures of miRNA expression may not only be used in the future as tumor biomarkers for diagnosis and patient risk stratification, but since hypermethylation was identified as an important mechanism of miRNA silencing, deregulated miRNAs may also represent novel targets for an anticancer therapy. In an approach taking advantage of disruption of the DNMT1 and DNMT3B gene in colon cancer cells (so-called double-knock out cells, DKO), CpG hypermethylation was identified as one important mechanism leading to a global loss of miRNA expression [27], mostly affecting those species that exhibit tumor suppressive functions (reviewed in [118]). Since generally, DNA hypermethylation is a reversible process e.g. by inhibitors of DNMTs, this drug class may also potentially revert a part of epigenetically silenced miRNAs in cancer diseases.
Table 1.
Differentially expressed miRNAs in human breast cancer.
| Name | Proposed target genes | Reference |
|---|---|---|
| Downregulateda | ||
| miR-9-1 | – | [116] |
| miR-29a-c | DNMT3A, DNMT3B | [91] |
| miR-148 | DNMT3B | [113] |
| Downregulatedb | ||
| let-7 | RAS | [19] |
| miR-15/16 | BCL2 | [122] |
| miR-17-5p | AIB1 | [123] |
| miR-27b | CYPB1 | [124] |
| miR-125a/b | HER2, HER3 | [125] |
| miR-126 | p85β | [126, 127] |
| miR-130a | GAX, HOXA5 | [128] |
| miR-143 | ERK5 | [129] |
| miR-145 | MAP3K3, MAP4K4, CBFB | [22] |
| miR-155 | SOCS1, APC | [22] |
| miR-200c | TCF8 | [130] |
| miR-205 | HER3 | [131] |
| miR-335 | PTPRN2, MERTK, SOX4 | [126] |
| Upregulatedb | ||
| miR-10b | HOXD10 | [132] |
| miR-18a | KRAS | [123] |
| miR-21 | TPM1, PDCD4 | [23] |
| miR-27a | ZBTB10 | [132] |
| miR-206 | ER-α | [92] |
amiRNAs silenced by or involved in epigenetic mechanisms
bmiRNAs deregulated by genetic or undetermined mechanisms
Recently, evidence was shown that miRNAs, besides aberrant DNA methylation, may also be deregulated due to copy number variations. One study describing differential expression patterns of miRNAs in breast cancer also investigated its expression changes in relation to chromosomal localization. Interestingly, the authors found several miR candidates that reside in chromosomal regions which are either frequently deleted or amplified in breast cancer, e.g. downregulation of miR-125b in the frequently deleted region 11q-23-24, or overexpression of miR-21 in 17q23, which is commonly amplified in breast cancer [22]. Furthermore, a study applying CGH arrays on different epithelial tumors demonstrated that 73% of miRNA genes in breast cancer reside in genomic regions affected by copy number variations [119], providing a further possible explanation for differential miRNA profiles in tumor tissues.
Another mechanism by which miRNA profiles may be altered in tumors lies within abnormalities in the miRNA processing machinery. It was the same study that observed significant changes in expression of Dicer and AGO1, both being involved in miRNA maturation processes [119]. In ovarian cancer, DICER1 is overexpressed in correlation with copy number gain in 25% of cases, and overexpression of AGO1 is attributable to gene amplification in 52% of tumors. A further defect in miRNA processing enzymes was recently described by Melo and colleagues. In this study the authors investigated mutational alterations in several genes of the miRNA processing machinery [120], including DICER1, DROSHA, DGCR8, TRPB, AGO1, AGO2, and AGO4. In microsatellite instable colorectal cancer they detected truncating frameshift mutations in TARBP2, encoding a member of the Dicer-containing complex [121]. TARBP2 mutant cells exhibited a 90% reduced efficiency of endogenous miRNA processing, and re-introduction of the wild-type protein restored processing capacity to normal levels. Moreover, the loss of functional TARBP2 protein also affected the expression of Dicer as a secondary effect, since Dicer expression was substantially reduced in the TARBP2-mutant, whereas its expression was restored after re-expression of the wild-type TARBP2 protein. Decreased Dicer expression was recently observed in breast cancer, where loss of expression represented an independent prognostic factor in the metastatic disease, and reduced expression of Dicer was associated with the highly aggressive mesenchymal phenotype [28]. Whether genetic lesions like TARBP2 mutations in colorectal cancer also account for impaired miRNA processing in breast cancer remains to be determined in future studies. It is, however, conclusive that besides DNA hypermethylation of miRNA genes, structural genetic alterations also contribute to the observed dramatic changes of miRNA expression profiles in human cancer.
Concluding Remarks
In cancer research, our knowledge on hypermethylated DNA sequences encoding either proteins or miRNAs has significantly increased over the last decade. Yet, the precise mechanisms initiating hypermethylation during tumor development and progression are still not clear. Apparently, cancer cells acquire a complex pattern of genetic as well as epigenetic lesions, which most intriguingly may even become inter-connected. One example is the hypermethylation of DNA repair genes, such as BRCA1 or MLH1, which favors the acquisition of further structural DNA lesions in the transformed cell. Conversely, structural aberrations in the DNMT3B gene, may be partly responsible for increased Dnmt3b expression and consequently hypermethylation of critical genes in human tumors. Adding complexity to this, such specific genetic-epigenetic relations may not necessarily be found in all tumor types: BRCA1 methylation, for instance, is observed in breast and ovarian cancer only, but almost absent in any other cancer type. Further investigations are needed in order to unravel the question of how the hypermethylome is established and maintained in a cancer cell.
Acknowledgement
Supported by the Health (FIS PI08 1345) and Education and Science (I+D+I MCYT08-03 and Consolider MEC09-05) Departments of the Spanish Government, the Health Department of the Catalan Government, the Spanish Association Against Cancer (AECC) and the European Union FP7 CANCERDIP HEALTH-F2-2007-200620.
Conflicts of interest
The authors of the article declare that there are no potential conflicts of interest including employment, consultancies, stock ownership, honoraria, paid expert testimony and patent applications/registrations related to the current manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 3′UTR
3′ untranslated region
- CDS
Coding sequence
- CpG
Cytosine-phosphate-guanine dinucleotide
- DNMT
DNA (cytosine-5)-methyltransferase
- ER
Estrogen receptor
- HDAC
Histone deacetylase
- ICF
Immunodeficiency, centromere instability and facial abnormalities
- HuR
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R)
- MBD
Methyl-CpG binding domain protein
- miRNA
microRNA
- PgR
Progesterone receptor
- RISC
RNA induced silencing complex
- UTR
Untranslated region
References
- 1.Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Kundu TK, Rao MR. CpG islands in chromatin organization and gene expression. J Biochem. 1999;125:217–222. doi: 10.1093/oxfordjournals.jbchem.a022276. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Greger V, Passarge E, Höpping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 6.Herman JG, Latif F, Weng Y, Lermann MI, Zbar B, Samid D, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 11.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 12.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 13.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 14.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 15.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 16.Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 17.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 18.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 19.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 20.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. AS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 28.Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 30.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 37.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 38.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 39.Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, et al. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet. 1993;2:731–735. doi: 10.1093/hmg/2.6.731. [DOI] [PubMed] [Google Scholar]
- 40.Tuck-Muller CM, Narayan A, Tsien F, Smeets DF, Sawyer J, Fiala ES, et al. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 41.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 42.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernadino J, Roux C, Almeida A, Vogt N, Gibaud A, Gerbault-Seureau M, et al. DNA hypomethylation in breast cancer: an independent parameter of tumor progression? Cancer Genet Cytogenet. 1997;97:83–89. doi: 10.1016/s0165-4608(96)00385-8. [DOI] [PubMed] [Google Scholar]
- 45.Soares J, Pinto AE, Cunha CV, André S, Barão I, Sousa JM, et al. Global DNA hypomethylation in breast carcinoma: correlation with prognostic factors and tumor progression. Cancer. 1999;85:112–118. [PubMed] [Google Scholar]
- 46.Alves G, Tatro A, Fanning T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene. 1996;176:39–44. doi: 10.1016/0378-1119(96)00205-3. [DOI] [PubMed] [Google Scholar]
- 47.Bera TK, Tsukamoto T, Panda DK, Huang T, Guzman RC, Hwang SI, et al. Defective retrovirus insertion activates c-Ha-ras protooncogene in an MNU-induced rat mammary carcinoma. Biochem Biophys Res Commun. 1998;248:835–840. doi: 10.1006/bbrc.1998.9059. [DOI] [PubMed] [Google Scholar]
- 48.Widschwendter M, Jiang G, Woods C, Müller HM, Fiegl H, Goebel G, et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 49.Costa FF, Paixão VA, Cavalher FP, Ribeiro KB, Cunha IW, Rinck JA, Jr, et al. SATR-1 hypomethylation is a common and early event in breast cancer. Cancer Genet Cytogenet. 2006;165:135–143. doi: 10.1016/j.cancergencyto.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 50.Narayan A, Ji W, Zhang XY, Marrogi A, Graff JR, Baylin SB, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 51.Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54:1766–1771. [PubMed] [Google Scholar]
- 52.Guo Y, Pakneshan P, Gladu J, Slack A, Szyf M, Rabbani SA. Regulation of DNA methylation in human breast cancer. Effect on the urokinase-type plasminogen activator gene production and tumor invasion. J Biol Chem. 2002;277:41571–41579. doi: 10.1074/jbc.M201864200. [DOI] [PubMed] [Google Scholar]
- 53.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 54.Sharma G, Mirza S, Parshad R, Srivastava A, Datta Gupta S, Pandya P, et al. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients. Clin Biochem. 2009 Oct 29. [Epub ahead of print] doi:10.1016/j.clinbiochem.2009.10.009. [DOI] [PubMed]
- 55.Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 56.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 57.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 58.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 59.Ehrlich M, Woods CB, Yu MC, Dubeau L, Yang F, Campan M, et al. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25:2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 61.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 62.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 63.Veeck J, Noetzel E, Bektas N, Jost E, Hartmann A, Knüchel R, et al. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol Cancer. 2008;7:83. doi: 10.1186/1476-4598-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci U S A. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;87:572–579. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 67.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 69.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Issa JP, Vertino PM, Wu J, Sazawal S, Celano P, Nelkin BD, et al. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst. 1993;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 71.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 72.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 73.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 74.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 75.Jin F, Dowdy SC, Xiong Y, Eberhardt NL, Podratz KC, Jiang SW. Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol Oncol. 2005;96:531–538. doi: 10.1016/j.ygyno.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 76.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nosho K, Shima K, Irahara N, Kure S, Baba Y, Kirkner GJ, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 79.Ostler KR, Davis EM, Payne SL, Gosalia BB, Expósito-Céspedes J, Le Beau MM, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 81.Soejima K, Fang W, Rollins BJ. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene. 2003;22:4723–4733. doi: 10.1038/sj.onc.1206510. [DOI] [PubMed] [Google Scholar]
- 82.Lin H, Yamada Y, Nguyen S, Linhart H, Jackson-Grusby L, Meissner A, et al. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 84.Sato M, Horio Y, Sekido Y, Minna JD, Shimokata K, Hasegawa Y. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene. 2002;21:4822–4829. doi: 10.1038/sj.onc.1205581. [DOI] [PubMed] [Google Scholar]
- 85.Oue N, Kuraoka K, Kuniyasu H, Yokozaki H, Wakikawa A, Matsusaki K, et al. DNA methylation status of hMLH1, p16(INK4a), and CDH1 is not associated with mRNA expression levels of DNA methyltransferase and DNA demethylase in gastric carcinomas. Oncol Rep. 2001;8:1085–1089. doi: 10.3892/or.8.5.1085. [DOI] [PubMed] [Google Scholar]
- 86.Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62:4992–4995. [PubMed] [Google Scholar]
- 87.Guo X, Zhang L, Wu M, Wang N, Liu Y, Er L, et al. Association of the DNMT3B polymorphism with colorectal adenomatous polyps and adenocarcinoma. Mol Biol Rep 2009 Jul 22. [Epub ahead of print]; doi:10.1007/s11033-009-9626-z. [DOI] [PubMed]
- 88.Singal R, Das PM, Manoharan M, Reis IM, Schlesselman JJ. Polymorphisms in the DNA methyltransferase 3b gene and prostate cancer risk. Oncol Rep. 2005;14:569–573. [PubMed] [Google Scholar]
- 89.Montgomery KG, Liu MC, Eccles DM, Campbell IG. The DNMT3B C-->T promoter polymorphism and risk of breast cancer in a British population: a case-control study. Breast Cancer Res. 2004;6:R390–R394. doi: 10.1186/bcr807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cebrian A, Pharoah PD, Ahmed S, Ropero S, Fraga MF, Smith PL, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27:1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- 91.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 94.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 95.Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, et al. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Denkert C, Weichert W, Pest S, Koch I, Licht D, Köbel M, et al. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–195. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 98.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, et al. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008;27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Denkert C, Weichert W, Winzer KJ, Müller BM, Noske A, Niesporek S, et al. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res. 2004;10:5580–5586. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 100.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 101.Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomäki K, Blomqvist C, et al. Prognostic role of HuR in hereditary breast cancer. Clin Cancer Res. 2007;13:6959–6963. doi: 10.1158/1078-0432.CCR-07-1432. [DOI] [PubMed] [Google Scholar]
- 102.Hostetter C, Licata LA, Witkiewicz A, Costantino CL, Yeo CJ, Brody JR, et al. Cytoplasmic accumulation of the RNA binding protein HuR is central to tamoxifen resistance in estrogen receptor positive breast cancer cells. Cancer Biol Ther. 2008;7:1496–1506. doi: 10.4161/cbt.7.9.6490. [DOI] [PubMed] [Google Scholar]
- 103.Brosens LA, Keller JJ, Pohjola L, Haglund C, Morsink FH, Iacobuzio-Donahue C, et al. Increased expression of cytoplasmic HuR in familial adenomatous polyposis. Cancer Biol Ther. 2008;7:424–427. doi: 10.4161/cbt.7.3.5417. [DOI] [PubMed] [Google Scholar]
- 104.Yoo PS, Sullivan CA, Kiang S, Gao W, Uchio EM, Chung GG, et al. Tissue microarray analysis of 560 patients with colorectal adenocarcinoma: high expression of HuR predicts poor survival. Ann Surg Oncol. 2009;16:200–207. doi: 10.1245/s10434-008-0209-3. [DOI] [PubMed] [Google Scholar]
- 105.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 106.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gouble A, Morello D. Synchronous and regulated expression of two AU-binding proteins, AUF1 and HuR, throughout murine development. Oncogene. 2000;19:5377–5384. doi: 10.1038/sj.onc.1203910. [DOI] [PubMed] [Google Scholar]
- 108.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Silanes IL, Gorospe M, Taniguchi H, Abdelmohsen K, Srikantan S, Alaminos M, et al. The RNA-binding protein HuR regulates DNA methylation through stabilization of DNMT3b mRNA. Nucleic Acids Res. 2009;37:2658–2671. doi: 10.1093/nar/gkp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 112.Lees-Murdock DJ, Shovlin TC, Gardiner T, De Felici M, Walsh CP. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev Dyn. 2005;232:992–1002. doi: 10.1002/dvdy.20288. [DOI] [PubMed] [Google Scholar]
- 113.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. MiR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 115.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 116.Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 117.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- 119.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 125.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 126.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 130.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 131.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]