Abstract
Amelogenins, the chief proteins in enamel matrix, undergo progressive degradation by matrix metalloproteinase-20 (MMP-20) to facilitate crystal growth. Proline is the most abundant residue in amelogenin and is located upstream to all MMP-20 cleavage sites in the amelogenin sequence. Pro41 is critical for amelogenin N-terminal processing, while the role of prolines at the amelogenin C terminus have not been determined. This study sought to elucidate the effect of the C-terminal prolines on apatite binding and MMP-20 hydrolysis. To compare apatite affinity, recombinant full-length human amelogenin (rh174) and mutated variants (P156T and P164T) were incubated with hydroxyapatite (HAP), and the unbound proteins were quantified by the Bradford assay. rh174 and mutants, as well as 3 oligopeptides, including wild-type peptide and peptides containing 2 mutations, were incubated with MMP-20 both in solution and on HAP. The digested products were analyzed by SDS-PAGE, reverse-phase high-performance liquid chromatography and mass spectrometry. Mutated amelogenins displayed a significantly lower affinity to HAP than the wild type (P156T < P164T < rH174). The proline mutation at amino acid location 164 significantly reduced the initial hydrolysis of either the amelogenins in solution or the proteins bound on HAP, which was confirmed by amelogenin oligopeptide assays. It was concluded that prolines at the amelogenin C terminus are essential for the initial processing of amelogenin and amelogenin-mineral interactions.
Key Words: Amelogenin, Proline, Tooth enamel, Hydroxyapatite, Matrix metalloproteinase 20
Introduction
Amelogenin, the predominant matrix protein in developing dental enamel, has been considered essential for proper enamel formation [Moradian-Oldak, 2001]. Amelogenin self-assembles into nanospheres that serve as a scaffold for the oriented elongation of enamel crystals. As enamel matures, amelogenin proteins are progressively processed by proteinases, including matrix metalloproteinase-20 (MMP-20) in the secretory stage of development and kallikrein 4 in maturation stage, allowing the crystals to grow in thickness.
Proteolysis experiments demonstrate that amelogenin is processed by MMP-20 predominantly at its N and C termini [Moradian-Oldak et al., 2001]. It is of interest to note that prolines are found adjacent and upstream to all identified MMP-20 cleavage sites in the amelogenin sequence [Ryu et al., 1999]. Proline is the only cyclic amino acid with a pyrrolidine ring that restricts the conformational range of adjacent residues. The regularly spaced prolines are presumably important in maintaining the extended chain conformation of proteins. The unique distribution and structural properties of prolines prompted us to investigate their potential functions in amelogenin. Pro41 is located upstream of the W44/L cutting site of MMP-20. The substitution of this proline with threonine, reported in a type of hypomineralized X-linked amelogenesis imperfecta, has been shown to inhibit amelogenin N-terminal processing by MMP-20 [Li et al., 2003].
The C-terminal end of amelogenin was verified to affect the protein self-assembly and apatite binding properties [Moradian-Oldak et al., 2002a]. The proline residues in amelogenin C terminus are highly conserved across many species, implying their functional importance. However, there has been no experimental evidence to substantiate it. By selectively mutating the C-terminal prolines of amelogenin, this study sought to understand if these proline residues contribute to maintain the properties of amelogenin C terminus.
Material and Methods
Preparations of Recombinant Human Amelogenin (rh174), Recombinant Human MMP-20 and Hydroxyapatite
Recombinant human amelogenin (rh174) and recombinant human MMP-20 (rhMMP-20) were expressed and purified by the methods described in our previous publications [Li et al., 2003]. Two amelogenin mutants (P156T and P164T) containing proline-to-threonine mutations at location 156 and 164, respectively, were generated with a QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, Calif., USA). Synthesized hydroxyapatite (HAP) [Mayer et al., 1997] was selected by sequential filtering through 30- and 60-μm meshes for experiments. The surface area of HAP was measured to be 80.8957 m2/g by the Brunauer-Emmett-Teller method (Micromeritics Analytical Services, Norcross, Ga., USA).
Role of Amelogenin C-Terminal Prolines in Apatite Binding
Two micrograms of amelogenin or mutants (P156T and P164T) were incubated with 1 μg HAP in 20 mM Tris/HCl (pH 7.5) for 1 h with constant shaking. After centrifugation at 5,000 rpm for 5 min, the unbound proteins remaining in the supernatant were estimated by the Bradford assay (Bio-Rad, Hercules, Calif., USA). The amount of protein adsorbed (PA) per square meter of HAP crystals was calculated by the following equation, where V is the volume of the solution, W is the weight of the adsorbent HAP, Ci is the initial protein concentration and Cf is the final protein concentration:
The Role of C-Terminal Prolines in the Initial Cleavage of Amelogenin by MMP-20
Equal amounts of amelogenin wild type and mutants were digested with MMP-20 at an enzyme-to-substrate ratio of 1/200 in reaction buffer at 37°C. After 0, 1, 2 and 6 h of digestion, each reaction was terminated by rapid freezing on dry ice/ethanol bath. The samples were then analyzed by SDS-PAGE and MALDI-TOF mass spectrometry. SDS-PAGE gels were scanned and the amount of substrate at each time point was quantified using NIH Image 1.22 (National Institutes of Health, Bethesda, Md., USA). The ratio of substrate at time t to substrate at time zero (St/S0) was estimated to determine the hydrolysis rate.
To compare digestion of HAP-bound proteins, 0.4 μg of wild-type or mutant amelogenin were incubated with 1 μg HAP. The protein amount used was less than the saturated binding level of each protein (fig. 1) to ensure the same amount of protein bound on HAP. The bound and unbound proteins were separated by centrifugation and quantified by Bradford protein assay. The bound wild-type and mutated amelogenins were digested by MMP-20 at an enzyme-to-substrate ratio of 1/200. The digested samples were separated by 12% SDS-PAGE. The ratio of St/S0 was determined using the method described above.
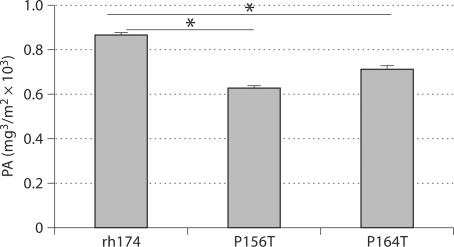
Fig. 1.
Different affinities of mutated and wild-type amelogenins for hydroxyapatite. Wild-type amelogenin (rh174) or mutants (P156T or P164T) were mixed with HAP with constant shaking. Both P156T and P164T amelogenin mutants have significantly reduced protein-absorbed amount per square meter of HAP, compared to the wild-type controls. * p < 0.001.
Roles of Proline in Amelogenin C-Terminal Processing by Amelogenin Oligopeptide Assay
Three peptides were synthesized at GenScript Corporation (Piscataway, N.J., USA). The peptide PP (MLPDLTLEAWPSTD) covers the sequence of rh174 from residue 154 to 168. Peptides TP (MLTDLTLEAWPSTD) and PT (MLPDLTLEAWTSTD) contain proline-to-threonine mutations at corresponding residues 156 and 164, respectively. Equal amounts of peptide were incubated with rhMMP-20 at a 1:200 (w/w) enzyme-to-substrate ratio at 37°C. After 30 min, proteolysis was terminated by addition of an equal volume of 0.1% trifluoroacetic acid. The digested peptide samples were analyzed by reverse-phase high-performance liquid chromatography on a C18 column (Varian, Lake Forest, Calif., USA). The decrease in the peak area of each peptide after hydrolysis was compared with the peak area of the corresponding control sample (incubation without enzyme). The degree of hydrolysis (DH) was calculated by using the following formula.
Statistical analysis was performed with two-way ANOVA, using Softmax software 4.7.1 (Molecular Devices, Sunnyvale, Calif., USA).
Results
Effects of Amelogenin C-Terminal Prolines on Protein Binding Affinity
To determine the influence of prolines on amelogenin binding capacity, we compared the amounts of wild-type and mutated amelogenins (per square meter) adsorbed to HAP. The saturated binding amounts of mutated amelogenins were remarkably lower than the wild-type protein [P156T (632 ± 9.3) < P164T (720 ± 10.8) < wild type (870 ± 9.6)] (fig. 1). Statistical analysis revealed significant differences among the groups (p < 0.001).
Effects of C-Terminal Prolines on Initial Processing by MMP-20
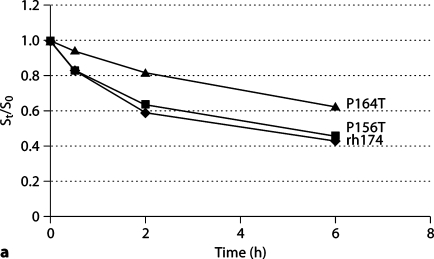
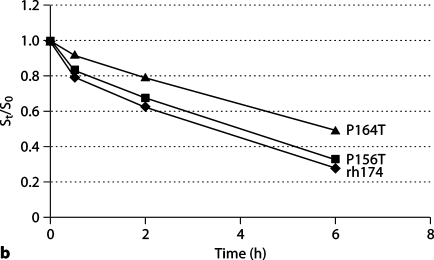
The initial cleavage of amelogenin by MMP-20 occurred at P164/S within the C terminus, releasing the last 11 amino acids teleopeptide (1307.7 Da; data not shown). The MMP-20 hydrolysis of amelogenin wild type and mutants (P156T and P164T) were evaluated both in solution and on HAP. As indicated in a time course analysis by SDS-PAGE (fig. 2), the P164T amelogenin was hydrolyzed at a dramatically slower rate both in solution and on HAP (p < 0.05), displaying a higher St/S0 ratio at each time point of hydrolysis, but the cleavage of P156T amelogenin had no significant difference when compared to rh174.
Fig. 2.
Comparative hydrolysis of wild-type and mutated amelogenins in solution or on HAP. Hydrolyzed wild-type amelogenin (rh174) and 2 mutants (P156T and P164T) were analyzed by SDS-PAGE and the protein bands were quantified using the NIH Image software. The digestion rates of the proteins, either in solution (a) or bound on HAP (b), were compared by the ratio of substrate at time t to substrate at time zero (St/S0). Both in solution and on HAP, the P164T amelogenin mutant had a much higher St/S0 ratio than the wild-type and P156T amelogenins.
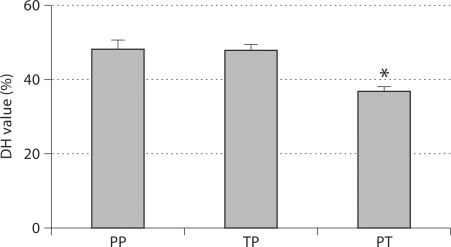
The studies using 3 oligopeptides further confirmed these results. As shown in figure 3, the peptides PP and TP were cleaved by the enzyme at approximately similar speed (DH was 48.5±1.8 and 47.8 ± 1.6%, respectively). In marked contrast, peptide PT was hydrolyzed at a significantly slower rate with a DH value of 36.9 ± 1.3% (p < 0.001).
Fig. 3.
Comparative peptide digestion by MMP-20. The degree of hydrolysis of the peptides PP, TP and PT was determined by measuring the changes in substrate peak area in high-performance liquid chromatography, showing that peptide PT was digested significantly more slowly than the other 2 peptides. * p < 0.001.
Discussion
A variety of studies suggest that the C-terminal tail of amelogenin plays important functions in enamel biomineralization [Moradian-Oldak et al., 2002a]. Though both N- and C-terminal domains of amelogenin are accessible to proteolytic actions [Moradian-Oldak et al., 2002b], the time-dependent mass-spectrometric analysis revealed that the proteolysis of amelogenin by MMP-20 was initiated at the P164/S site in the C-terminal region [unpubl. data]. This processing event released the hydrophilic C-terminal telopeptide and simultaneously created a ‘23K’ product. The truncated 23K amelogenin is one of the most predominant proteolytic amelogenin components, comprising 11% of the enamel protein matrix. Using recombinant proteins and synthetic peptides, the present study demonstrated that this processing event was regulated by these C-terminal proline residues. However, the influence of proline seems to vary between different locations. Proline residues 156 and 164 of human amelogenin correspond to positions 9 and 1 upstream to the cleavage site P164/S, respectively. The proteolytic cleavage at P164/S was only impeded by the replacement of proline with threonine at position-1. The presence of proline at position 1 (Pro164) may hold the downstream sequence in a more ‘turn-like’ structure which provides a close enzyme-substrate fit with less entropic cost [Truckses et al., 1996]. Pro164 may also serve as a docking site for MMP-20, because residue 1 relative to the cleavage site has been proposed to be part of the recognition site for the processing enzyme [Perlman and Halvorson, 1983]. Pro164 substantially influences the MMP20 processing efficiency of amelogenin at its C-terminal end, while Pro156 may be too far away to affect the cleavage at P164/S. This observation, along with the abundance of prolines near the MMP-20 cleavage sites, suggests that proline is a critical amino acid in MMP-20 hydrolysis of amelogenin. The binding capacity of the C terminus was associated with the protein conformation. In the present study, substitution of proline with threonine at position 156 or 164 displayed a significantly lower affinity to HAP, suggesting that these 2 C-terminal prolines are important for optimal adsorption of amelogenin protein to HAP. The prolines appear to be essential conformational determinants that alter the accessibility of amelogenin C terminus to apatite, which is related to the growth of apatite crystals and enamel development.
Acknowledgement
This research was supported by NIH NIDCR grant R01 DE015821.
Abbreviations used in this paper
- HAP
hydroxyapatite
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- MMP-20
matrix metalloproteinase-20
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
- Li W., Gao C., Yan Y., P. DenBesten. X-linked amelogenesis imperfecta may result from decreased formation of tyrosine rich amelogenin peptide (TRAP) Arch Oral Biol. 2003;48:177–183. doi: 10.1016/s0003-9969(02)00170-x. [DOI] [PubMed] [Google Scholar]
- Mayer I., Schlam R., Featherstone J.D. Magnesium-containing carbonate apatites. J Inorg Biochem. 1997;66:1–6. doi: 10.1016/s0162-0134(96)00145-6. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Bouropoulos N., Wang L., Gharakhanian N. Analysis of self-assembly and apatite binding properties of amelogenin proteins lacking the hydrophilic C-terminal. Matrix Biol. 2002a;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Gharakhanian N., Jimenez I. Limited proteolysis of amelogenin: toward understanding the proteolytic processes in enamel extracellular matrix. Connect Tissue Res. 2002b;43:450–455. doi: 10.1080/03008200290000835. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J., Jimenez I., Maltby D., Fincham A.G. Controlled proteolysis of amelogenins reveals exposure of both carboxy- and amino-terminal regions. Biopolymers. 2001;58:606–616. doi: 10.1002/1097-0282(200106)58:7<606::AID-BIP1034>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H.O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Ryu O.H., Fincham A.G., Hu C.C., Zhang C., Qian Q., Bartlett J.D., Simmer J.P. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- Truckses D.M., Somoza J.R., Prehoda K.E., Miller S.C., Markley J.L. Coupling between trans/cis proline isomerization and protein stability in staphylococcal nuclease. Protein Sci. 1996;5:1907–1916. doi: 10.1002/pro.5560050917. [DOI] [PMC free article] [PubMed] [Google Scholar]