Abstract
Introduction
The amelogenin proteins secreted by ameloblasts during dental enamel development are required for normal enamel structure. Amelx null (KO) mice have hypoplastic, disorganized enamel similar to that of human patients with mutations in the AMELX gene, and provide a model system for studies of the enamel defect amelogenesis imperfecta. Because many amelogenin proteins are present in developing enamel due to RNA alternative splicing and proteolytic processing, understanding the function of individual amelogenins has been challenging.
Purpose
Our objective was to better understand the role of LRAP, a 59 amino acid leucine-rich amelogenin peptide, in the development of enamel.
Approach
Teeth from transgenic mice that express LRAP under control of the Amelx regulatory regions were analyzed for mechanical properties, and transgenic males were mated with female KO mice. Male offspring with a null background that were transgene positive or transgene negative were compared to determine phenotypic differences using microcomputed tomography (microCT) and scanning electron microscopy (SEM).
Results
Nanoindentation revealed no differences between LRAP transgenic and wild-type murine enamel. Using microCT, LRAPKO enamel volume and density measurements were similar to those from KO mice. However, in etched samples examined by SEM, the organization of the enamel rod pattern was altered by the presence of the LRAP transgene.
Conclusions
The presence of LRAP leads to changes in enamel appearance compared to enamel from KO mice. Expression of a combination of amelogenin transgenes in KO mice may lead to rescue of the individual characteristics of normal enamel.
Key Words: Enamel, Amelogenin, Leucine-rich amelogenin peptide, Mineral, Null mice, Transgenic mice
Introduction
During development of enamel in the unerupted tooth, ameloblasts produce various enamel proteins that provide the organic milieu in which enamel structure forms. The abundant, highly conserved amelogenins are expressed from the X-chromosomal amelogenin gene in mice [Chapman et al., 1991; Delgado et al., 2007]. The primary RNA transcript can be alternatively spliced to form at least 15 mRNAs [Hu et al., 1997; Li et al., 1998, 2006], which are translated into amelogenin proteins that vary in abundance. The amelogenin referred to as LRAP, or leucine-rich amelogenin peptide, includes 59 amino acids, and is relatively plentiful in developing bovine enamel [Fincham et al., 1983]. The LRAP protein produced by alternative splicing of the primary amelogenin RNA transcript lacks exon 4 and most of exon 6 [Gibson et al., 1991].
During enamel development, amelogenins have been proposed to assemble into structures called nanospheres in order to guide mineral crystal growth, and similar structures can be produced in vitro using recombinant 20-kD amelogenins [Fincham et al., 1995; Moradian-Oldak et al., 2000]. In vitro, while the recombinant human 175 amino acid amelogenin forms nanospheres that lead to mineral crystal growth, synthetic LRAP proteins can assemble into nanospheres, but are unable to participate in growth of mineral crystals [Habelitz et al., 2006]. However, properties related to intercellular signaling have also been described for LRAP, as gene expression changes were observed when it was added to organ or cell culture or injected into tissues [Tompkins and Veis, 2002; Boabaid et al., 2004].
Amelogenin null (KO) mice were previously generated, and in these mice the amelogenin proteins are not detected [Gibson et al., 2001]. Transgenic (Tg) mice had previously been generated that express bovine LRAP [Gibson et al., 1991; Chen et al., 2003]. Male TgLRAP mice were mated with KO females and male offspring that were transgene positive (TgLRAPKO) or transgene negative (KO) were compared, using microcomputed tomography (microCT) and scanning electron microscopy (SEM), to evaluate effects of the transgene on KO enamel.
Materials and Methods
Animals
Mice were housed in an AAALAC-accredited facility and treated using procedures approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Genetically altered mice were previously described [Gibson et al., 2001; Chen et al., 2003]. Female KO and male Tg mice were mated and offspring genotyped for each experiment.
DNA Analysis
High molecular weight genomic DNA was isolated from mouse tails. The Qiagen multiplex PCR kit (Valencia, Calif., USA) was used for PCR, and products were analyzed on a 4% Nusieve 3:1 agarose gel. Primers were designed to distinguish wild-type from KO background [Li et al., 2008] and to detect the transgene [Chen et al., 2003].
Nanoindentation
Mandibles were dissected and teeth were mounted in room temperature cure epoxy (Allied High Tech Products Inc., Rancho Dominguez, Calif., USA). Teeth were ground from the mesial side using a 400-grit silicon carbide paper and the exposed interior was polished by 1,500-grit silicon paper followed by ultramicrotoming with a 2.5-mm-wide and 45° angle diamond knife (Diatome Inc., Hatfield, Pa., USA) fitted on an MT 6000-XL ultramicrotome (Bal-Tec RMC Inc., Tucson, Ariz., USA). Measurements were made using a Triboscope™ nanoindentation unit (Hysitron Inc., Minneapolis, Minn., USA) attached to an Autoprobe CP scanning probe microscope (Veeco Inc., Santa Barbara, Calif., USA) in air. Hardness and elastic modulus were calculated by the software accompanying the nanoindentation unit [Oliver and Pharr, 1992].
Microcomputed Tomography
Samples were scanned (MicroPhotonics, Allentown, Pa., USA) under maximum voltage and power under saline, through 180° of rotation, with exposure time of 420 ms. The images were processed by three-dimensional reconstruction software and analyzed to determine density and volume for enamel and dentin. The mandibular first molar was analyzed at the position of the mesial root apex.
Scanning Electron Microscopy
Mandibles were fixed in 4% paraformaldehyde, embedded in epoxy resin, cut in half, and cut surfaces were polished to 0.25-μm finish, followed by a 20-second etch with 20% phosphoric acid. Some incisors were also fractured transversely for SEM. All samples were mounted on Al stubs and coated with AuPd for analysis by SEM (JEOL JSM T330A; JEOL Inc., Peabody, Mass., USA).
Results
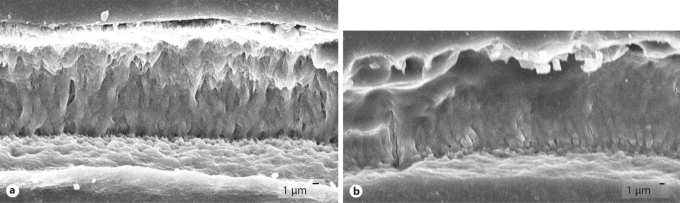
TgLRAP murine enamel was subjected to nanoindentation in order to determine whether changes would be detected in hardness or elastic modulus of the enamel or dentin layers compared to wild-type samples, but no differences were observed (table 1). SEM images of enamel rods in TgLRAP incisors indicate a normal phenotype compared to the aprismatic KO enamel (fig. 1). These results are similar to those obtained from overexpressing TRAP, the N′-terminal fragment of amelogenin in Tg mice, leading to no obvious difference in properties [Paine et al., 2004].
Table 1.
Nanoindentation approach for measurement of hardness and elastic modulus of enamel and dentin for TgLRAP
| Lower left incisors, crosssection, approx. 1 mm from the tip of the tooth | Enamel |
Dentin |
||
|---|---|---|---|---|
| hardness GPa | elastic modulus GPa | hardness GPa | elastic modulus GPa | |
| 4-week LRAP (n = 1)1 | 2.8±0.4 | 83±4 | 0.8±0.1 | 22±3 |
| 6-week LRAP (n = 2)1 | 2.9±0.7 | 89±4 | 0.7±0.3 | 22±2 |
Both measures are similar to wild-type enamel and dentin [Li et al., 2008].
Number of teeth tested. Each value is represented by a total of 20 measurements per tissue per tooth.
Fig. 1.
SEM images of fractured incisors. a TgLRAP incisor showing enamel rods characteristic of normal enamel structure. b KO incisor with aprismatic enamel lacking rod structure.
Female KO mice were mated with Tg males to compare male littermates with or without the transgene on the KO genetic background. PCR analysis of tail DNA was used to determine both genetic background and transgene status.
MicroCT measurements for enamel volume and density for the TgLRAPKO teeth indicated an enamel layer is not apparent in either molars or incisor teeth (table 2), and the measurements are indistinguishable from those for KO mice. The molar dentin volume and density were unaltered by the presence of the LRAP transgene on the KO genetic background (table 2). TgLRAPKO incisors have slightly less dentin volume and an increase in average dentin density over molars from the same mice.
Table 2.
MicroCT analysis for volume and density of enamel and dentin from TgLRAPKO mice
| Genotype | Enamel |
Dentin |
n | ||
|---|---|---|---|---|---|
| volume voxels | density g/m3 | volume voxels | density g/m3 | ||
| TgLRAPKO molar | 0 | 0 | 42,447±4,019 | 2.066±0.07 | 6 |
| TgLRAPKO incisor | 0 | 0 | 38,749±2,241 | 2.316±0.05 | 6 |
Zero volume and density values were also obtained for KO enamel [Li et al., 2008].
An etching procedure was employed to reveal changes unrelated to volume or density in the LRAPKO enamel samples. The LRAPKO and KO enamel layers etched readily compared to wild-type samples, due to the reduced mineral content or differences in composition or structure. In the KO incisors and molars, a thin enamel layer can be discerned, without obvious organization into rod structures (fig. 2). However, in both incisors and molars from TgLRAPKO mice there was an organization in the enamel rod structure not evident in the samples from KO mice (fig. 3, 4).
Fig. 2.
SEM images of etched KO teeth. Incisor (a) and molar (b), with a layer of aprismatic enamel adjacent to the junction with dentin.
Fig. 3.
SEM images of etched TgLRAPKO teeth. Incisor (a) and molar (b), both of which reveal some prismatic organization in the enamel layer.
Fig. 4.
SEM image of etched TgLRAPKO incisor, showing some prismatic structure at higher magnification.
Discussion
The phenotypes of the TgLRAP and KO mice had been previously described, and a rescue experiment had been attempted [Gibson et al., 2001; Chen et al., 2003]. When the gross phenotype of the TgLRAPKO incisor had been examined, there was little obvious change in the appearance of the enamel surface, which remained highly irregular [Chen et al., 2003]. Recently, we reexamined TgLRAPKO samples using the etch/SEM approach, which revealed a greater degree of rod organization when LRAP was present in TgLRAPKO compared to KO enamel.
Structures known as nanospheres are thought to form in vivo by assembly of amelogenins, and these structures are processed by proteases while directing the growth of enamel mineral crystals [Robinson et al., 1979; Fincham et al., 1995]. The greater definition of rod structure seen in TgLRAPKO compared to KO enamel seems to be in agreement with in vitro experiments where LRAP and M180 were compared for ability to form nanospheres and contribute to mineral formation [Habelitz et al., 2006]. Using this approach, both recombinant proteins were able to assemble into nanospheres; however, only the M180 amelogenin enhanced mineral formation. LRAP may function cooperatively with amelogenins such as M180 or with other amelogenins, and perhaps in vivonanospheres require additional amelogenins for normal function. In addition, amelogenins other than LRAP are required for proper thickness of the enamel layer, which could explain the biological importance of alternative splicing of Amelx RNA.
The bovine LRAP cDNA has 6 amino acid differences from murine LRAP within the coding region, and it could be that this may affect a cooperative function with other amelogenins, although none of the differences are found at conserved amino acids [Delgado et al., 2007]. The C-terminal 18 amino acids are identical between bovine and mouse LRAP.
A signaling function for the LRAP proteins, with and without exon 4, has been described, as changes in gene expression and cellular phenotype were observed when recombinant proteins were injected into tissue or added to cell or organ cultures [Tompkins and Veis, 2002; Boabaid et al., 2004]. However, in the KO mice, dentin structure and odontoblast morphology are relatively unaffected by absence of amelogenin. Intriguingly, the Amelx gene is active at a low level in cells other than ameloblasts, including odontoblasts [Nagano et al., 2003; Papagerakis et al., 2003] and other tissues during development [Deutsch et al., 2006; Li et al., 2006; Haze et al., 2007], so that roles related to signaling during normal development as well as in enamel mineral formation can be proposed, and perhaps are related to the level of expression.
Acknowledgements
We gratefully acknowledge discussions with C. Robinson, J.-Y. Sire and E. Golub. We thank S. Labadessa for technical assistance and the School of Dental Medicine vivarium personnel for excellent care of the mice. This work was supported by NIDCR grant DE011089 (C.W.G.).
Abbreviations used in this paper
- KO
amelogenin null
- LRAP
leucine-rich amelogenin peptide
- microCT
microcomputed tomography
- PCR
polymerase chain reaction
- SEM
scanning electron microscopy
- Tg
transgenic
References
- Boabaid F., Gibson C.W., Kuehl M.A., Berry J.E., Snead M.L., Nociti F.H., Jr., Katchburian E., Somerman M.J. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- Chapman V.M., Keitz B.T., Disteche C.M., Lau E.C., Snead M.L. Linkage of amelogenin (Amel) to the distal portion of the mouse X chromosome. Genomics. 1991;10:23–28. doi: 10.1016/0888-7543(91)90479-x. [DOI] [PubMed] [Google Scholar]
- Chen E., Yuan Z.A., Wright J.T., Hong S.P., Li Y., Collier P.M., Hall B., D'Angelo M., Decker S., Piddington R., Abrams W.R., Kulkarni A.B., Gibson C.W. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tiss Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- Delgado S., Ishiyama M., Sire J.-Y. Validation of amelogenesis imperfecta inferred from amelogenin evolution. J Dent Res. 2007;86:326–330. doi: 10.1177/154405910708600405. [DOI] [PubMed] [Google Scholar]
- Deutsch D., Haze-Filderman A., Blumenfeld A., Dafni L., Leiser Y., Shay B., Gruenbaum-Cohen Y., Rosenfeld E., Fermon E., Zimmermann B., Haegewald S., Bernimoulin J.P., Taylor A.L. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci. 2006;114(suppl 1):183–189. doi: 10.1111/j.1600-0722.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Belcourt A.B., Termine J.D., Butler W.T., Cothran W.C. Amelogenins; sequence homologies in enamel-matrix proteins from three mammalian species. Biochem J. 1983;211:149–154. doi: 10.1042/bj2110149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham A.G., Moradian-Oldak J., Diekwisch T.G., Lyaruu D.M., Wright J.T., Bringas P., Jr., Slavkin H.C. Evidence for amelogenin nanospheres as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Gibson C.W., Golub E., Ding W.D., Shimokawa H., Young M., Termine J., Rosenbloom J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Commun. 1991;174:1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- Habelitz S., DenBesten P.K., Marshall S.J., Marshall G.W., Li W. Self-assembly and effect on crystal growth of the leucine-rich amelogenin peptide. Eur J Oral Sci. 2006;114(suppl 1):315–319. doi: 10.1111/j.1600-0722.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- Haze A., Taylor A.L., Blumenfeld A., Rosenfeld E., Leiser Y.L., Dafni B., Shay Y., Gruenbaum-Cohen E., Fermon S., Haegewald J.P., Bernimoulin D., Duetsch D. Amelogenin expression in long bone and cartilage cells and in bone marrow progenitor cells. Anat Rec. 2007;290:455–460. doi: 10.1002/ar.20520. [DOI] [PubMed] [Google Scholar]
- Hu C.C., Ryu O.H., Qian Q., Zhang C.H., Simmer J.P. Cloning, characterization and heterologous expression of exon-4-containing amelogenin mRNAs. J Dent Res. 1997;76:641–647. doi: 10.1177/00220345970760020401. [DOI] [PubMed] [Google Scholar]
- Li W., Mathews C., Gao C., DenBesten P.K. Identification of two additional exons at the 3′ end of the amelogenin gene. Arch Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- Li Y., Yuan Z.A., Aragon M.A., Kulkarni A.B., Gibson C.W. Comparison of body weight and gene expression in amelogenin null and wild-type mice. Eur J Oral Sci. 2006;114(suppl 1):190–193. doi: 10.1111/j.1600-0722.2006.00286.x. [DOI] [PubMed] [Google Scholar]
- Li Y., Suggs C., Wright J.T., Yuan Z.A., Aragon M., Fong H., Simmons D., Daly B., Golub E.E., Harrison G., Kulkarni A.B., Gibson C.W. Partial rescue of the amelogenin null dental enamel phenotype. J Biol Chem. 2008;283:15056–15062. doi: 10.1074/jbc.M707992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian-Oldak J., Paine M.L., Lei Y.P., Fincham A.G., Snead M.L. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- Nagano T., Oida S., Ando H., Gomi K., Arai T., Fukae M. Relative levels of mRNA encoding enamel proteins in enamel organ epithelia and odontoblasts. J Dent Res. 2003;82:982–986. doi: 10.1177/154405910308201209. [DOI] [PubMed] [Google Scholar]
- Oliver W., Pharr G. An improved technique for determining hardness and elastic modulus using load and displacement sensing indentation experiments. J Mater Res. 1992;4:1564–1586. [Google Scholar]
- Paine M.L., Zhu D.H., Luo W., Snead M.L. Over-expression of TRAP in the enamel matrix does not alter the enamel structural hierarchy. Cells Tissues Organs. 2004;176:7–16. doi: 10.1159/000075023. [DOI] [PubMed] [Google Scholar]
- Papagerakis P., MacDougall M., Hotton D., Bailleul-Forestier I., Oboeuf M., Berdal A. Expression of amelogenin in odontoblasts. Bone. 2003;32:228–240. doi: 10.1016/s8756-3282(02)00978-x. [DOI] [PubMed] [Google Scholar]
- Robinson C., Briggs H.D., Atkinson P.J., Weatherell J.A. Matrix and mineral changes in developing enamel. J Dent Res. 1979;58:871–882. doi: 10.1177/00220345790580024101. [DOI] [PubMed] [Google Scholar]
- Tompkins K., Veis A. Polypeptide translated from alternatively spliced transcripts of the amelogenin gene, devoid of the exon 6a, b, c region, have specific effects on tooth germ development in culture. Connect Tissue Res. 2002;43:224–231. doi: 10.1080/03008200290001096. [DOI] [PubMed] [Google Scholar]