Abstract
Transcription in mammalian cells can be assessed at a genome-wide level, but it has been difficult to reliably determine whether individual transcripts are derived from the Plus- or Minus-strands of chromosomes. This distinction can be critical for understanding the relationship between known transcripts (sense) and the complementary antisense transcripts that may regulate them. Here we describe a technique that can be used to (i) identify the DNA strand of origin for any particular RNA transcript and (ii) quantify the number of sense and antisense transcripts from expressed genes at a global level. We examined five different human cell types and in each case found evidence for antisense transcripts in 2900 to 6400 human genes. The distribution of antisense transcripts was distinct from that of sense transcripts, was non-random across the genome, and differed among cell types. Antisense transcripts thus appear to be a pervasive feature of human cells, suggesting that they are a fundamental component of gene regulation.
The DNA in each normal human cell is virtually identical. The key to cellular differentiation therefore lies in understanding the gene products – transcripts and proteins – that are derived from the genome. For more than a decade, it has been possible to measure the levels of transcripts in a cell at the whole genome level (1). The word "transcriptome" was coined to denote this genome-wide assessment (2). However, it has been difficult to determine which of the two strands of the chromosome (Plus or Minus) serves as template for transcripts in a global fashion. Sense transcripts of protein-encoding genes produce functional proteins while antisense transcripts are often thought to have a regulatory role (3–7).
Several unequivocal examples of antisense transcripts, such as those corresponding to imprinted genes, have been described (reviewed in (3–7)). However, estimates of the fraction of genes associated with antisense transcripts in mammalian cells vary from less than 2% to more than 70% of the total genes (8–18). We have developed a technique called ASSAGE (Asymmetric Strand Specific Analysis of Gene Expression) that allows unambiguous assignment of the DNA strand coding for a transcript. The key to this approach is the treatment of RNA with bisulfite, which changes all cytidine residues to uridine residues. The sequence of a bisulfite-treated RNA molecule can only be matched to one of the two possible DNA template strands (fig. S1). After generating cDNA from bisulfite-treated RNA with reverse transcriptase (RT), sequencing of the RT-PCR product can be used to establish whether a particular RNA was transcribed from the Plus- or Minus-strand. To identify the DNA strands of origin for the entire transcriptome, cDNA fragments derived from bisulfate-treated RNA are ligated to adapters and the sequence of one end of each fragment determined through sequencing-by-synthesis. The number and distribution of the sequenced tags provide information about the level of transcription of each gene in the analyzed cell population as well as the strand from which each transcript was derived.
We used ASSAGE to study transcription in normal human peripheral blood mononuclear cells (PBMC). Several quality controls were performed to evaluate the library of tags derived from this RNA source. First, we calculated the bisulfite conversion efficiency from the sequences of the tags and found that 95% of the C residues in the original RNA had been converted to U residues (19). Second, we determined whether the bisulfite treatment altered the distribution of tags by preparing libraries without bisulfite treatment. We found a good correlation between the number of sense tags in a gene derived from ASSAGE data and the number of tags derived from RNA-seq data from the same cells (R2= 0.59). We also found a correlation between the relative expression levels determined by ASSAGE and those assessed by hybridization to microarrays (R2= 0.45; (19) ).
From the PBMC tag library, four million experimental tags could be unambiguously assigned to a specific genomic position in the converted genome (table S1). Of the 4 million tags, 47.5% had the sequence of the Plus-strand, meaning that the template of these transcripts had been the Minus-strand, while 52.5% had the sequence of the Minus-strand. This is consistent with the expected equal distribution of sense transcripts from the two strands (20). 90.3% of the 4 million tags could be assigned to known genes while the remaining tags were in unannotated regions of the genome (table S1). The fraction of unannotated tags (9.7%) is consistent with data from other sources indicating that there are likely to be actively transcribed genes in human cells that have not yet been discovered or annotated (6, 21–24). Of the informative tags in annotated regions, 11% were antisense and 89% were sense (table S1).
We next assessed the expression of each gene by counting the total number of tags matching a gene or by counting tags with identical sequence matching a gene only once (distinct tags). On average, there were three total tags for each distinct tag, but this number varied widely and reflected the level of expression of the corresponding transcript. With respect to antisense transcription, genes could be divided into three main classes. S genes were defined as those in which sense tags predominated (≥ 5:1 ratio of distinct sense:distinct antisense tags). AS genes were defined as those in which antisense tags predominated (≥ 5:1 ratio of distinct antisense:distinct sense tags). The SAS class included the remaining genes, all of which contained both sense and antisense tags. In PBMC, we identified 329 (2.5%) AS genes, 2061 (15.9%) SAS genes and 10586 (81.6%) S genes among the 12,976 Ensembl genes in which at least five distinct tags were observed (Table 1 and table S2). There were 6,457 genes in which at least two distinct antisense tags were found.
Table 1. Classification of genes with respect to antisense tags.
We classified only those genes whose sum of distinct sense and antisense tags was five or more. S genes contained only sense tags or had a sense/antisense tag ratio of five or more; AS genes contained only antisense tags or had a sense/antisense tag ratio of 0.2 or less; SAS genes contained both sense and antisense tags and had a sense/antisense ratio between 0.2 and 5. Samples were derived from the following sources: PBMC, peripheral blood mononuclear cells isolated from a healthy volunteer; Jurkat, a T-cell leukemia line; HCT116, a colorectal cancer cell line; MiaPaCa2, a pancreatic cancer line; MRC5, fibroblast cell line derived from normal lung.
| Cell type: | PBMC | Jurkat | HCT116 | MiaPaCa2 | MRC5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All genes | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | |
| S genes | 10586 | 81.60% | 9928 | 89.60% | 11176 | 88.00% | 9500 | 89.50% | 10165 | 89.30% | |
| AS genes | 329 | 2.50% | 240 | 2.20% | 203 | 1.60% | 155 | 1.50% | 212 | 1.9% | |
| SAS genes | 2061 | 15.9% | 908 | 8.2% | 1327 | 10.4% | 959 | 9% | 1002 | 8.8% | |
| Total | 12976 | 11076 | 12706 | 10614 | 11379 | ||||||
| Coding genes | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | |
| S genes | 10375 | 81.30% | 9778 | 89.50% | 10770 | 87.60% | 9348 | 89.40% | 10029 | 89.20% | |
| AS genes | 325 | 2.50% | 239 | 2.20% | 201 | 1.60% | 154 | 1.50% | 210 | 2% | |
| SAS genes | 2055 | 16.1% | 907 | 8.3% | 1325 | 10.8% | 959 | 9.2% | 1000 | 8.9% | |
| Total | 12755 | 10924 | 12296 | 10461 | 11239 | ||||||
| Noncoding genes | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | # of genes | Fraction | |
| S genes | 211 | 95.50% | 150 | 98.70% | 406 | 99.00% | 152 | 99.30% | 136 | 97.10% | |
| AS genes | 4 | 1.80% | 1 | 0.70% | 2 | 0.50% | 1 | 0.70% | 2 | 1.4% | |
| SAS genes | 6 | 2.7% | 1 | 0.70% | 2 | 0.50% | 0 | 0% | 2 | 1.4% | |
| Total | 221 | 152 | 410 | 153 | 140 | ||||||
When normalized by length, there was an obvious concentration of antisense tags in exons compared to the entire genome or to introns (p < 0.0001; Fig. 1). Within promoter regions, there was a concentration of antisense tags near the transcription initiation site of the sense transcripts which gradually tapered off upstream (p < 0.01; Fig. 1 and fig. S2). There were also clear differences between the relative distributions of sense and antisense tags, with a higher proportion of antisense tags than sense tags within promoter and terminator regions of genes (p <0.0001; Fig. 1). Examples of the distribution of sense and antisense tags derived from S and AS genes are shown in Fig. 2 and fig. S3. The prediction of AS transcripts could be confirmed by ASSAGE using gene specific primers (fig. S4).
Fig. 1.
ASSAGE tag densities in PBMC. The density of distinct sense and antisense tags in the indicated regions were normalized to the overall genome tag density. The promoter and terminator regions were defined as the 1 kb of sequence that was upstream or downstream, respectively, of the transcript start and end sites.
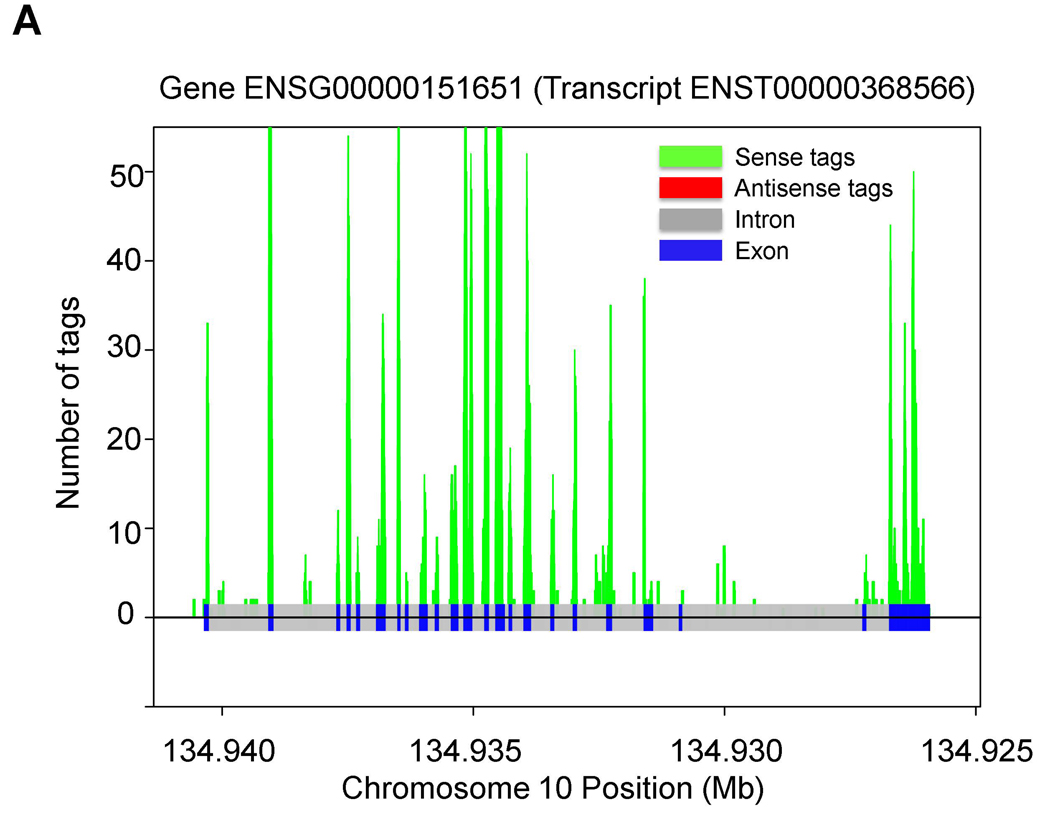
Fig. 2.
Tag distribution in the indicated S (A) and AS (B) genes in PBMC.
To determine whether the patterns described above were particular to PBMC, we used ASSAGE to study four additional human cell types. In all cases, the patterns observed were similar to those in PBMCs including the proportion of S, AS, and SAS genes found in each of the four lines (Table 1 and table S1). However, the identity of the S, AS, and SAS genes varied among the cell lines, suggesting that the expression of antisense tags may be regulated in a cell or tissue-specific manner (fig. S5 and tables S2 and S3). These differences were not related to inter-experimental variation as repeat experiments performed with independently generated ASSAGE libraries from the same RNA sample were highly correlated (fig. S6 and table S2) and differential expression could be confirmed by strand specific-PCR from RNA (fig. S7). In every sample, there was a concentration of both sense and antisense tags within exons compared to the whole genome or to intronic regions and a preferential concentration in promoter and terminator regions (p < 0.01; fig. S2 and S8).
To determine whether splicing of antisense transcripts occurred, we constructed new libraries from Jurkat and MRC5 cells and determined the sequences of both ends of each cDNA fragment (“paired-end sequencing”). As expected, transcripts levels assessed with this paired-end ASSAGE and the original ASSAGE were highly correlated (fig. S9). The size-selected transcript fragments used to construct these libraries were, on average, ~175 bp in length. A cDNA fragment whose ends were located at genomic positions more than three times this distance (>600 bp apart) would be expected to represent spliced transcripts. By this criterion, more than 20% of sense strand cDNA fragments were spliced (fig. S10). In contrast, only ~1% of antisense fragments exhibited this spliced pattern. Sequencing of five putative spliced antisense transcripts confirmed the splicing and comparison with genomic DNA revealed the splice site consensus sequences at the expected locations (fig. S11 and S12).
Our results raise many questions about the genesis and metabolism of antisense transcripts. It has been hypothesized that antisense transcripts are widely and promiscuously expressed, perhaps due to weak promoters distributed throughout the genome (reviewed in (25, 26)). Our data argue against this hypothesis in human cells: promiscuous expression would lead to a uniform distribution of antisense tags across the genome, while the observed distribution was non-random, localized to genes and within particular regions of genes, much like sense transcripts (Fig. 1 and fig. S2, S8). This distribution is consistent with a model wherein many antisense transcripts initiate and terminate near the terminators and promoters, respectively, of the sense transcripts. Some of the apparent antisense transcripts from a gene on the Plus-strand could actually be sense transcripts originating from unterminated transcription of a downstream gene on the Minus-strand (or vice versa). However, this idea is not generally supported by the poor correlation between antisense tag density within a gene and the density of sense tags from the closest downstream gene (fig. S13). One explanation for the higher density of antisense tags in transcribed regions is that transcription of the sense transcripts from correct initiation sites would reduce nucleosome density throughout the entire transcribed region, thereby increasing DNA accessibility and hence the likelihood of non-specific transcription (26). This is unlikely given that genes with high sense tag densities did not generally have high antisense densities. There is substantial evidence that sense transcripts can be negatively regulated by antisense transcripts (3–7). Such regulation can occur either by transcriptional interference or through post-transcriptional mechanisms involving splicing or RISC-like processes. Our data support the possibility that antisense-mediated regulation affects a large number of genes.
Supplementary Material
References and Notes
- 1.Brown PO, Botstein D. Nat Genet. 1999;21:33. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu VE, et al. Cell. 1997;88:243. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 3.Lapidot M, Pilpel Y. EMBO Rep. 2006;7:1216. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. J Cell Sci. 2007;120:2755. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 5.Timmons JA, Good L. Biochem Soc Trans. 2006;34:1148. doi: 10.1042/BST0341148. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Willingham AT, Gingeras TR. Nat Rev Genet. 2007;8:413. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 7.Yazgan O, Krebs JE. Biochem Cell Biol. 2007;85:484. doi: 10.1139/O07-061. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, et al. Nucleic Acids Res. 2004;32:4812. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahey ME, Moore TF, Higgins DG. Comp Funct Genomics. 2002;3:244. doi: 10.1002/cfg.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehner B, Williams G, Campbell RD, Sanderson CM. Trends Genet. 2002;18:63. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 11.Shendure J, Church GM. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelin R, et al. Nat Biotechnol. 2003;21:379. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 13.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Genome Res. 2003;13:1324. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipman DJ. Nucleic Acids Res. 1997;25:3580. doi: 10.1093/nar/25.18.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmichael GG. Nat Biotechnol. 2003;21:371. doi: 10.1038/nbt0403-371. [DOI] [PubMed] [Google Scholar]
- 16.Katayama S, et al. Science. 2005;309:1564. [Google Scholar]
- 17.Okazaki Y, et al. Nature. 2002;420:563. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 18.Kampa D, et al. Genome Res. 2004;14:331. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See supporting material on Science Online.
- 20.Simpson AJ, de Souza SJ, Camargo AA, Brentani RR. Comp Funct Genomics. 2001;2:169. doi: 10.1002/cfg.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters BA, et al. Genome Res. 2007;17:287. doi: 10.1101/gr.5486607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sultan M, et al. Science. 2008;321:956. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Wu JQ, et al. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Trends Genet. 2005;21:93. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Struhl K. Nat Struct Mol Biol. 2007;14:103. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 27.We thank Wayne Yu for assistance with microarrays. Supported by The Virginia and D.K. Ludwig Fund for Cancer Research and NIH grants CA57345, CA 43460, CA 62924 and CA121113. Under a licensing agreement between the Johns Hopkins University and Genzyme, technologies related to SAGE were licensed to Genzyme for commercial purposes, and B.V., V.V. and K.W.K. are entitled to a share of the royalties received by the university from the sales of the licensed technologies. The university and researchers (B.V. and K.W.K.) own Genzyme stock, which is subject to certain restrictions under university policy. The terms of these arrangements are being managed by the university in accordance with its conflict of interest policies.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.