Abstract
The catalytic subunit of the mitochondrial (mt) RNA polymerase (RNAP) is highly homologous to the bacteriophage T7/T3 RNAP. Unlike the phage RNAP, however, the mtRNAP relies on accessory proteins to initiate promoter-specific transcription. Rpo41, the catalytic subunit of the Saccharomyces cerevisiae mtRNAP, requires Mtf1 for opening the duplex promoter. To elucidate the role of Mtf1 in promoter-specific DNA opening, we have mapped the structural organization of the mtRNAP using site-specific protein-DNA photo-cross-linking studies. Both Mtf1 and Rpo41 cross-linked to distinct sites on the promoter DNA, but the dominant cross-links were those of the Mtf1, which indicates a direct role of Mtf1 in promoter-specific binding and initiation. Strikingly, Mtf1 cross-linked with a high efficiency to the melted region of the promoter DNA, based on which we suggest that Mtf1 facilitates DNA melting by trapping the non-template strand in the unwound conformation. Additional strong cross-links of the Mtf1 were observed with the −8 to −10 base-paired region of the promoter. The cross-linking results were incorporated into a structural model of the mtRNAP-DNA, created from a homology model of the C-terminal domain of Rpo41 and the available structure of Mtf1. The promoter DNA is sandwiched between Mtf1 and Rpo41 in the structural model, and Mtf1 closely associates mainly with one face of the promoter across the entire nona-nucleotide consensus sequence. Overall, the studies reveal that in many ways the role of Mtf1 is analogous to the transcription factors of the multisubunit RNAPs, which provides an intriguing link between single- and multisubunit RNAPs.
Keywords: DNA/Protein Interaction, DNA/Transcription, Protein/Binding/DNA, Protein/Nucleic Acid Interaction, Transcription, Transcription/Initiation Factors, Transcription/Promoter, Transcription/Yeast
Introduction
The mitochondrial (mt)2 RNA polymerase (RNAP) is closely related to the single-subunit bacteriophage T7/T3 RNAP (1). Their C-terminal ∼800 amino acids show ∼30% sequence identity to T7 RNAP (1–3). Despite this similarity, the mtRNAP depends on transcription factors for promoter-specific initiation (4–7). The core RNAP subunit of the Saccharomyces cerevisiae, Rpo41, requires one major factor, Mtf1 (or sc-MtfB) (6, 8–11), whereas the human core mtRNAP requires two factors, mtTFB1/2 and mtTFA (12–17).
Rpo41 by itself does not recognize and melt the mt promoter or initiate RNA synthesis unless the promoter is pre-melted around the transcription site (18–20). Similarly, there is no evidence that Mtf1 by itself interacts with the promoter (7, 18). When Mtf1 and Rpo41 are present together, the complex (21) melts the promoter from −4 to +2 without requiring initiating NTPs (18). Based on these results, it has been suggested that Rpo41 lacks the mechanism for melting/stabilizing the open promoter and relies on Mtf1 for promoter-specific binding, melting, and stabilizing of the melted promoter.
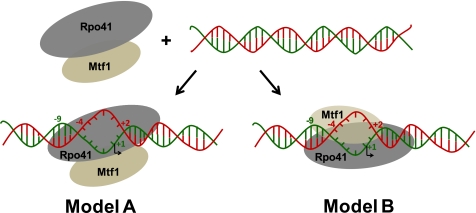
There are two general ways in which Mtf1 can facilitate promoter opening (Fig. 1). In Model A, only the Rpo41 protein within the mtRNAP complex (Rpo41·Mtf1) interacts with the promoter DNA, and Mtf1 acts allosterically. Through protein-protein interactions, Mtf1 causes conformational changes within some or all the components of the ternary complex that allow Rpo41 to bind and melt the DNA. In Model B, the Mtf1 protein interacts directly with the promoter DNA to initiate/stabilize the formation of a DNA bubble in the open complex. In Model-B, both Rpo41 and Mtf1 contact the promoter DNA.
FIGURE 1.
Proposed models of Mtf1 mechanism in promoter melting. Two models of promoter melting by Mtf1 are shown. Rpo41 (gray) and Mtf1 (brown-gray) can assemble in the absence of promoter DNA to form a complex (mtRNAP) (21). In the presence of promoter DNA (the red strand denotes the non-template strand, and the green strand denotes the template strand) two scenarios can be visualized. In model A, Mtf1 interacts only with Rpo41; binding of Mtf1 to Rpo41 causes a change in the conformation of Rpo41 and/or Mtf1 to facilitate DNA melting, where the DNA is melted from −4 to +2. In Model B, Mtf1 directly interacts with the promoter DNA and/or Rpo41 to facilitate promoter melting.
Protein-DNA photo-cross-linking approaches have been successfully used to map the structural organization of bacterial, eukaryotic, and archaeal transcription complexes (22–29). Here, we have used similar methods to investigate the structural organization of the S. cerevisiae mtRNAP open complex and to understand the mechanism of promoter melting by Mtf1. Our results show that (a) Mtf1 is in direct physical proximity to the promoter DNA, (b) Mtf1 contacts the −10 to +4 region of the promoter, (c) Mtf1 has a major presence near the melted nucleotides of the non-template strand and the −8 to −10 region in the open complex, and (d) there is a small rearrangement in the transcriptional bubble upon transition of the open complex to the initially transcribing complex with ATP without changes in the upstream contacts of Mtf1/Rpo41.
EXPERIMENTAL PROCEDURES
Expression and Purification of His-Rpo41 and His-Mtf1
Expression and purification of His-Rpo41 and His-Mtf1 was carried out as described (18).
Steady State RNA Synthesis
Transcription assays (22 °C) were carried out in reaction buffer (50 mm Tris acetate, pH 7.5, 100 mm potassium glutamate, 10 mm magnesium acetate, 0.05% Tween 20, 1 mm dithiothreitol) and contained ATP, UTP, and GTP (Sigma) (50 μm each) and 0.8 μCi of [γ-32P]ATP (6000 mCi/mmol, 10 mCi/ml from PerkinElmer Life Sciences) and an equimolar mix of Rpo41 and Mtf1 (0.5 μm each) and promoter DNA (1 μm). Reactions were stopped after 3 min with 100 mm EDTA and formamide dye (98%, 0.025% bromphenol blue, 10 mm EDTA), heated at 95 °C for 5 min, and loaded on an 18% sequencing gel (19:1 acrylamide to bisacrylamide) with 7 m urea to resolve RNA products. The gel was exposed to a phosphor screen and scanned on a Typhoon 9410 PhosphorImager instrument (Amersham Biosciences) to quantify RNA products.
Preparation of Derivatized Promoter DNA
Oligodeoxynucleotides phosphorothioated at a single defined site (Integrated DNA Technologies) were purified by urea-denatured PAGE before use. The bold region is the nona-nucleotide promoter sequence: 45NT, 5′-ccataatttatttattattatataagtaataaataattgttttatatcc; 45T, 5′-ccggatataaaacaattatttattacttatataataataaataaattat; −4 + 2JBNT, 5′-ccataatttatttattattatatgcagcttaaataattgttttatatcc. The underlined nucleotides indicate the bases that are mismatched within the promoter region on the NT strand.
Derivatization was carried out in the dark for 3 h at 37 °C in a 100-μl reaction volume containing 50 μm DNA strand (phosphorothioated at a single defined site), 10 mm 4-azidophenacylbromide (Sigma) (dissolved initially in methanol), and 50 mm potassium phosphate buffer, pH 7.0. The DNA strand was ethanol-precipitated, washed with 80% ethanol, and dried under a vacuum. The derivatized DNA was analyzed by reverse-phase high performance liquid chromatography using a C-18 column (supplemental Fig. S1). The mobile phase consisted of a gradient from 5% acetonitrile, 0.1 m triethylammonium acetate to 30% acetonitrile, 0.1 m triethylammonium acetate. The derivatized DNA was annealed to the complementary strand and purified by native PAGE.
Protein-DNA Cross-linking Reaction
In a 15-μl reaction, 100 nm Rpo41 or Mtf1 or Rpo41 and Mtf1 were incubated with excess radiolabeled derivatized DNA (200 nm) at room temperature for 10 min. A 50-fold excess unmodified promoter DNA (trap) was added to this reaction and incubated for 5 min before UV-irradiation for 3 min with 366 nm light (UVP Inc. Model UVGL-25) shone from a distance of 2 inches (supplemental Fig. S4). After adding the SDS containing loading buffer (100 mm Tris·Cl, pH 6.8, 200 mm dithiothreitol, 4% SDS, 0.2% bromphenol blue, 20% glycerol) to the reactions, the samples were loaded on a 6.5% SDS-PAGE gel or 4–15% PAGE gels (Bio-Rad). After electrophoresis, the gel was dried and exposed to a phosphor screen and scanned on Typhoon 9410 PhosphorImager instrument (Amersham Biosciences) to visualize the bands. The bands were quantified, and the cross-linking efficiency (CE) was calculated using Equation 1,
where R, M, or D are the intensities of Rpo41, Mtf1, or DNA bands on the gel, and F is fraction of derivatized DNA calculated from the percent modification (supplemental Fig. S1). Cross-linking to Rpo41 or Mtf1 alone contributed less than 14% to the signal observed with the Rpo41·Mtf1 complex (supplemental Fig. S4F).
Molecular Modeling
A three-dimensional model of Rpo41 was constructed using Geno3D (30) and M4T (31, 32). Rpo41 residues 416–1214 were used as the query sequence, for which Geno3D generated a list of homologous proteins with known three-dimensional structures. Template hits included T7 RNAP structures in their initiation (Protein Data Bank (PDB) code 1QLN) and elongation conformations (1MSW). Among the different templates generated, 1QLN was selected as the target structure, based on which the three-dimensional model of Rpo41 was constructed by Geno3D (30). Rpo41 residues 416–1214 were also submitted to M4T, a completely automated three-dimensional model building server (31, 32). M4T automatically selected 1QLN as the target sequence and generated a three-dimensional model of Rpo41 in the initiation conformation. Because user-defined template selection is not allowed on M4T, we could not obtain a model of Rpo41 in the elongation complex (EC) using this server.
The quality of the obtained models was evaluated using PROCHECK (33, 34), PROSA (35), VERIFY3D (36), and ERRAT (37). The secondary structure predictions were performed using PSIPred (38) and PORTER (39). The assessment indicated that the models obtained from both servers were of a good quality (see the supplemental data and Figs. S8–S11 for a full report).
The three-dimensional co-ordinates of the Rpo41 model (both the initiation complex (IC) and elongation complex (EC) structures) are included in the supplemental information (Rpo41_IC.pdb and Rpo41_EC.pdb). The model of Rpo41 was then superimposed onto the structure of T7RNAP (PDB code 1QLN) in the program UCSF Chimera (40) using the command MATCHMAKER. T7RNAP (1QLN) was selected as the reference chain, and the Rpo41 model was selected as the query chain for structure comparison using Smith-Waterman alignment and other default parameters.
RESULTS
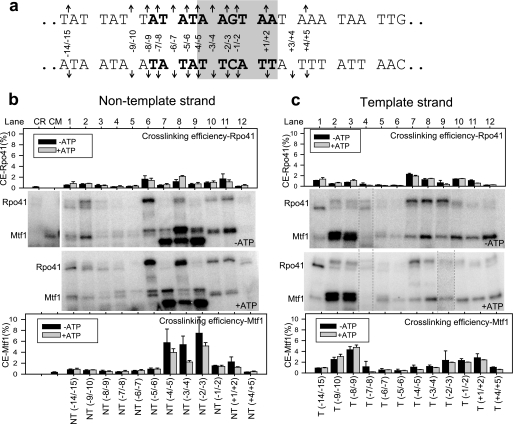
The structural organization of the Rpo41 and Mtf1 proteins within the mtRNAP-DNA complex was characterized using a series of phenyl azide-derivatized promoters in which the photoactivable probe was attached to the phosphodiester backbone of the promoter DNA (27). The phenyl azide probe on the DNA is unreactive in the dark, but when exposed to UV irradiation it forms a highly reactive nitrene species that is capable of forming covalent bonds with nucleophiles on the protein within its van der Waals contact distance. Each of the 24 derivatized double-stranded DNAs contained the probe at a single defined site on the non-template (NT) or the template (T) strand within and flanking the nona-nucleotide promoter sequence of the natural 14S rRNA promoter (Fig. 2a). The derivatized strands were annealed to the complementary strands, and the double-stranded DNAs were purified using native PAGE to remove any extra single-stranded DNA that might cross-link to the proteins (supplemental Fig. S1). The derivatized double-stranded DNAs were analyzed by transcription reactions (supplemental Fig. S2), which showed that, except for T(+1/+2) and T(+3/+4), most were effective substrates of mtRNAP and supported synthesis of abortive products (2–7/8 nt) and a specific 20-nt run-off product. All the unmodified phosphorothioated DNAs supported the transcription reaction (supplemental Fig. S3).
FIGURE 2.
Photo-cross-linking of Rpo41·Mtf1 in the initial open complex and initial transcribing complex with ATP. a, a 45-bp DNA with the 14 S rRNA promoter consensus promoter (in bold) was used in the cross-linking studies. The melted region in the initial open complex is highlighted in gray, and the arrows indicate the sites of derivatization. The nomenclature NT(−8/−9) indicates that the backbone phosphate between positions −8 and −9 on the NT was phosphorothioated and derivatized with 4-azidophenacylbromide. The nomenclature T(−8/−9) indicates a DNA in which the complementary position of NT(−8/−9) on the T was phosphorothioated and derivatized with 4-azido-phenacylbromide. b, cross-linking reactions were carried out with an equimolar mixture of Rpo41 and Mtf1 (100 nm each) and 300 nm radiolabeled DNA (derivatized at a single defined site) in the absence of ATP (−ATP panel) or in the presence of 1 mm ATP (+ATP panel). Cross-linked protein-DNA complexes were resolved on a 4–15% SDS-PAGE gel. Protein-DNA cross-linked products of Rpo41·Mtf1 complex with each of the derivatized DNAs on the non-template (NT) strand (lanes 1–12). Lanes CR and CM are control reactions with Rpo41 alone on NT(−5/−6) or Mtf1 alone on (NT +1/+2), respectively. The CE of Rpo41 and Mtf1 was determined using Equation 1 and plotted using the same y scale for better comparison. Error bars were calculated from three independent experiments. c, protein-DNA cross-linked products of Rpo41·Mtf1 to the T strand (lanes 1–12). The calculations are similar to those of the NT strand. Dashed lines within c (−ATP and +ATP panels) indicate the incorporation of additional lanes.
Rpo41 and Mtf1 Cross-link to the Nona-nucleotide Promoter Region
Rpo41·Mtf1 complex was assembled on the photoactivable promoter DNA in the dark, treated with excess trap DNA (underivatized promoter DNA of the same length) for 5 min, and cross-linked using 366 nm UV light for 3 min at 22 °C (supplemental Fig. S4). Under these conditions, the underivatized DNA forms a stable open complex where the bases from −4 to +2 are in the single-stranded conformation (18). The cross-linking experiments show that both Rpo41 and Mtf1 get cross-linked to the promoter DNA (Fig. 2b), but the major cross-links are those of the Mtf1. Visual inspection reveals differential cross-links of the two proteins to the various positions on the promoter. At some locations on the promoter, Mtf1 cross-links are dominant at some Rpo41s and at some both. Two closely spaced protein-DNA cross-link bands for Rpo41 and three for Mtf1 are observed at some positions. The origin of the multiple cross-linked species is discussed below.
To quantify the relative presence of the two proteins at each of the positions on the promoter, we calculated the cross-linking efficiency of each protein using Equation 1. Because the intensity of free DNA varies slightly from one lane to another, the cross-linking efficiency is a better representation of reactivity than visual inspection alone. However, for most lanes the cross-linking efficiency agrees well with the conclusions made from visual inspection of band intensities (Fig. 2, b and c). The CE of Mtf1 and Rpo41 are summarized in Fig. 4a.
FIGURE 4.
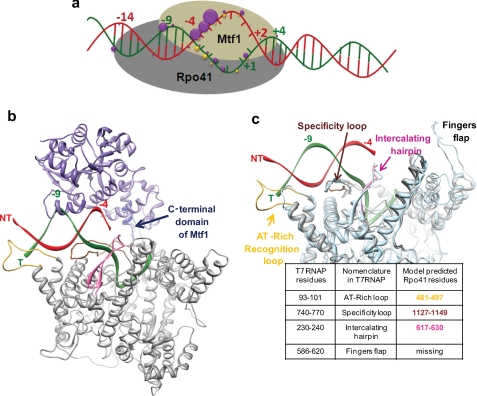
A structural model of the S. cerevisiae mitochondrial RNAP open complex. a, a summary of the protein-DNA cross-linking results is shown. The purple spheres represent Mtf1 cross-links, and the yellow ones represent those of the Rpo41 (Fig. 2). The size of the sphere is proportional to the CE (supplemental Fig. S7). b, a structural model of the mtRNAP open complex is shown. The Rpo41 (416–1214) structure is based on homology modeling to T7 RNAP (PDB code 1QLN) (supplemental information). This region of Rpo41 represents just over half of the protein and shows the highest sequence similarity to T7 RNAP. The DNA is in the same conformation as in 1QLN. The non-template strand (NT) is shown in red, and the template strand (T) is in green. The orientation of Mtf1 is speculative. The placement of Mtf1 (PDB code 1I4W, shown in purple) is based on the cross-linking results from our studies and the proximity of the C-terminal tail of Mtf1 (in dark blue) to the promoter (42). The model shows the promoter sandwiched between Rpo41 and Mtf1 and Mtf1 contacting mainly one face of the promoter at −8/−10, the unwound NT strand, and the transcription start site region. c, shown is comparison of the T7 RNAP structure with the homology model of Rpo41. The three-dimensional structure of Rpo41, 416–1214 (in gray) is aligned with the three-dimensional structure of T7 RNAP, 6–883 residues (PDB code 1QLN) (light blue). The superimposition highlights the model-predicted DNA binding structural elements in Rpo41 that are homologous to the structural elements of T7 RNAP, namely the AT-rich recognition loop (yellow), the specificity loop (brown), and the intercalating hairpin (pink).
Cross-linking of Mtf1
The most striking result we noted was the strong Mtf1 cross-links to the NT strand, specifically with the positions (−4/−5 to +1/+2) that have been shown to be melted in the open complex (18). Within this region, three positions, NT(−4/−5), NT(−3/−4), and NT(−2/−3), showed a very high (6–8%) cross-linking efficiency to Mtf1 (Fig. 2b, −ATP panel, lanes 7–9). Another strong cross-link of Mtf1 was observed to the upstream end of the nona-nucleotide sequence at T(−8/−9) and T(−9/−10) (Fig. 2c, −ATP panel, lanes 2 and 3). Additionally, Mtf1 cross-links were found near the transcription start site at T(+1/+2) and as far downstream as T(+4/+5) (Fig. 2c, −ATP panel, lanes 11 and 12). These results indicate that Mtf1 is in direct physical proximity to the promoter and likely interacts with the entire promoter region from −10 to +4. Additionally, Mtf1 has a major presence near the unwound region of the NT strand of the promoter.
Cross-linking of Rpo41
In general, Rpo41 cross-links were weaker than those of Mtf1. In the melted region of the promoter, Rpo41 cross-links mirrored those of the Mtf1 cross-links. Whereas Mtf1 cross-links within this region were dominant on the NT strand, Rpo41 cross-links dominated on the T strand from T(−4/−5) to T(−1/−2) (Fig. 2c, −ATP panel). A strong Rpo41 cross-link was observed at NT(−5/−6), which is the upstream edge of the initial transcription bubble, and there was negligible Rpo41 cross-linking at the corresponding position on the T strand (compare Fig. 2, b and c, lanes 6). Rpo41 like Mtf1 cross-linked efficiently to the upstream end of the nona-nucleotide sequence at T(−9/−10), but its contacts extended further up to −14.
The 4–15% SDS-PAG allowed us to resolve the closely spaced 2 bands for Rpo41 cross-links and 3 bands for Mtf1 cross-links. The multiple bands were position-specific and reproducible. They varied in intensity and showed clustering around certain positions (supplemental Fig. S5). We concluded after further analysis of the cross-linked complexes, which included DNase I treatment and relabeling of the protein-shortened DNA complex, that the multiple bands arise from cross-linking of the relatively long derivatized DNA (45 bp) perhaps to different amino acid positions on the protein that confers slightly different mobilities of the cross-linked species on the SDS-polyacrylamide gel (supplemental Fig. S6). The phenyl azide photoreactive probe when fully extended has a maximum reach of 11 Å (from backbone phosphate phosphorus atom to the cross-linking target) (27). The photochemical probe potentially could cross-link to a number of amino acids on the protein within its vicinity.
Interactions of Rpo41 and Mtf1 with the Promoter in the Initially Transcribing Complex with ATP
To determine whether Mtf1 cross-links are altered in the initially transcribing complex, we carried out protein-DNA cross-linking studies in the presence of ATP, which serves as the +1 and +2 nucleotides for 2-nt RNA synthesis. We used a high concentration of ATP (1 mm), which assured that the nucleotide was not exhausted within the time span of the cross-linking reaction. The cross-linking efficiencies of Rpo41 and Mtf1 with the promoter were compared with and without ATP (Fig. 2, b and c, +ATP panels). Notable changes were observed within the unwound region of the promoter at NT(−3/−4) with ATP. At this position, the cross-linking efficiency of Rpo41 increased substantially with ATP, and that of Mtf1 decreased (compare Fig. 2b, lane 8, −ATP and +ATP panels). A small decrease in Mtf1 cross-linking efficiency was observed also at NT(−4/−5) and NT(−2/−3) without any change in the band pattern. The results indicate that whereas the upstream contacts of Mtf1/Rpo41 are unaltered in the presence of ATP, there is rearrangement of the transcription bubble upon transition of the open complex to the initially transcribing complex. This is consistent with 2-aminopurine fluorescence studies (18), which indicated that the size of the bubble does not change in the presence of ATP, but that specific bases are more unstacked.
Interactions of Rpo41 and Mtf1 with the Pre-melted Promoter
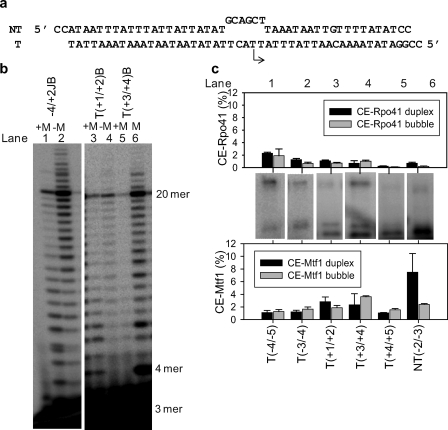
Rpo41 carries out promoter-specific transcription initiation from a pre-melted promoter without requiring Mtf1 (19). We derivatized the pre-melted promoter (mismatches from −4 to +2 in the NT strand, Fig. 3a) at the following positions to investigate its interactions with Mtf1 and Rpo41: T(−4/−5)B, T(−3/−4)B, T(+1/+2)B, T(+3/+4)B, T(+4/+5)B, and NT(−2/−3)B. The modifications at T(+1/+2) and T(+3/+4) positions, which were transcriptionally inactive in the duplex promoter, were active in the pre-melted promoter (Fig. 3b). Rpo41 and Mtf1 cross-linked to the pre-melted DNA (Fig. 3c) with efficiencies similar to the duplex DNA, except for NT(−2/−3)B, which showed a lower cross-linking efficiency to both proteins as compared with NT(−2/−3) in the duplex promoter. The cross-linking efficiency of Rpo41 to the pre-melted promoters in the absence of Mtf1 was very low (supplemental Fig. S4). This indicates that Rpo41 alone, although active on the pre-melted promoters, forms a weaker complex that is chased away by the trap added before UV irradiation. In the absence of Mtf1, we observed longer than the expected 20-nt runoff RNA products (Fig. 3b, lanes 2 and 6). These products could arise from transcription initiating from alternative start sites or by the nontemplated addition of NTPs.
FIGURE 3.
Photo-cross-linking of Rpo41·Mtf1 with the pre-melted promoters. a, the sequence of the pre-melted promoter is shown with a non-consensus NT sequence from −4 to +2 to create mismatches in that region. The arrow indicates the location and direction of transcription initiation. b, shown is RNA synthesis from derivatized pre-melted DNAs. The suffix B (bubble) after each position indicates derivatization of that position in the pre-melted DNA. 500 nm Rpo41 (lanes 2, 4, and 6) or an equimolar mixture of Rpo41 and Mtf1 (500 nm each) (lanes 1, 3, and 5) were mixed with derivatized pre-melted DNA (1 μm), ATP, UTP, and GTP (250 μm each) spiked with [γ-32P]ATP for 3 min at 22 °C. The RNA products were resolved on an 18% polyacrylamide sequencing gel containing 7 m urea. The first C base in the DNA sequence is encountered at +21. Exclusion of CTP, therefore, results in a runoff product of 20 nt. c, a 4–15% PAGE gel show the results of the cross-linking reaction with each of the derivatized DNAs. The cross-linking efficiencies for duplex DNA and pre-melted DNA are shown for comparison.
DISCUSSION
Mtf1 Contacts the Promoter DNA and the Unwound Non-template Strand
The most striking observation from the protein-DNA cross-linking studies reported here is that Mtf1 is closely associated with the unwound region of the promoter (Fig. 4a). The results clearly support Model B of Mtf1 action (Fig. 1). Mtf1 by itself does not interact stably with the promoter (7, 18). Thus, Mtf1-promoter interactions are activated or stabilized by its binding to Rpo41 or to the Rpo41-promoter complex. The close proximity of Mtf1 to the unwound region of the promoter indicates that Mtf1 may stabilize/drive open complex formation by binding to and trapping the unwound non-template strand. In addition to the cross-links within the melted region of the DNA, Mtf1 also cross-linked strongly to the upstream end of the nona-nucleotide promoter sequence at T(−9/−10) and T(−8/−9), which are in the duplexed conformation (18). These upstream interactions may provide an anchor to efficiently open the promoter. Mtf1 cross-links were found downstream of the nona-nucleotide sequence at the transcription start sites, indicating that Mtf1 may be involved in transcription start selection (41). A related work has reported 4-thio-dTMP-derivatized promoter cross-linking to the yeast mtRNAP (42). These cross-linking studies showed that the C-terminal region of Mtf1 (amino acids 319–341), which is disordered in the crystal structure (8), is closely associated with T(−3) and T(−4).
Structural Model of mtRNAP-DNA Open Complex
The C-terminal domain residues (416–1214) of the Rpo41 show significant sequence similarity to T7 RNAP (supplemental information). This similarity allowed us to create a homology model of the C-terminal domain of Rpo41 using the initiation conformation of T7 RNAP (PDB code 1QLN) as the template (43). A structural model of Rpo41·Mtf1·DNA open complex (Fig. 4b) was created by positioning the crystal structure of Mtf1(8) on the structural model of Rpo41-DNA using the following criteria: (a) Mtf1 is close to the unwound non-template strand of the promoter, (b) Mtf1 is close to the −8 to −9 template strand, and (c) the C-terminal amino acids of Mtf1 are close to the −3/−4 template strand (42). This simple placement showed that Mtf1 protein is large enough to span the entire length of the nona-nucleotide consensus sequence from the upstream end of the promoter to the downstream transcription start site.
In T7 RNAP, three elements, namely the AT-rich recognition loop, specificity loop, and β-hairpin are involved in promoter binding/melting (43, 44). The homology modeling suggests the presence of similar elements in Rpo41 (Fig. 4c, supplemental Figs. S8–S11). Based on homology modeling to the initiation conformation of T7 RNAP, we propose that the amino acids 481–497 in Rpo41 are analogous to the AT-rich recognition loop of T7 RNAP that interacts with the −14 region of the T7 promoter. Amino acids 1127–1149 are analogous to the specificity loop that interacts with the promoter from −9 to −1. The 630–660 residues are analogous to the intercalating β-hairpin of T7 RNAP that interacts with the upstream edge of the bubble at −5/−6 in the T7 promoter. We propose that the above-mentioned amino acids in Rpo41 serve a similar function of promoter binding in the mtRNAP. Although there is no experimental proof for the AT-rich recognition and intercalating β-hairpin-like elements in Rpo41, a recent study has shown the presence of a specificity loop-like element in Rpo41 that contacts the promoter from −8 to +6 (45). We also obtained a reasonable fit of the C-terminal domain of Rpo41 to the elongation conformation of T7 RNAP (supplemental Fig. S9), suggesting that perhaps Rpo41 undergoes protein conformational changes during transition from initiation to elongation analogous to T7 RNAP (46, 47).
Possible Mechanism of DNA Melting by Rpo41·Mtf1
It is intriguing that the Rpo41 protein, although highly homologous to T7 RNAP, is unable to melt the promoter on its own. One possible explanation could be that the large N-terminal domain of Rpo41, absent in T7 RNAP, represses the promoter-specific melting activity of the catalytic domain. Alternatively, Rpo41 may be lacking critical residues or elements for promoter melting, which are then provided by the closely associated Mtf1 protein. The homology model (Fig. 4c) reveals one noticeable difference between T7 RNAP and the Rpo41-modeled structure and that is the absence of the fingers-flap domain (residues 593–610 in T7 RNAP) in Rpo41. The exact role of the fingers flap is not known, but these residues in the T7 RNAP contact the promoter DNA downstream of the transcription bubble (48, 49) and have been speculated to aid in stabilizing the downstream end of the transcription bubble.
T7 RNAP by itself is not efficient at melting the promoter and requires GTP, the +1 and +2 nucleotides, for efficient open complex formation (50). Mtf1 may play a role similar to the initiating GTPs with regard to stabilizing the open complex. T7 RNAP uses the binding energy of GTPs to stabilize the open complex. The mtRNAP may use the binding energy of Mtf1-DNA to stabilize the initially open complex. The Mtf1-DNA binding energy may be used to nucleate bubble formation or optimally bend the promoter to initiate melting, as observed with the GTPs in T7 RNAP (51, 52). Once formed, the initiation bubble may be further stabilized by the proposed intercalating β-hairpin motif of Rpo41, as in T7 RNAP (53, 54).
Similarity to the Transcription Factors of the Multisubunit RNAP
Mtf1 has no sequence or structural homology to the initiation factors of the multi-subunit RNAP, yet it appears to be functionally similar to those initiation factors. Like the general transcription factors of the eukaryotes (23–26, 55), archaeal (28, 29), or the σ factors of bacteria (22, 56), Mtf1 action involves contacting the promoter DNA and stabilizing the melted DNA. Several aromatic and basic residues in the σ region 2.3, for instance, interact with the promoter on the NT strand and are implicated in the nucleation and propagation of the transcription bubble (56–59), and mutant proteins with changes in these residues require supercoiled templates for transcription (60, 61). Similarly, mutations within a basic region of Mtf1 (region encompassing residues 178–192) were shown to have a similar effect (62, 63), suggesting that these residues may contribute to the promoter melting function of Mtf1 either by direct physical interaction with the promoter and/or by interacting with Rpo41.
Does Mtf1 have functions other than helping in DNA melting? We were surprised to see Mtf1 cross-links near the transcription start site on the T strand. Although an unexpected result, it is not unprecedented in transcription factor studies. General transcription factors have been observed to be near the transcription start site in eukaryotic and archaeal initiation complexes. Cross-linking studies of the bacterial transcription open-complex have implicated σ factor region 3.2 in direct contact with the initiating nucleotide near the RNAP active center (64). Similarly, TFIIB in S. cerevisiae nuclear RNAP II preinitiation complex (26) and its archaeal homolog TFB1 (from Pyrococcus furiosus RNAP) (29) also cross-link close to the transcription start site. In our work cross-linking of Mtf1 close to the transcription start site on the T strand points to a potential function of Mtf1 in the transcription initiation process, such as nucleotide binding (64, 65). Thus, the dependence of T7 phage-like single-subunit RNAP on transcription factors for initiation represents an added level of complexity that might be required for regulating the efficiency of transcription initiation and hence gene expression in mitochondria.
Supplementary Material
Acknowledgments
We thank Guo-Qing Tang for detailed insights into this study. We thank Manjula Pandey for the initial help with the preparation of the derivatized DNA, Vikas Nanda for consultations on homology modeling, and the Patel laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51966 (to S. S. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11 and Models Rpo41_IC.pdb and Rpo41_EC.pdb.
- mt
- mitochondrial
- RNAP
- RNA polymerase
- CE
- cross-linking efficiency
- nt
- nucleotide(s)
- NT
- non-template
- T
- template.
REFERENCES
- 1.Masters B. S., Stohl L. L., Clayton D. A. (1987) Cell 51, 89–99 [DOI] [PubMed] [Google Scholar]
- 2.Hedtke B., Börner T., Weihe A. (1997) Science 277, 809–811 [DOI] [PubMed] [Google Scholar]
- 3.Cermakian N., Ikeda T. M., Cedergren R., Gray M. W. (1996) Nucleic Acids Res. 24, 648–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas T. K., Edwards J. C., Rabinowitz M., Getz G. S. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1954–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osinga K. A., De Haan M., Christianson T., Tabak H. F. (1982) Nucleic Acids Res. 10, 7993–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schinkel A. H., Koerkamp M. J., Touw E. P., Tabak H. F. (1987) J. Biol. Chem. 262, 12785–12791 [PubMed] [Google Scholar]
- 7.Schinkel A. H., Groot Koerkamp M. J., Tabak H. F. (1988) EMBO J. 7, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubot F. D., Chen C. J., Rose J. P., Dailey T. A., Dailey H. A., Wang B. C. (2001) Protein Sci. 10, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkley C. S., Keller M. J., Jaehning J. A. (1985) J. Biol. Chem. 260, 14214–14223 [PubMed] [Google Scholar]
- 10.Ticho B. S., Getz G. S. (1988) J. Biol. Chem. 263, 10096–10103 [PubMed] [Google Scholar]
- 11.Xu B., Clayton D. A. (1992) Nucleic Acids Res. 20, 1053–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 13.Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. (1992) J. Biol. Chem. 267, 3358–3367 [PubMed] [Google Scholar]
- 14.Fisher R. P., Clayton D. A. (1988) Mol. Cell. Biol. 8, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parisi M. A., Clayton D. A. (1991) Science 252, 965–969 [DOI] [PubMed] [Google Scholar]
- 16.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 17.McCulloch V., Shadel G. S. (2003) Mol. Cell. Biol. 23, 5816–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang G. Q., Paratkar S., Patel S. S. (2009) J. Biol. Chem. 284, 5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga M., Jaehning J. A. (2004) J. Biol. Chem. 279, 44239–44242 [DOI] [PubMed] [Google Scholar]
- 20.Mangus D. A., Jaehning J. A. (1996) Methods Enzymol. 264, 57–66 [DOI] [PubMed] [Google Scholar]
- 21.Mangus D. A., Jang S. H., Jaehning J. A. (1994) J. Biol. Chem. 269, 26568–26574 [PubMed] [Google Scholar]
- 22.Naryshkin N., Revyakin A., Kim Y., Mekler V., Ebright R. H. (2000) Cell 101, 601–611 [DOI] [PubMed] [Google Scholar]
- 23.Kim T. K., Ebright R. H., Reinberg D. (2000) Science 288, 1418–1422 [DOI] [PubMed] [Google Scholar]
- 24.Kim T. K., Lagrange T., Wang Y. H., Griffith J. D., Reinberg D., Ebright R. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12268–12273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagrange T., Kim T. K., Orphanides G., Ebright Y. W., Ebright R. H., Reinberg D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B. S., Mandal S. S., Hampsey M. (2004) Biochemistry 43, 12741–12749 [DOI] [PubMed] [Google Scholar]
- 27.Chen Y., Ebright R. H. (1993) J. Mol. Biol. 230, 453–460 [DOI] [PubMed] [Google Scholar]
- 28.Renfrow M. B., Naryshkin N., Lewis L. M., Chen H. T., Ebright R. H., Scott R. A. (2004) J. Biol. Chem. 279, 2825–2831 [DOI] [PubMed] [Google Scholar]
- 29.Bartlett M. S., Thomm M., Geiduschek E. P. (2004) J. Biol. Chem. 279, 5894–5903 [DOI] [PubMed] [Google Scholar]
- 30.Combet C., Jambon M., Deléage G., Geourjon C. (2002) Bioinformatics 18, 213–214 [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Fuentes N., Madrid-Aliste C. J., Rai B. K., Fajardo J. E., Fiser A. (2007) Nucleic Acids Res. 35, W363–W368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rykunov D., Steinberger E., Madrid-Aliste C. J., Fiser A. (2009) J. Struct. Funct. Genomics 10, 95–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski R. A., M. W. M., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 34.Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 35.Wiederstein M., Sippl M. J. (2007) Nucleic Acids Res. 35, W407–W410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisenberg D., Lüthy R., Bowie J. U. (1997) Methods Enzymol. 277, 396–404 [DOI] [PubMed] [Google Scholar]
- 37.Colovos C., Yeates T. O. (1993) Protein Sci. 2, 1511–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 39.Pollastri G., McLysaght A. (2005) Bioinformatics 21, 1719–1720 [DOI] [PubMed] [Google Scholar]
- 40.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 41.Amiott E. A., Jaehning J. A. (2006) J. Biol. Chem. 281, 34982–34988 [DOI] [PubMed] [Google Scholar]
- 42.Savkina M., Temiakov D., McAllister W. T., Anikin M. (2010) J. Biol. Chem. 285, 3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheetham G. M., Steitz T. A. (1999) Science 286, 2305–2309 [DOI] [PubMed] [Google Scholar]
- 44.Cheetham G. M., Jeruzalmi D., Steitz T. A. (1999) Nature 399, 80–83 [DOI] [PubMed] [Google Scholar]
- 45.Nayak D., Guo Q., Sousa R. (2009) J. Biol. Chem. 284, 13641–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandwar R. P., Ma N., Emanuel S. A., Anikin M., Vassylyev D. G., Patel S. S., McAllister W. T. (2007) J. Biol. Chem. 282, 22879–22886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandwar R. P., Tang G. Q., Patel S. S. (2006) J. Mol. Biol. 360, 466–483 [DOI] [PubMed] [Google Scholar]
- 48.Guo Q., Nayak D., Brieba L. G., Sousa R. (2005) J. Mol. Biol. 353, 256–270 [DOI] [PubMed] [Google Scholar]
- 49.Guo Q., Sousa R. (2006) J. Mol. Biol. 358, 241–254 [DOI] [PubMed] [Google Scholar]
- 50.Stano N. M., Levin M. K., Patel S. S. (2002) J. Biol. Chem. 277, 37292–37300 [DOI] [PubMed] [Google Scholar]
- 51.Tang G. Q., Patel S. S. (2006) Biochemistry 45, 4936–4946 [DOI] [PubMed] [Google Scholar]
- 52.Ujvári A., Martin C. T. (2000) J. Mol. Biol. 295, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 53.Stano N. M., Patel S. S. (2002) J. Mol. Biol. 315, 1009–1025 [DOI] [PubMed] [Google Scholar]
- 54.Brieba L. G., Sousa R. (2001) Biochemistry 40, 3882–3890 [DOI] [PubMed] [Google Scholar]
- 55.Bushnell D. A., Westover K. D., Davis R. E., Kornberg R. D. (2004) Science 303, 983–988 [DOI] [PubMed] [Google Scholar]
- 56.Schroeder L. A., Choi A. J., DeHaseth P. L. (2007) Nucleic Acids Res. 35, 4141–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helmann J. D., deHaseth P. L. (1999) Biochemistry 38, 5959–5967 [DOI] [PubMed] [Google Scholar]
- 58.Campbell E. A., Muzzin O., Chlenov M., Sun J. L., Olson C. A., Weinman O., Trester-Zedlitz M. L., Darst S. A. (2002) Mol. Cell 9, 527–539 [DOI] [PubMed] [Google Scholar]
- 59.Juang Y. L., Helmann J. D. (1994) J. Mol. Biol. 235, 1470–1488 [DOI] [PubMed] [Google Scholar]
- 60.Aiyar S. E., Helmann J. D., deHaseth P. L. (1994) J. Biol. Chem. 269, 13179–13184 [PubMed] [Google Scholar]
- 61.Aiyar S. E., Juang Y. L., Helmann J. D., deHaseth P. L. (1994) Biochemistry 33, 11501–11506 [DOI] [PubMed] [Google Scholar]
- 62.Karlok M. A., Jang S. H., Jaehning J. A. (2002) J. Biol. Chem. 277, 28143–28149 [DOI] [PubMed] [Google Scholar]
- 63.Shadel G. S., Clayton D. A. (1995) Mol. Cell. Biol. 15, 2101–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulbachinskiy A., Mustaev A. (2006) J. Biol. Chem. 281, 18273–18276 [DOI] [PubMed] [Google Scholar]
- 65.Kennedy W. P., Momand J. R., Yin Y. W. (2007) J. Mol. Biol. 370, 256–268 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.