Abstract
Green Fluorescent Protein (GFP) is a widely utilized molecular marker of gene expression. However, its use in in vivo imaging has been restricted to transparent tissue mainly due to the tissue penetrance limitation of optical imaging. Here, we report a novel approach to detect GFP with Magnetization Transfer Contrast (MTC) Magnetic Resonance Imaging (MRI). MTC is an MRI methodology currently utilized to detect macromolecule changes such as decrease in myelin and increase in collagen content. We employed MTC MRI imaging to detect GFP both in vitro and in in vivo mouse models. We demonstrated that our approach produces values that are protein specific, and concentration dependent. This approach provides a flexible, non-invasive in vivo molecular MRI imaging strategy that is dependent upon the presence and concentration of the GFP reporter.
Keywords: Magnetization Transfer, Green Fluorescent Protein (GFP), Magnetic Resonance Imaging (MRI), gene reporter
Introduction
To visualize gene expression via any imaging modality, a reporter system needs to be developed to cause sufficient contrast between the tissues where the gene is expressed and where it is not expressed. For in vitro cell work, many techniques are available that can detect expression at both the RNA and protein stages including fluorescence based methods for both RNA and protein, immunoblot based techniques for proteins and reverse transcriptase polymerase chain reaction (RT-PCR) techniques for RNA detection. However, it is much more difficult to track gene expression in vivo in small animal models without sacrificing the animal due to current limitations in imaging technology.
Green Fluorescent Protein (GFP) is an 11-strand beta-barrel wrapped around a central helix that is widely utilized as a fluorescent marker of gene expression (Tsien, 1998). GFP is detected by optical spectroscopy through its fluorescent properties, it has a major excitation peak at 395 nm which results in an emission peak at 508 nm (Tsien, 1998). Because it is detected optically, it can only be measured in vivo if the tissue is transparent or expression is near the tissue surface: less than 100 μm for traditional fluorescence microscopy (Helmchen and Denk, 2005) and less than 1mm for 2-photon based methods (Helmchen and Denk, 2005). New alternatives such as Fluorescence Lifetime Imaging Microscopy (FLIM) that focus upon the length of the fluorescent signal instead of the intensity may be able to increase penetrance up to maximum of 2 cm in homogeneous phantoms with improved fluorophores (Roy et al., 2007). Even then, due to this depth limitation among other reasons, most fluorescence experiments focus upon in vitro cell culture, ex vivo histology slices, or transparent animal models (Shaner et al., 2007; Tavaré et al., 2001). GFP based experiments and associated methods are undoubtedly very useful but they have yet to reach their full potential.
The ability to detect GFP in vivo in small animal models in a non-invasive manner would be a tremendous technical advance. Even though fluorescent probes can be seen in transparent tissue such as in C. elegans and Xenopus, these animal models may be too far removed from humans for some applications. Moreover, while new near-infrared fluorescent probes (Ralph Weissleder et al., 1999; Becker et al., 2001) may be an improvement over GFP, they still exhibit many of the same limitations, namely limited tissue penetrance which impedes the utility of such probes for in vivo use. In the case of live, fluorescence based, animal brain imaging another barrier is the presence of the skull. Bone can deflect light forcing the use of a thinned skull (Grutzendler et al., 2002) or a glass window (Trachtenberg et al., 2002) to be able to detect fluorescent reporters in vivo.
It would be extremely useful to utilize magnetic resonance imaging (MRI) to detect gene expression. MRI is a very useful imaging technology that is utilized both clinically and in the lab for in vivo tissue and organ imaging. In addition to the relatively high resolution that can be obtained with MRI (tens of μm), the data is collected not only in vivo but also can be collected at multiple time points and in three-dimensional (3D) datasets. To date, there are at least five MRI based reporters of gene expression: ferritin (Cohen et al., 2007), transferrin (Moore et al., 2001), tyrosinase (Enochs et al., 1997), MagA (Zurkiya et al., 2008) and a β-galactosidase activated gadolinium chelate (Louie et al., 2000). The first four strategies are genes that increase the amount of intracellular iron ions, by either trapping it or increasing its transport. While no changes in cell viability have been reported, the side effects of altered iron homeostasis could potentially prove detrimental (Edison et al., 2008). The fifth strategy is a more complex design that takes advantage of the common reporter β-galactosidase to activate a modified gadolinium (Gd3+) chelate. All of these alternatives require both a reporter and a substrate creating readouts dependent upon the concentrations of both. Even though the MRI reporters could be used in theory as gene reporters, their use is currently still limited for in vivo applications.
Magnetization Transfer Contrast (MTC) MRI, is an imaging method that evolved from NMR spectroscopy (Wolff and Balaban, 1989; McConnell and Thompson, 1957). In tissue imaging, MTC relies upon the interaction of less mobile protons associated with macromolecules such as proteins and their interactions with protons freely associated with water (Henkelman et al., 2001). The premise is that in a system where molecules move and exchange position, whether it be a change in spatial position in asymmetrical molecules or an exchangeable proton between a molecule and water, the magnetization state will also move and be transferred.
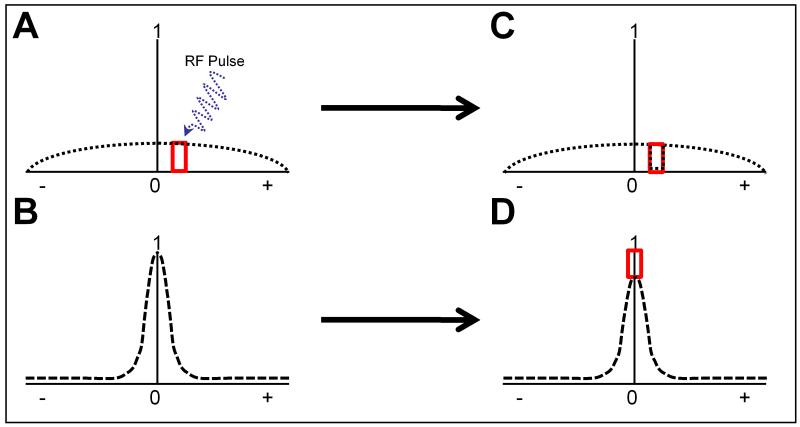
A two pool model can be utilized to illustrate the theory behind MTC MRI (Fig. 1). Conventional MRI detects only the free water pool while the macromolecular pool remains mostly undetected. Both the macromolecular (Fig. 1A) and free water pools (Fig. 1B) are centered around the same frequency but the macromolecular pool is shallower and wider. Saturation is achieved by applying an off-resonance radio frequency (RF) pulse specific to a peak in the macromolecular pool before excitation at the center frequency (Fig. 1B). The RF pulse saturates the signal from the section (Fig. 1C) leading to ideally no signal at the off-resonance frequency. Since both pools interact this saturation is transferred to the free water pool (Fig. 1D). While it is not possible to detect the changes in the macromolecular pool directly, it can be assumed that the loss in signal intensity of the free water pool corresponds to the changes in the macromolecular pool.
Figure 1.
Overview of Magnetization Transfer Contrast (MTC). Panel A shows the signal from protons associated with macromolecules. Panel B shows the signal from protons associated with the free water pool. When an RF pulse (blue wavyarrow) is applied off-frequency (red box), it leads to saturation of those macromolecular protons. Panel C is the saturated macromolecular pool. Notice the black dotted line drops in the area of the red square. Because this macromolecular pool is associated with the free water pool, the saturation can be transferred as is seen in Panel D.
Ideally, an increase is preferable to a decrease in signal intensity since it is easier to visualize changes in brightness over changes in darkness. To achieve this type of image, a Magnetization Transfer Ratio (MTR) is calculated using a base image without saturation to measure the relative loss of signal intensity in a pixel by pixel basis:
To date, most MTC experiments have focused upon intrinsic markers including collagen (Harel et al., 2008; Laurent et al., 2006) and myelin (Zaaraoui et al., 2008). Specifically, MTC of myelin is clinically useful to grade multiple sclerosis lesions (Rovira and León, 2008). MTC is very similar in function to a novel type of imaging known as chemical exchange saturation transfer (CEST) developed in part by Balaban and colleagues (Ward et al., 2000). The theory behind CEST tries to explain a limited part of magnetization transfer by linking it to chemical exchange systems. There are various example of CEST reporters such as one based on an artificial lysine rich–protein (Gilad et al., 2007). This molecular and gene reporter, for which no mouse models have yet been made, focuses on the rapid exchange of amide (NH) protons between lysine and water. The prospect for CEST agents is intriguing and has great potential as a class of molecular imaging agents.
Here, we present a novel approach to detect GFP with MTC MRI both in vitro and in vivo in three separate mouse strains differentially expressing GFP. The detection proved to be protein specific and concentration dependent. The presented strategy provides a flexible, non-invasive in vivo molecular imaging system exclusively dependent upon the concentration of the reporter GFP. This approach also has the advantage of utilizing readily accessible resources as multiple cell lines and mouse lines that express GFP are already available.
Materials and Methods
In Vitro Phantoms
Bovine Serum Albumin (BSA) was purchased from Sigma (St. Louis, MO) while eGFP was purchase from Alpha Diagnostic International (San Antonio, TX). Solutions were used at a concentration of 0.1 mg/mL in Phosphate buffered saline (PBS). Further dilutions of the GFP solution were generated by diluting the 0.1mg/mL GFP solution with PBS in the following ratios: 1:2, 1:4, and 1:8.
Animals
GFP-expressing Inhibitory Neurons (GIN) mice (Oliva et al., 2000), originally made by Dr. John Swann’s lab, were purchased from The Jackson Laboratory (Bar Harbor, ME) and express GFP under the control of a Gad1 promoter. Dr. Michael T. Lewis and Dr. Huda Y. Zoghbi generously donated Actin-GFP mice (Hadjantonakis et al., 1998), originally made by Dr. Andras Nagy’s lab, and Cerebellar L7-GFP (Tomomura et al., 2001) mice, originally made by Dr. Michisuke Yuzaki’s lab, respectively. The Actin GFP mice were originally in a mixed 129X1/SvJ x 129S1/Sv background (Hadjantonakis et al., 1998) and introgressed into a C57Bl6/J background by serial backcrossing in Dr. Lewis’ lab. GIN and Cerebellar GFP mice were both on an FVB background. The number of animals used per group varied from 4 to 9 and is indicated in the figures with the variable, “n”. All the animals utilized in this study were handled in compliance with institutional and national regulations and policies. The protocols were approved by the Institutional Animal Subjects Committee at Baylor College of Medicine.
MRI Imaging Protocol
Animals were initially anesthetized with the gaseous anesthesia, isoflurane, at 5% in oxygen and placed into a mouse holder within the magnet where they were subsequently maintained at 2% isoflurane mixed with oxygen. Imaging was performed utilizing a Bruker Avance Biospec, 9.4 T spectrometer, 21 cm bore horizontal imaging system (Bruker Biospin, Billerica, MA) with a 35 mm volume resonator. During the imaging, the animal body temperature was maintained at 37.0°C continually monitored with a rectal probe using an animal warmed air heating system (SA Instruments, Stony Brook, NY). Protein solutions were imaged within the same instrument using the same coil and imaging parameters.
Pilot T1 weighted images were taken to locate the appropriate MTC slice placement depending upon the mouse model assessed. The MTC imaging pulse sequence comprised a pre-saturation square pulse at the designated offset frequency followed by a spin echo sequence with TE/TR=10.25/1457 msec. The matrix size was 256×256, Field of View = 3×3 cm, slice thickness = 2mm, and number of averages = 1. Pre-saturation off-resonance pulses ranged from 0 to 20 kHz. Specific MTC parameters were as follows: pulse length = 40 msec, number of pulses = 36, pulse strength = 12 μT, and saturation time = 1440 msec.
Fluorescence Imaging
After MR imaging was completed, animals were fixed by intracardial perfusion of 4% paraformaldehyde and the brains were dissected out. The fixed brain was cut on a cryostat into 50-micrometer sections and placed on microscopy slides without any further preparation. Slides were wet mounted using Fluoro-Gel mounting media (Electron Microscopy Sciences, Hatfield, PA). Fluorescence microscopy Images were obtained on a Nikon Eclipse Ti-U inverted microscope with a 4x objective using their NIS Elements v.2.35 software. Imaging parameters were first optimized for GFP expressing sections and then controls were imaged under the same settings.
Image and Data Analysis
Magnetization Transfer Ratios (MTR) were calculated from averaged regions of interest (ROI) using Paravision software (Bruker Biospin, Billerica, MA). Graphs and the in vitro statistical analyses were conducted using Prism (GraphPad Software, San Diego, CA). The statistics for the in vivo models were conducted on SAS 9.2 (SAS Institute, Cary, NC) using Proc Mixed. Data in bar graphs are shown as Mean + SEM. Pixel by pixel MTR calculations were performed in Amira (Visage Imaging, San Diego, CA) and MATLAB (The Mathworks, Natick MA) to generate pseudocolored images. Images are shown cropped and in the case of the mouse images, additionally aligned via an affine transformation using 4-6 reference points along the outside border of the brain.
Statistical analysis
For the in vitro analysis, comparison between groups was performed using ANOVA followed by a Dunnet’s post-test. Each offset frequency and each model were compared independently. For the in vivo models, a mixed model ANOVA was used to test for a group effect, an offset effect and an interaction while accounting for the repeated measures nature of the experiment, in which, within group, the same mice were studied at each offset. Having found either significant group effects or a significant group by offset interaction, we used linear contrasts to explicitly test for the effect of group within an offset set level.
Results
In Vitro Phantoms
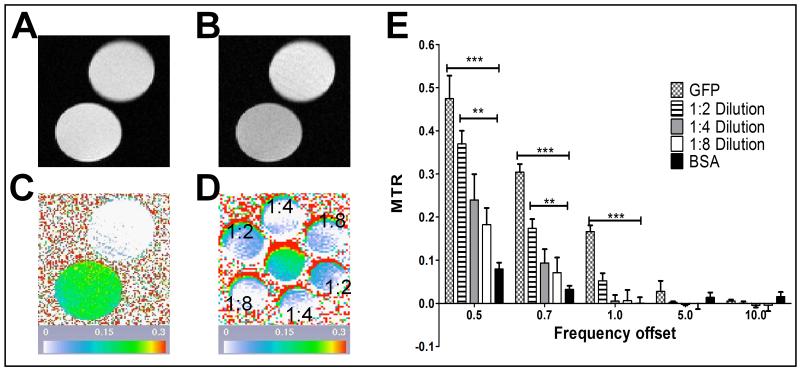
First, we compared MTC profiles of eGFP and Bovine Serum Albumin (BSA). Both proteins were at equal concentrations of 0.1 mg/ml in 1 ml vials. BSA was chosen as a non-specific control to compare against GFP as it is a fairly generic protein. The goal was to determine the frequency at which there is the largest difference between GFP and the BSA control. Both phantoms were imaged first without (Fig. 2A) and then with a saturation pulse (Fig.2B). Five MTC datasets at 0.5, 0.7, 1, 5, and 10 kHz were acquired. Pseudocolored pixel by pixel MTR calculations at the 1kHz offset (Fig. 2C) visually show a clear difference between the GFP and BSA phantoms. The same analysis was performed with dilutions of the initial GFP solution at ratios of 1:2, 1:4, and 1:8 in 0.2 mL vials. Pseudocolored pixel by pixel MTR calculations of the initial GFP solution and dilutions at the 1kHz offset (Fig 2D) show a loss of MTR signal in the more diluted samples. From the images, the MTR was calculated for all offset frequencies for all samples (Fig 2E). The largest significant differences between the initial GFP and BSA samples occurred at 0.5, 0.7, and 1 kHz with a p-value <0.001. The diluted GFP samples also show a significant difference against the BSA sample at 0.5 and 0.7kHz with a p- value <0.01 for the 1:2 dilution. All p-values were determined from an Analysis of Variance (ANOVA) followed by a Dunnet post-test.
Figure 2.
GFP compared with BSA protein phantoms. For panels A-C top phantom is BSA and bottom phantom is GFP, both at a concentration of 0.1mg/mL. Panel A shows the unsaturated image while Panel B shows the saturated image at the 1 kHz offset frequency. Panel C is the pseudocolored pixel by pixel MTR calculation for the 1 kHz offset frequency. Panel D is also a pseudocolored pixel by pixel MTR calculation for the 1 kHz offset frequency of GFP at different concentrations. The central phantom is the initial 0.1 mg/mL concentration whereas the surrounding phantoms contain different GFP dilutions, indicated on the panel. Panel E shows the region based MTR calculations for the different frequency offsets and GFP concentrations. Significance was assessed using an ANOVA with a Dunnet post-test within each offset with ***p<0.001 and **p<0.01.
GFP Mouse Models
We assessed three different strains of GFP expressing mice that provide three different models of GFP expression: ubiquitous, cerebellar specific, and cortex specific. The ubiquitous model expresses eGFP ubiquitously under the control of the β-actin promoter (Hadjantonakis et al., 1998). The cerebellar specific model expresses eGFP under the control of the L7 promoter in purkinje cells in the cerebellum (Tomomura et al., 2001). Last, the cortex specific model (GIN mice) expresses eGFP under the control of the Gad1 promoter in GABAergic interneurons (Oliva et al., 2000). Corresponding age and strain matched controls were imaged for all groups. Animals were imaged first without and then with a saturation pulse. Five MTC datasets at 0.5, 1, 5, 10, and 20 kHz offset frequencies were acquired. Region based MTR calculations as well as pixel by pixel MTR maps were calculated for all groups. Additionally, expression of GFP was confirmed ex vivo through fluorescence microscopy. We focused upon the 1 kHz offset as this proved to be the offset resonance frequency that gave the most consistent results and optimal differences between control and GFP expressing animals.
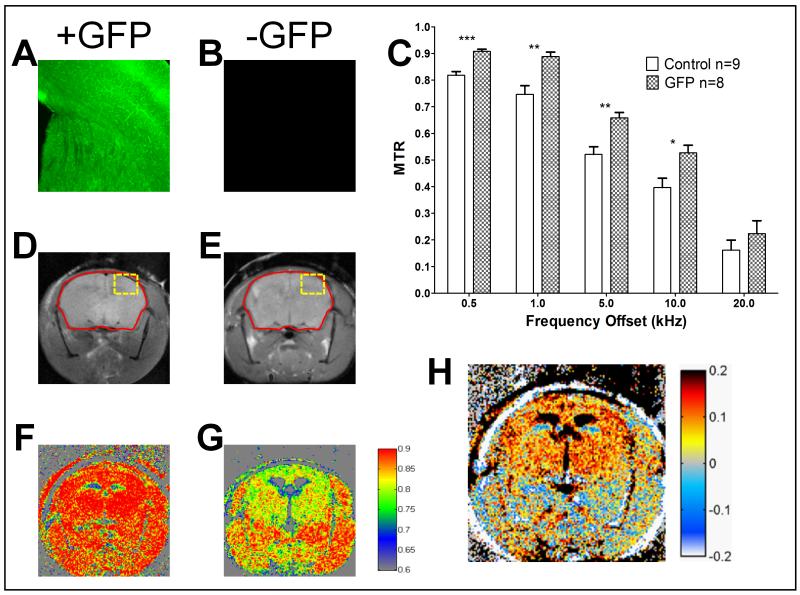
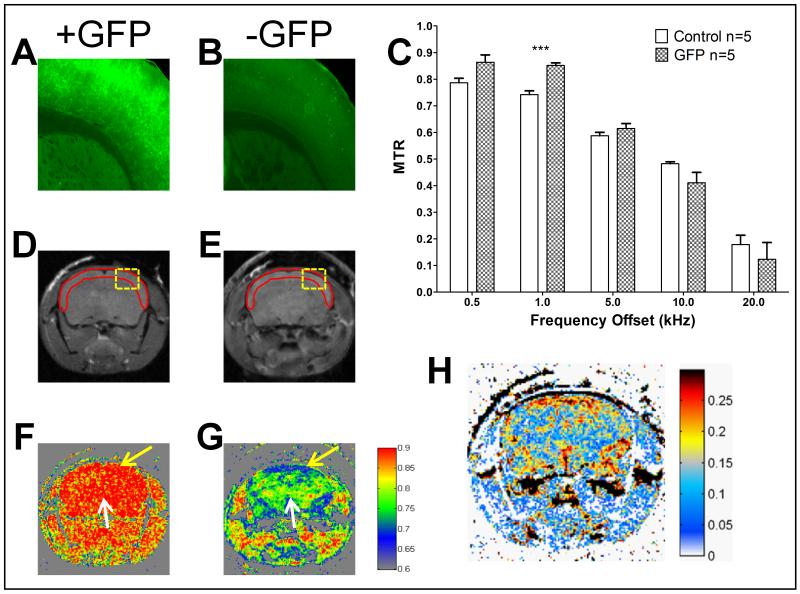
For the ubiquitous model (Fig. 3), to keep the analysis consistent, we focused primarily on brain expression. Fluorescence micrographs of a GFP expressing animal and control (Fig. 3A and B respectively) at exactly the same settings confirm GFP expression only in the experimental group. The region of the brain assessed for fluorescence microscopy is depicted as a yellow box in Fig. 3C and D. The control animal did not give any appreciable signal at this setting (Fig. 3B). Region based MTR calculations for all the frequency offsets were performed focusing on the whole brain as the region of interest (Fig. 3C). For all offset frequencies, the GFP expressing animals (n=8) exhibited a larger MTR than controls (n=9). We performed a mixed model ANOVA to test for a group effect, an offset effect and an interaction while accounting for the repeated measures nature of the experiment. The offset effect was very significant with a p-value <.0001. More importantly the group effect was significant with a p-value <.01. We then used linear contrasts to identify at which offsets the groups were significantly different. We determined that the largest difference occurred at the 1 kHz offset giving a difference of 0.14 (19% difference increased MTR) compared to control and a p-value <0.01 (see Supplementary Table 1). However, it should be noted that other offset frequencies also provided significant MTR differences compared to control: 0.5 kHz with a P-value <0.0001, 5 kHz with a P-value <0.01, and 10 kHz with a P-value <0.05 (p = .0113).
Figure 3.
Ubiquitous expression GFP mouse model. Panels A and B are representative fluorescence images of the cortex and midbrain from GFP expressing (A) versus control (B) mice. Control sample (B) does not give appreciable signal at the settings used for GFP positive animals. Panel C shows the region based MTR calculations for the different frequency offsets for GFP vs. Control. Significance was assessed with a mixed model ANOVA followed by linear contrasts for each offset frequency with ***p<0.001, **p<0.01 and * p<0.05 (p=0.0113). Panels D and E contain the unsaturated images from GFP (D) and control (E) mice. The red outline depicts the region of interest for Panel C. Panel F and G show the pseudocolored pixel by pixel MTR calculation for 1 kHz offset from GFP (F) and Control (G) mice. Panel H is the subtraction of the MTR images in the form of GFP positive minus GFP negative MTR.
A visual representation is more useful to assess the spatial distribution of signal changes compared to a single ROI analysis. The unsaturated images for a representative GFP expressing and control animal (Fig. 3D and E respectively) demonstrated similar signal intensities. However, the MTR maps with the same colormap (Fig. 3F and G) demonstrated an enhancement in the GFP expressing animals. Whereas some differences were appreciable at this stage, subtracting the control image from the GFP positive image provided a better representation of the GFP positive regions (Fig. 3H). Even though this model ubiquitously expresses GFP, we observed the most MTR contrast in the brain region most likely because there is less background there as is seen in the control image (Fig. 3G).
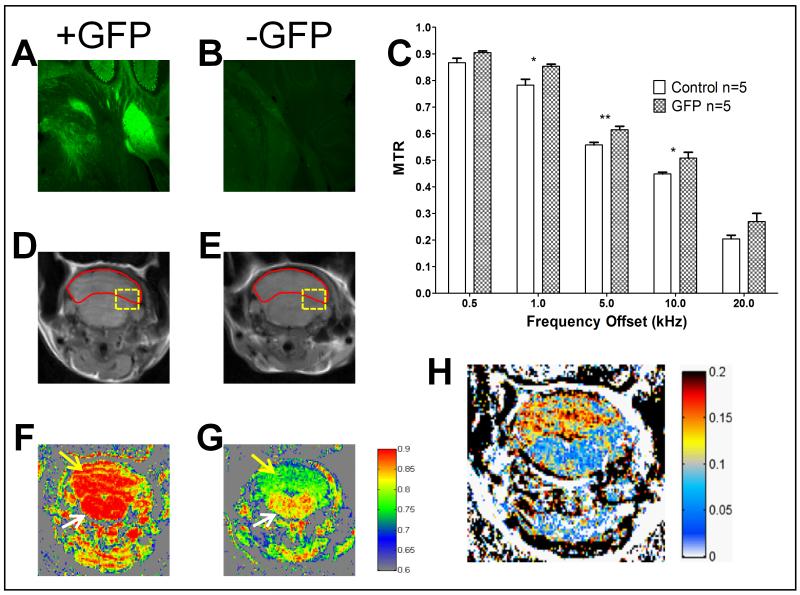
For the cerebellar specific model (Fig. 4), we focused our attention upon the cerebellum in contrast with the brainstem. Fluorescence micrographs of a GFP expressing animal and control (Fig. 4A and B respectively) at exactly the same settings confirm GFP expression in the GFP expressing animals that is limited to the cerebellum. The region of the brain assessed for fluorescence microscopy is depicted as a yellow box in Fig. 4C and D. In the GFP animals, we observe a brighter signal from the cerebellum versus the brainstem that is not seen in the control animals. Region based MTR calculations for all the frequency offsets were performed focusing on the cerebellum as the region of interest (Fig. 4C). For all data points, the GFP expressing animals (n=5) exhibited a larger MTR than controls (n=5). We performed a mixed model ANOVA to test for a group effect, an offset effect and an interaction while accounting for the repeated measures nature of the experiment. The offset effect was very significant with a p-value <.0001. More importantly the group effect was significant with a p-value <.01. We then used linear contrasts to identify at which offsets the groups were significantly different.
Figure 4.
Cerebellar expression GFP mouse model. Panels A and B are representative fluorescence images of the cerebellum and brainstem from GFP expressing (A) versus control (B) mice. Control sample (B) does not give appreciable signal at the settings used for GFP positive animals. Panel C shows the region based MTR calculations for the different frequency offsets for GFP vs. Control. Significance was assessed a mixed model ANOVA followed by linear contrasts for each offset frequency with **p<0.01 and * p<0.05 (p= 0.0171 and 0.0321 for 1, and 10 kHz respectively). Panels D and E contain the unsaturated images from GFP (D) and control (E) mice. The red outline depicts the region of interest for Panel C. Panel F and G show the pseudocolored pixel by pixel MTR calculation for 1 kHz offset from GFP (F) and Control (G) mice. Yellow arrows point to the cerebellum while white arrows point to the brainstem.Panel H is the subtraction of the MTR images in the form of GFP positive minus GFP negative MTR.
The largest difference was observed at 1 kHz giving a difference of .07 (a 9.04% difference increased MTR) compared to control with a p-value of 0.0171 (see Supplementary Table 2). However, it should be noted that the 5 kHz and 10 kHz offset frequencies also provided a significant MTR difference compared to control with a p-value <0.01 and 0.0321 respectively.
The difference between the groups was also identified via a visual representation. The unsaturated images for a representative GFP expressing and control animal (Fig. 4D and E respectively) showed similar signal intensities. However, the MTR maps with the same colormap (Fig. 4F and G) demonstrated an enhancement in the GFP expressing animals. Whereas some differences were appreciable at this stage, subtracting the control image from the GFP positive image again provided a better representation of the GFP positive regions (Fig. 4H). We observed a markedly high MTR signal difference in the cerebellum, as shown by red areas in Fig. 4H, but not in the brainstem, which corresponds with the model.
For the cortex specific model (Fig. 5), we focused our attention on the differences between the cortex versus the midbrain. Fluorescence micrographs of a GFP expressing animal and control (Fig. 5A and B respectively) at exactly the same settings confirmed GFP expression in the GFP expressing animals that was evident in the cortex and not the midbrain. The region of the brain assessed for fluorescence microscopy is depicted as a yellow box in Fig. 5C and D. Region based MTR calculations for all the frequency offsets were performed focusing on the cortex as the region of interest (Fig. 5C). Here we observed an interesting trend where the GFP expressing animals (n=5) exhibited a larger MTR at the lower offsets while the controls (n=5) seemed slightly higher at the higher offsets. We performed a mixed model ANOVA to test for a group effect, an offset effect and an interaction while accounting for the repeated measures nature of the experiment. The offset effect was very significant with a p-value <.0001. The group effect was not significant but th interaction of group and offset effects was significant with a p-value <.0001. There is no general effect for group in this experiment because Control is lower than GFP for low offsets, and higher than GFP for high offsets. We then used linear contrasts to identify at which offsets the groups were significantly different. The only significant difference appeared to be at 1 kHz giving a difference of .11 (14.71% difference increased MTR) compared to control with a p-value <0.001 (see Supplementary Table 3).
Figure 5.
Cortex expression GFP mouse model. Panels A and B are representative fluorescence images of the cortex and midbrain from GFP expressing (A) versus control (B) mice. The control sample (B) does not give appreciable signal at the settings used for GFP positive animals. Panel C shows the region based MTR calculations for the different frequency offsets for GFP vs. Control. Significance was assessed by a mixed model ANOVA followed by linear contrasts for each offset frequency with *** p<0.001. Panels D and E contain the unsaturated images from GFP (D) and control (E) mice. The red outline depicts the region of interest for Panel C. Panel F and G show the pseudocolored pixel by pixel MTR calculation for 1 kHz offset from GFP (F) and Control (G) mice. Panel H is the subtraction of the MTR images in the form of GFP positive minus GFP negative MTR. Yellow arrows point to the cortex while white arrows point to the midbrain. Panel H is the subtraction of the MTR images in the form of GFP positive minus GFP negative MTR.
An interesting pattern was observed in the visual representation of the data. The unsaturated images for a representative GFP expressing and control animal (Fig. 5D and E respectively) showed similar signal intensities. However, the MTR maps with the same colormap (Fig. 5F and G) demonstrated an enhancement in the GFP expressing animals. Whereas some differences were appreciable at this stage, subtracting the control image from the GFP positive image provided a better representation of the GFP positive regions (Fig. 5H). Areas of high MTR signal difference, as shown by red areas in Fig. 5H, were primarily observed in the outer cortex forming a band in the outer edge of the brain. This is a very similar pattern to the GFP expression originally described for this mouse line (Oliva et al., 2000).
Discussion
MTC MR imaging has been previously utilized to distinguish intrinsic macromolecule concentration changes. The use of extrinsic protein marker provides an added flexibility. The main advantages of detecting GFP with MTC MRI over other MRI based reporters includes that there are multiple GFP mouse lines available and it poses no toxicity. Additionally, GFP allows for ex vivo validation through simple optical techniques that have been extensively characterized. The advantage of MRI over fluorescent methods is the ability to conduct studies in vivo non-invasively. This advantage is even clearer for the study of the brain which is not easily accessible by fluorescent methods and has the added interference from the skull. MRI in itself has the intrinsic advantage of providing phenotypical analysis beyond what is possible with optical techniques in vivo highlighting detailed brain anatomy and some functionality.
The work reported here assessed detection of GFP through MTC MRI both in vitro and in vivo in three different mouse models of GFP expression. Collectively, our results point towards the 1 kHz offset as the most robust offset frequency for the MTC detection of GFP. While the different mouse models provided multiple offsets at which there was a significant enhancement of the MTR, the 1 kHz offset was the only one that was common in all 3 models and consistently provided the largest difference (see Supplementary Tables 1-3).
A larger difference in MTR makes it easier to identify positive results while the main purpose of the percent difference is as a comparison against other methods. Our results at 1 kHz show an enhancement ranging from 9 to 19% difference in vivo in mouse brain. For comparison, a similar CEST reporter based on a poly-L-lysine peptide demonstrated an 8% enhancement over brain baseline (Gilad et al., 2007). In this study, the work entailed transplanting glioma cells labeled with their poly-L-lysine peptide into the brain and glioma cells labeled with a control protein into the other hemisphere. As an additional note, the control cells in that study gave a 3% enhancement (Gilad et al., 2007).
An additional consideration is that because the proteome of each tissue is different, the background MTR varies from tissue to tissue. This was evident in the ubiquitous model of GFP driven by the β-actin promoter. Our results showed that we obtained what appeared to be a better enhancement when we compared brain tissue versus the surrounding muscle tissue in part because the brain tissue had a lower background MTR (Fig. 3g). In addition, while the results are consistent they are not exactly the same for all models. We conclude that the 1 kHz offset is the most consistent offset but for different applications, the offset frequency can be fine-tuned to provide optimal results.
A drawback to this technique lies in its sensitivity, which is an ongoing challenge in MRI molecular imaging. The MTR value is considerable in control animals, so a small increase may not be statistically significant. Even in vitro, where the GFP MTR signal is isolated, a concentration of 25μg/mL (~926nM) is needed to differentiate GFP from the control protein BSA. Though we did not determine what level of GFP expression is needed in vivo, it would be expected that the smallest detectable concentration would be higher than in vitro because the background MTR is higher.
Future work entails assessing additional fluorescent proteins to determine whether they can be used in conjunction with GFP without overlap. It would then be possible to recapitulate some of the bi-fluorescent experiments performed in cells into animal models. The approach described presents a strategy to expand current molecular reporter technology into in vivo small animal models that could mimic and complement what has already been performed in vitro and ex vivo. Detection of GFP with MTC MR imaging provides a potential broad set of applications while minimizing the number of animals used. Fluorescent experiments that currently require sacrificing animals at each time point could now be performed using the same animals for all time points in a non-invasive manner.
Supplementary Material
Acknowledgements
Interdepartmental Program in Translational Biology and Molecular Medicine is funded in part by the Howard Hughes Medical Institutes Med into Grad Initiative. This work was funded in part by a BCM Cancer Center SPORE Development Grant (RGP), an Anonymous Foundation (RGP) and the Dan L. Duncan Cancer Center core grant (P30CA125123)(SGH). We would also like to thank the Biostatistics and Informatics Shared Resource at the Dan L. Duncan Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Citations
- Becker A, Hessenius C, Licha K, Ebert B, Sukowski U, Semmler W, Wiedenmann B, Grötzinger C. Receptor-targeted optical imaging of tumors with near-infrared fluorescent ligands. Nat Biotechnol. 2001;19:327–31. doi: 10.1038/86707. [DOI] [PubMed] [Google Scholar]
- Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benjamin LE, Neeman M. MRI detection of transcriptional regulation of gene expression in transgenic mice. Nat Med. 2007;13:498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- Edison ES, Bajel A, Chandy M. Iron homeostasis: new players, newer insights. European Journal of Haematology. 2008;81:411–424. doi: 10.1111/j.1600-0609.2008.01143.x. [DOI] [PubMed] [Google Scholar]
- Enochs WS, Petherick P, Bogdanova A, Mohr U, Weissleder R. Paramagnetic metal scavenging by melanin: MR imaging. Radiology. 1997;204:417–23. doi: 10.1148/radiology.204.2.9240529. [DOI] [PubMed] [Google Scholar]
- Gilad AA, McMahon MT, Walczak P, Winnard PT, Raman V, van Laarhoven HWM, Skoglund CM, Bulte JWM, van Zijl PCM. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25:217–9. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan W. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Harel A, Eliav U, Akselrod S, Navon G. Magnetization transfer based contrast for imaging denatured collagen. J Magn Reson Imaging. 2008;27:1155–63. doi: 10.1002/jmri.21353. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Meth. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Henkelman RM, Stanisz GJ, Graham SJ. Magnetization transfer in MRI: a review. NMR Biomed. 2001;14:57–64. doi: 10.1002/nbm.683. [DOI] [PubMed] [Google Scholar]
- Laurent D, O’Byrne E, Wasvary J, Pellas TC. In vivo MRI of cartilage pathogenesis in surgical models of osteoarthritis. Skeletal Radiol. 2006;35:555–64. doi: 10.1007/s00256-006-0133-1. [DOI] [PubMed] [Google Scholar]
- Louie AY, Hüber MM, Ahrens ET, Rothbächer U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–5. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- McConnell HM, Thompson DD. Molecular Transfer of Nonequilibrium Nuclear Spin Magnetization. J. Chem. Phys. 1957;26:958–959. [Google Scholar]
- Moore A, Josephson L, Bhorade RM, Basilion JP, Weissleder R. Human transferrin receptor gene as a marker gene for MR imaging. Radiology. 2001;221:244–50. doi: 10.1148/radiol.2211001784. [DOI] [PubMed] [Google Scholar]
- Oliva AA, Jiang M, Lam T, Smith KL, Swann JW. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J Neurosci. 2000;20:3354–68. doi: 10.1523/JNEUROSCI.20-09-03354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A, León A. MR in the diagnosis and monitoring of multiple sclerosis: an overview. Eur J Radiol. 2008;67:409–414. doi: 10.1016/j.ejrad.2008.02.044. [DOI] [PubMed] [Google Scholar]
- Roy R, Godavarty A, Sevick-Muraca EM. Fluorescence-enhanced three-dimensional lifetime imaging: a phantom study. Physics in Medicine and Biology. 2007;52:4155–4170. doi: 10.1088/0031-9155/52/14/009. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–60. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- Tavaré JM, Fletcher LM, Welsh GI. Using green fluorescent protein to study intracellular signalling. J Endocrinol. 2001;170:297–306. doi: 10.1677/joe.0.1700297. [DOI] [PubMed] [Google Scholar]
- Tomomura M, Rice DS, Morgan JI, Yuzaki M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur J Neurosci. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Tung C, Mahmood U, Bogdanov A. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotech. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med. 1989;10:135–44. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- Zaaraoui W, Deloire M, Merle M, Girard C, Raffard G, Biran M, Inglese M, Petry KG, Gonen O, Brochet B, Franconi J, Dousset V. Monitoring demyelination and remyelination by magnetization transfer imaging in the mouse brain at 9.4 T. MAGMA. 2008;21:357–62. doi: 10.1007/s10334-008-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magnetic Resonance in Medicine. 2008;59:1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.