Abstract
Kinetic intermediates that appear early during protein folding often resemble the relatively stable molten globule intermediates formed by several proteins under mildly denaturing conditions. Molten globules have a substantial amount of secondary structure but lack virtually all tertiary side-chain packing characteristics of natively folded proteins. Due to exposed hydrophobic groups, molten globules are prone to aggregation, which can have detrimental effects on organisms. The molten globule that is observed during folding of α-β parallel flavodoxin from Azotobacter vinelandii is a remarkably non-native species. This folding intermediate is helical and contains no β-sheet and is kinetically off-pathway to the native state. It can be trapped under native-like conditions by substituting residue Phe44 for Tyr44. To characterize this species at the residue level, in this study, use is made of interrupted hydrogen/deuterium exchange detected by NMR spectroscopy. In the molten globule of flavodoxin, the helical region comprising residues Leu110-Val125 is shown to be better protected against exchange than the other ordered parts of the folding intermediate. This helical region is better buried than the other helices, causing its context-dependent stabilization against unfolding. Residues Leu110–Val125 thus form the stable core of the helical molten globule of α-β parallel flavodoxin, which is almost entirely structured. Non-native docking of helices in the molten globule of flavodoxin prevents formation of the parallel β-sheet of native flavodoxin. Hence, to produce native α-β parallel protein molecules, the off-pathway species needs to unfold.
Keywords: Isotope exchange, NMR, Protein conformation, Protein denaturation, Protein folding, H/D exchange, NMR, Flavodoxin, Molten globule, Off-pathway
Introduction
Non-native protein conformations initially drew attention by their importance for the understanding of the process of protein folding (1–3). Now it is recognized that these conformations also yield important information about protein misfolding and aggregation (4). Partially folded states of proteins with exposed hydrophobic surfaces, such as molten globules, are prone to aggregation and can be precursors of amyloid fibril formation. This aggregation phenomenon can have devastating effects on organisms (4). The resemblance between early kinetic intermediates and molten globules (5–8) suggests that these molten globules can be considered as models of transient intermediates (9). This resemblance has been demonstrated for α-lactalbumin (10–12), apomyoglobin (13, 14), RNase H (15), T4 lysozyme (16), Im7 (17), and flavodoxin (18). Understanding the formation and conformation of these molten globules offers insights into factors responsible for protein misfolding and, potentially, for numerous debilitating pathologies (19).
Upon descending the folding funnel, proteins encounter folding energy landscapes that are rough (20, 21). As a result, partially folded intermediates, which may be on- or off-pathway to the native state, are populated. When the intermediate is on-pathway, as is observed for the majority of proteins studied to date, it has a native-like topology and is productive for folding. In contrast, when the intermediate is off-pathway, it is trapped in such a manner that the native state cannot be reached without substantial reorganizational events (9). Several kinetic studies have revealed involvement of off-pathway intermediates during protein folding (18, 22, 23). A decrease of the folding rate due to the presence of an off-pathway molten globule, which is kinetically trapped and partially folded, increases the likelihood of protein aggregation.
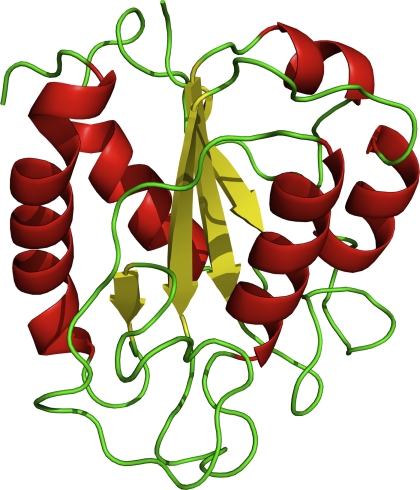
Here, using H/D exchange2 experiments, we report the characterization of the off-pathway molten globule folding intermediate of a 179-residue flavodoxin from Azotobacter vinelandii. Flavodoxins are monomeric proteins involved in electron transport and contain a non-covalently bound FMN cofactor. The proteins consist of a single structural domain and adopt the flavodoxin-like or α-β parallel topology (Fig. 1), which is characterized by a parallel β-sheet that is surrounded by α-helices and is widely prevalent in nature.
FIGURE 1.
Schematic diagram of native flavodoxin from A. vinelandii (Protein Data Bank code 1YOB (45)). The protein adopts the flavodoxin-like or α-β parallel topology and is characterized by a parallel β-sheet surrounded by α-helices at either side of the sheet. The FMN cofactor is not shown.
We showed that the in vitro folding of flavodoxin occurs spontaneously and involves two folding intermediates (18, 24–26). One of these intermediates, Ion, is a productive species that lies on a direct route from unfolded to native protein and is highly unstable and thus is hardly populated. The other intermediate, Ioff, is a relatively stable off-pathway species that populates heavily during refolding and needs to unfold to produce native protein and thus acts as a trap. Approximately 90% of folding molecules fold via Ioff and ultimately form native flavodoxin. The kinetic folding of apoflavodoxin (i.e. flavodoxin without the FMN cofactor) is described by the reaction, Ioff ↔ U ↔ Ion ↔ N, where U is unfolded protein and N is native apoflavodoxin, which is structurally identical to flavodoxin except for some dynamic disorder in the flavin-binding region (27, 28). Apoflavodoxin folds autonomously, and the subsequent binding of FMN is the last step in flavodoxin folding (24).
Spectroscopic data show that the off-pathway folding species of flavodoxin is molten globule-like; its hydrodynamic radius is closer to the native state than to the unfolded state, its three tryptophans are solvent-exposed, and it has severely broadened NMR resonances due to exchange between different conformers on the micro- to millisecond time scale (18, 29, 30). An off-pathway intermediate is experimentally observed for all other α-β parallel proteins of which the kinetic folding has been investigated (i.e. apoflavodoxin from Anabaena (22), CheY (31), cutinase (32), and UMP/CMP kinase (33)).
We showed that at biologically relevant protein concentrations or by mimicking molecular crowding conditions present in cells, severe aggregation of the flavodoxin off-pathway species occurs (29, 30). To further the understanding of the conformational properties of the flavodoxin molten globule, in this study, H/D exchange detected by NMR spectroscopy is used as a powerful technique that gives detailed information about a protein at the level of single amino acids. Use is made of Tyr44-flavodoxin, in which Phe44 is substituted for Tyr44. This substitution leads to severe destabilization of native apoflavodoxin against unfolding and only marginally affects the stability of the molten globule (34). As a consequence, Tyr44-apoflavodoxin forms the molten globule at low salt concentration (i.e. in 10 mm potassium pyrophosphate (KPPi), whereas in 100 mm KPPi, 88% of the protein molecules are native). Now it is possible to characterize an off-pathway molten globule in the absence of denaturant. We use the interrupted H/D exchange methodology (13, 35, 36) to reveal the stable core of the molten globule of Tyr44-flavodoxin and discuss why this species needs to unfold to produce native flavodoxin.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The single cysteine at position 69 in wild-type A. vinelandii (strain ATCC 478) flavodoxin II was replaced by an alanine to avoid covalent dimerization of apoflavodoxin. This protein variant is largely similar to wild-type flavodoxin regarding both redox potential of holoprotein and stability of apoprotein (37, 38). Subsequently, in this protein variant, the phenylalanine at position 44 was substituted for a tyrosine using site-directed mutagenesis. The latter protein variant is referred to as Tyr44-flavodoxin. Uniformly 15N-labeled Tyr44-flavodoxin was obtained from transformed Escherichia coli cells grown on 15N-labeled algae medium (Silantes) and purified as described (38).
To obtain native Tyr44-apoflavodoxin, Tyr44-flavodoxin was denatured in 6 m GdnHCl. Subsequently, both FMN and denaturant were removed via gel filtration on a Superdex 75 column, which is loaded with 100 mm KPPi, pH 6.0. During this removal step, the protein folds to native Tyr44-apoflavodoxin.
H/D Exchange
To follow H/D exchange of the molten globule of Tyr44-flavodoxin, 1.0-ml samples were prepared of 86 μm native Tyr44-apoflavodoxin in 100 mm KPPi in H2O at pH 6.0. Subsequently, the molten globule was populated, and H/D exchange was simultaneously initiated by mixing each of these samples with 14 ml of 3.6 mm KPPi in 96% D2O. The resulting solutions contained 10 mm KPPi, pD 6.72 (uncorrected pH meter reading). Under these conditions, the molten globule of Tyr44-flavodoxin is fully populated, and protein concentration was 5.7 μm. After 10 s to 10 min, H/D exchange was quenched by adding 1 ml of 1.6 mm FMN in 1.45 m KPPi, pH 6.0, in H2O. The resulting solution initially contained 100 μm free FMN in 100 mm KPPi, at pD 6.55 (uncorrected pH meter reading). Under the latter conditions, Tyr44-flavodoxin is rapidly formed. The following exchange periods were used: 10 s, 11 s, 20 s, 30 s, 1 min, 2 min, 5 min, and 10 min.
To detect the backbone amides that are slowly exchanging in Tyr44-flavodoxin, a reference experiment with an exchange period of 10 min was performed with the same procedure except that Tyr44-flavodoxin instead of Tyr44-apoflavodoxin was used. During this experiment, no molten globule was formed, and the protein remained holoprotein throughout the 10-min H/D exchange period and also during the subsequent steps. This experiment revealed that some H/D exchange occurs for the backbone amides of Asn37 and Leu84 of Tyr44-flavodoxin. In total, 66 of the 179 backbone amides of the molten globule of Tyr44-flavodoxin showed no observable exchange in Tyr44-flavodoxin during the 10-min exchange period.
Immediately after H/D exchange, each sample was concentrated by reducing its volume to 500 μl using Amicon centrifugal filter units. Subsequently, 50 μl of 1 mm 2,2-dimethyl-2-silapentane-5-sulfonic acid in D2O was added, and each sample was kept frozen at −80 °C. The NMR samples contained either 91 or 10% D2O, and pD was 6.55 (uncorrected pH meter reading). All H/D exchange experiments were performed at 25 °C.
NMR Spectroscopy
Gradient-enhanced 1H-15N heteronuclear single quantum coherence (HSQC) spectra were recorded on a Bruker AMX 500-MHz machine. Sample temperature was 25 °C. In the 1H dimension, 2048 complex data points were acquired, whereas in the indirect 15N dimension, 256 complex data points were collected. With the number of scans set to 64, each experiment lasted 6 h and 7 min.
Spectra were processed, and maximal intensities of cross-peaks of backbone amides were determined per individual HSQC spectrum. To correct the cross-peak intensities for differences in protein concentration between the different NMR samples, after acquisition of each HSQC spectrum, one-dimensional proton NMR spectra were acquired (4096 complex data points with 1024 scans). Subsequently, the integral of the resonance at −1 ppm, which corresponds to a non-exchangeable methyl resonance of Tyr44-flavodoxin, was determined. This integral is a direct measure of the protein concentration in the corresponding NMR sample. All one-dimensional proton NMR spectra were acquired three times to estimate the error in the integral mentioned. To obtain relative cross-peak intensities, each cross-peak intensity was divided by the corresponding integral of the resonance at −1 ppm.
Hydrogen Exchange Data Analysis
A single exponential decay function was fitted to the time-dependent, relative cross peak intensities,
In Equation 1, t is the time between initiation of H/D exchange and quenching of this exchange, I(∞) is the peak intensity at infinite time, C is the pre-exponential factor, and kex is the amide proton exchange rate.
Peak intensities at infinite time, I(∞), are above zero for all backbone amides. For some residues, I(∞) equals the maximum noise level in the corresponding cross-peak area of the HSQC spectrum. For other residues, I(∞) is larger than the noise level due to the presence of 9% H2O in the samples, causing incomplete exchange of the corresponding backbone amides with D2O.
Model for H/D Exchange
Quantitative interpretation of H/D exchange is possible using a simple model (39),
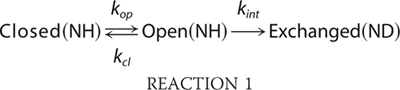 |
In this model, the open or exchange-competent form and the closed or exchange-incompetent form of a protein at the site of a particular amide proton interconvert with rate constants for opening, kop, and closing, kcl. From the open state, exchange takes place with the intrinsic rate constant kint. The time course of exchange is monitored at the residue level by NMR spectroscopy, because amide protons give rise to a 1H NMR signal, and replacement of the proton by a deuteron leads to disappearance of this signal.
Under conditions favoring the closed state (i.e. kop ≪ kcl), the observed exchange rate, kex, is as follows.
 |
Depending on the ratio of kcl and kint, two limiting cases may be reached. If kcl ≪ kint, Equation 2 reduces to the following.
Under this so-called EX1 condition, kex of a certain amide proton informs about the rate constant for local conversion between the closed and the open state of its microenvironment. If kcl ≫ kint, Equation 2 reduces to the following.
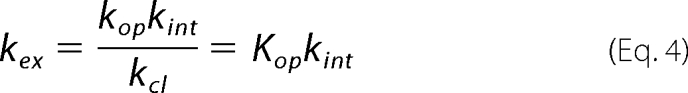 |
Under this so-called EX2 condition, the ratio of kex and kint provides the equilibrium constant Kop for local opening of the protein structure.
RESULTS
To probe hydrogen exchange rates of protein species that are marginally stable and thus have rapidly exchanging amide protons, as is the case for the molten globule of flavodoxin, amide proton exchange needs to be interrupted. In the interrupted H/D exchange methodology (13, 35, 36), an unstable protein species of interest is kept in D2O for a variable time period, t, during which exchange of amide protons for solvent deuterons occurs. After this period, conditions are changed such that the protein arrives into circumstances in which no, or very slow, hydrogen exchange occurs and hence exchange is effectively quenched. Now, by using NMR spectroscopy, exchange rates, kex, of amide protons in the species of interest can be inferred.
Interrupted H/D Exchange Tracks 68 Backbone Amides of the Molten Globule of Flavodoxin
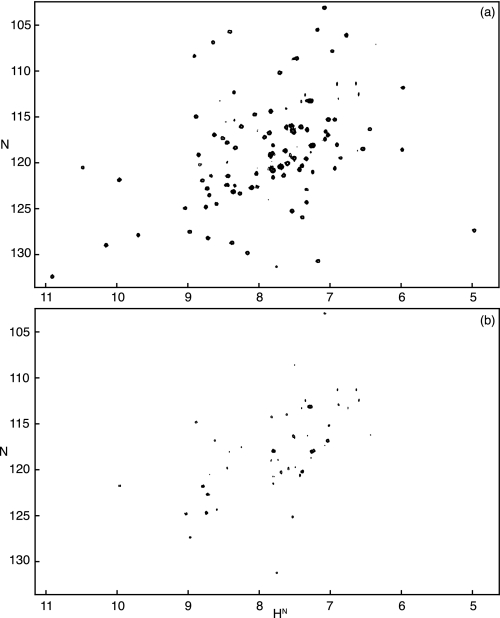
Induction of the molten globule folding intermediate of flavodoxin and simultaneous initiation of H/D exchange was achieved in the following manner. A solution that contained native Tyr44-apoflavodoxin and 100 mm KPPi in water was diluted 10-fold through the addition of an appropriate volume of D2O. This procedure led to a 10 mm KPPi solution containing the molten globule of flavodoxin in D2O. This molten globule has a low stability against unfolding of ∼2 kcal/mol (34). In 10 mm KPPi, the molten globule was fully populated within the dead time of the experiment (data not shown). After time periods, t, of 10 s to 10 min, H/D exchange was quenched by increasing the salt concentration and by the concomitant addition of an excess of FMN. This quenching led to rapid reconstitution of native flavodoxin (see “Experimental Procedures”), in which exchange of many amide protons was very slow (half-times of exchange, 1/kex, of more than 100 days) (28).3 Subsequently, 1H-15N HSQC spectra of the different flavodoxin samples were recorded (Fig. 2). The series of HSQC spectra, as a function of exchange time, t, allowed the determination of the exchange rates of backbone amides of the flavodoxin molten globule.
FIGURE 2.
Demonstration of the effects of H/D exchange of backbone amides of the molten globule of flavodoxin. a, HSQC spectrum of Tyr44-flavodoxin in D2O. b, HSQC spectrum of the Tyr44-flavodoxin sample obtained from an interrupted H/D exchange experiment during which the molten globule of flavodoxin was allowed to exchange its amide protons for solvent deuterons for a period of 2 min (see “Experimental Procedures”).
A prerequisite for successful use of the interrupted exchange experiment is that native flavodoxin is obtained after quenching of H/D exchange. To verify the latter, in a reference experiment, the interrupted H/D exchange methodology described above was used, except that in all steps, H2O instead of D2O was used, and thus no exchange occurred. This experiment indeed demonstrated that properly folded Tyr44-flavodoxin was obtained, because the corresponding HSQC spectrum is indistinguishable from the one of freshly purified Tyr44-flavodoxin (data not shown).
The only amide probes capable of reporting on properties of the molten globule in the interrupted H/D exchange experiment are those that are slowly exchanging in flavodoxin. In addition, cross-peaks of these amides in HSQC spectra of flavodoxin should not suffer from overlap. Due to these limitations, time-dependent amide proton exchange curves and corresponding exchange rates could be determined for 68 of the 179 backbone amides of the molten globule of Tyr44-flavodoxin (see Table 1 and “Experimental Procedures”). Examples of four typical exchange curves are shown in Fig. 3.
TABLE 1.
H/D exchange data of backbone amides of the molten globule of Tyr44-flavodoxin in 10 mm KPPi, pD 6.72, at 25 °C
Data are obtained from interrupted H/D exchange experiments (see “Experimental Procedures”). Shown are rates of H/D exchange, kex, and corresponding S.D. values as well as protection factors and corresponding S.D. values. PF, protection factor.
| Residue | Number | kex | S.D. kex | PF | S.D. PF |
|---|---|---|---|---|---|
| s−1 | s−1 | ||||
| Ile | 3 | 3.07e−02 | 9.20e−03 | 7.04 | 2.11 |
| Gly | 4 | 6.88e−02 | 2.75e−02 | 14.03 | 5.6 |
| Leu | 5 | 2.83e−02 | 9.16e−03 | 12.12 | 3.93 |
| Phe | 6 | 2.02e−02 | 6.68e−03 | 15.44 | 5.1 |
| Phe | 7 | 2.68e−02 | 8.42e−03 | 21.67 | 6.8 |
| Gly | 8 | 8.06e−02 | 4.64e−02 | 23.35 | 13.44 |
| Thr | 14 | ≥0.5a | |||
| Arg | 15 | ≥0.5a | |||
| Lys | 16 | 7.03e−02 | 2.80e−02 | 18.96 | 7.55 |
| Val | 17 | 9.84e−02 | 3.19e−02 | 2.35 | 0.76 |
| Ala | 18 | 5.01e−02 | 1.62e−02 | 12.74 | 4.12 |
| Lys | 19 | 1.08e−01 | 4.01e−02 | 7.42 | 2.75 |
| Ser | 20 | 6.48e−02 | 2.28e−02 | 41.96 | 14.74 |
| Ile | 21 | 8.53e−02 | 3.34e−02 | 3.84 | 1.5 |
| Lys | 22 | 1.10e−01 | 5.84e−02 | 4.31 | 2.3 |
| Lys | 23 | 4.79e−02 | 1.13e−02 | 22.08 | 5.19 |
| Phe | 25 | 5.22e−02 | 1.61e−02 | 16.11 | 4.98 |
| Met | 30 | 5.28e−02 | 3.54e−02 | 25.83 | 17.32 |
| Leu | 34 | 4.07e−02 | 3.08e−02 | 5.68 | 4.3 |
| Asn | 37 | 1.87e−01 | 1.56e−01 | 10.52 | 8.77 |
| Phe | 49 | 9.21e−03 | 2.24e−03 | 87.22 | 21.19 |
| Leu | 50 | 5.10e−03 | 2.30e−03 | 52.18 | 23.51 |
| Ile | 51 | 2.47e−03 | 1.80e−03 | 40.91 | 29.84 |
| Leu | 52 | 7.75e−03 | 3.12e−03 | 17.6 | 7.09 |
| Gly | 53 | 2.44e−02 | 1.12e−02 | 41.37 | 18.97 |
| Thr | 54 | 4.77e−02 | 1.40e−02 | 23.25 | 6.81 |
| Glu | 76 | 4.23e−02 | 6.43e−02 | 4.55 | 6.91 |
| Phe | 77 | 1.45e−02 | 4.29e−03 | 24.68 | 7.29 |
| Leu | 78 | 5.59e−03 | 1.90e−03 | 47.54 | 16.14 |
| Ile | 81 | 1.33e−02 | 4.58e−03 | 16.22 | 5.57 |
| Glu | 82 | 4.62e−02 | 1.29e−02 | 3.47 | 0.97 |
| Leu | 84 | 2.37e−02 | 1.56e−02 | 14.47 | 9.54 |
| Val | 91 | 4.13e−03 | 1.64e−03 | 67.42 | 26.84 |
| Ala | 92 | 1.48e−02 | 4.16e−03 | 43.03 | 12.09 |
| Leu | 93 | 9.63e−03 | 3.15e−03 | 24.03 | 7.85 |
| Phe | 94 | 8.63e−03 | 2.57e−03 | 36.19 | 10.76 |
| Gly | 95 | 5.67e−02 | 1.80e−02 | 33.19 | 10.53 |
| Leu | 96 | 2.34e−02 | 1.25e−02 | 14.64 | 7.8 |
| Asp | 98 | ≥0.5a | |||
| Val | 100 | 2.32e−02 | 1.21e−02 | 11.98 | 6.24 |
| Gly | 101 | 1.71e−01 | 1.65e−01 | 6.95 | 6.69 |
| Tyr | 102 | 4.22e−02 | 2.06e−02 | 16.58 | 8.08 |
| Tyr | 106 | 5.03e−02 | 1.60e−02 | 19.62 | 6.25 |
| Asp | 108 | 3.19e−02 | 7.35e−03 | 8.54 | 1.97 |
| Leu | 110 | 2.73e−03 | 1.22e−03 | 84.83 | 37.99 |
| Gly | 111 | 7.64e−03 | 2.36e−03 | 132.22 | 40.87 |
| Ser | 115 | 1.29e−02 | 2.27e−03 | 179.64 | 31.61 |
| Phe | 116 | 3.47e−03 | 1.37e−03 | 291.64 | 115.24 |
| Lys | 118 | 2.97e−03 | 1.16e−03 | 310.19 | 121.12 |
| Arg | 120 | 1.06e−02 | 2.82e−03 | 66.09 | 17.6 |
| Ala | 122 | 1.17e−02 | 1.79e−03 | 111.1 | 16.98 |
| Lys | 123 | 1.55e−02 | 4.14e−03 | 51.9 | 13.88 |
| Val | 125 | 5.14e−03 | 1.53e−03 | 20.12 | 6 |
| Trp | 128 | 6.17e−02 | 2.93e−02 | 11.07 | 5.25 |
| Ala | 140 | 5.99e−02 | 1.95e−02 | 10.4 | 3.38 |
| Val | 141 | 4.53e−02 | 1.04e−02 | 3.88 | 0.89 |
| Val | 142 | 3.46e−02 | 1.02e−02 | 3.68 | 1.09 |
| Phe | 146 | 7.01e−02 | 3.36e−02 | 9.53 | 4.57 |
| Val | 147 | 2.64e−02 | 1.16e−02 | 7.65 | 3.38 |
| Gly | 148 | 8.12e−02 | 2.56e−02 | 14.62 | 4.61 |
| Ala | 150 | 3.87e−02 | 1.68e−02 | 14.02 | 6.1 |
| Leu | 151 | 1.66e−02 | 6.20e−03 | 13.96 | 5.21 |
| Gln | 156 | 4.85e−02 | 1.98e−02 | 43.51 | 17.78 |
| Arg | 163 | 3.23e−02 | 1.34e−02 | 23.16 | 9.59 |
| Val | 164 | 2.61e−02 | 5.75e−03 | 11.18 | 2.47 |
| Trp | 167 | 1.54e−02 | 3.45e−03 | 22.27 | 4.99 |
| Leu | 168 | 7.72e−03 | 1.75e−03 | 23.27 | 5.26 |
| Gln | 170 | 2.05e−02 | 3.77e−03 | 49.31 | 9.07 |
| Ile | 171 | 1.50e−02 | 3.79e−03 | 17.37 | 4.4 |
| Ala | 172 | 1.26e−02 | 2.20e−03 | 41.24 | 7.21 |
| Phe | 175 | 4.72e−02 | 1.07e−02 | 7.59 | 1.72 |
a A cross-peak was observed for the corresponding backbone amide in a reference H/D exchange experiment using flavodoxin, during which the protein remained flavodoxin throughout a 10-min exchange period (see “Experimental Procedures”). However, in a regular interrupted H/D exchange experiment using the molten globule, with the shortest achievable exchange period of 10 s, no cross-peak was observed for this backbone amide. Consequently, the amide must have exchanged within 10 s, and the corresponding kex value must be ≥0.5 s−1.
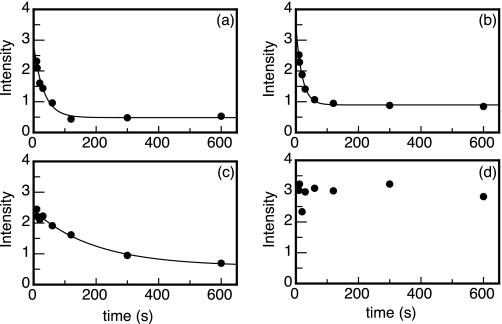
FIGURE 3.
Typical H/D exchange curves obtained for the molten globule folding intermediate of Tyr44-flavodoxin. Shown are time-dependent decreases of maximal intensities of cross-peaks arising from backbone amides of Ile3 (kex = 0.031 ± 0.009 s−1) (a), Ala18 (kex = 0.050 ± 0.016 s−1) (b), Leu50 (kex = 0.0051 ± 0.0023 s−1) (c), and Tyr114 (kex not determined) (d), respectively. Data are extracted from HSQC spectra like the ones shown in Fig. 2.
Most backbone amides of the molten globule of flavodoxin fully exchanged within 10 min (Fig. 3, a and b). Partial exchange occurred for the backbone amides of Leu50 (Fig. 3c), Ile51, Val91, Val100, Gly111, Phe116, Lys118, and Val125. The backbone amide of Tyr114 did not exchange at all within 10 min, and its average maximal intensity in this time period was 2.3 ± 0.3 (Fig. 3d). Similar maximal intensities, extrapolated to an exchange period of 0 s, were observed for the HSQC cross-peaks of the other detectable backbone amides.
Exchange Rates and Protection Factors
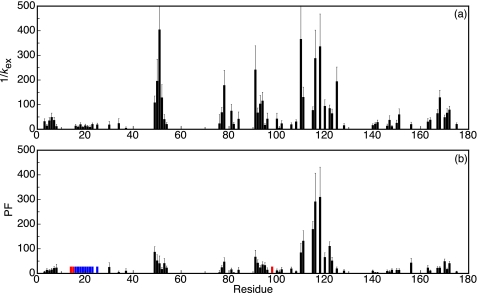
Half-times of exchange of the backbone amides of the molten globule of flavodoxin are shown in Fig. 4a. Due to the low stability against unfolding of the molten globule, the corresponding observable amide protons exchanged much faster than those of flavodoxin. Half-times of exchange of the molten globule did not exceed 500 s (except for the backbone amide of Tyr114).
FIGURE 4.
Protection against H/D exchange of the backbone amides of the molten globule of Tyr44-flavodoxin. a, half-times of exchange, 1/kex, could be determined for 68 residues. The corresponding protection factors (PF) are shown in b. Residues with backbone amide protons that fully exchange within the 10-s dead time of the interrupted H/D exchange experiment are indicated in red. Error bars, S.D. No protection factor can be determined for residues Lys16–Phe25 (indicated in blue), because the corresponding backbone amides exchange according to an EX1 mechanism.
Because interconversion between unfolded flavodoxin and the molten globule is fast (rate constant exceeds 400 s−1 (18)), exchange is expected to occur according to an EX2 mechanism of exchange (see “Experimental Procedures”). Consequently, protection factors against exchange can be calculated by dividing the intrinsic amide proton exchange rates, kint, by the measured amide proton exchange rates. The kint values are determined by using free peptide exchange rates, which are corrected for the effects of local amino acid sequence and calibrated for the pH and temperature of the exchange experiment according to Bai et al. (40). Protection factors against exchange of the molten globule are shown in Fig. 4b. With the exception of Tyr114, these protection factors are low and do not exceed a value of ∼300 (Table 1). These protection factors are comparable with those observed for the molten globules of apomyoglobin (36), apoleghemoglobin (41), and RNase HI (42). In contrast, backbone amides of native apoflavodoxin have protection factors up to 2 × 106.3
Remarkably, the half-times of exchange of residues Lys16–Phe25 are equal within error (1/kex = 14.4 ± 4.7 s) (Fig. 4a). This equality suggests that the corresponding backbone amides become exposed to solvent in a cooperative unfolding event and that exchange occurs before these residues refold to a structured element that is protected against exchange (i.e. exchange occurs according to an EX1 mechanism of exchange; see “Experimental Procedures”). As a result, no protection factor could be determined for residues Lys16–Phe25.
Residues residing in region Leu110–Val125 of the molten globule have the highest protection factors (Figs. 4b and 5b). In this region, five residues (Gly111, Ser115, Phe116, Lys118, and Ala122) have protection factors that exceed a value of 100. In the case of the backbone amide of Tyr114, the protection factor could not be determined, because this amide does not exchange at all during a 10-min exchange period (Fig. 3d). Consequently, the corresponding protection factor must be well above 300.
FIGURE 5.
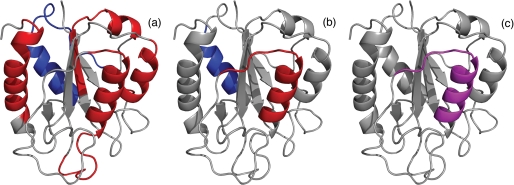
Several folding properties of flavodoxin are highlighted in schematic diagrams of the native protein. a, the four regions of unfolded flavodoxin in 3.4 m GdnHCl that have restricted flexibility on the (sub)nanosecond time scale (44) are colored red; assembly of these structured elements leads to formation of the ordered core of the flavodoxin molten globule (43). The region of the molten globule that remains random coil down to a GdnHCl concentration of 1.58 m (i.e. residues Lys13–Val36) (43) is colored blue. b, residues Leu110–Val125 of the molten globule of flavodoxin have the highest protection factors against H/D exchange and are colored red; the region of the molten globule that probably exchanges its backbone amides via an EX1 mechanism, as determined in this study, is colored blue. In unfolded flavodoxin in 3.4 m GdnHCl, residues Glu104–Lys118 transiently form an α-helix (44). c, partially unfolded form PUF4 of apoflavodoxin as detected by native state H/D exchange (25). In PUF4, only the amides of the residues that are colored purple are protected against H/D exchange, and all other parts of the protein are unfolded.
Using heteronuclear NMR spectroscopy and GdnHCl-induced unfolding, residues Leu110–Val125 have been identified to be part of the ordered core of the flavodoxin molten globule (43). In addition, these residues have restricted flexibility in the unfolded protein (44) (Fig. 5a). The protection factors show that residues Phe49–Thr54, Phe77–Leu84, Val91–Gly95, and Thr160–Leu179 are much less protected against exchange than Leu110–Val125 (Fig. 4b), although these residues have also been identified to be part of the ordered core of the flavodoxin molten globule (43).
DISCUSSION
Recently, by using heteronuclear NMR spectroscopy, we revealed native and non-native secondary structure and non-native hydrophobic interactions in unfolded flavodoxin (43, 44). The unfolded protein contains four transiently ordered regions with restricted flexibility on the (sub)nanosecond time scale (Fig. 5a). This ordering is due to α-helix formation and local and non-local hydrophobic interactions. The ordered regions comprise residues Ala41–Gly53, Glu72–Gly83, Gln99–Ala122, and Thr160–Gly176 (44). These structured elements in the unfolded protein transiently interact and subsequently form the ordered core of the off-pathway molten globule. The residues of the flavodoxin molten globule that have the highest midpoints against unfolding by GdnHCl roughly coincide with those residues that are transiently ordered in unfolded flavodoxin. Assembly of helices in unfolded flavodoxin through hydrophobic interactions leads to formation of the ordered core of the flavodoxin molten globule (43). As a consequence, the molten globule has a drastically different topology compared with native flavodoxin; it is helical and contains no β-sheet (34). Thus, structure formation in unfolded flavodoxin does not direct folding to the native state but instead causes formation of a misfolded folding species, which is common for proteins with a flavodoxin-like topology (26).
The Molten Globule Folding Intermediate of Flavodoxin Is Almost Entirely Structured
The hydrodynamic radius of the molten globule of flavodoxin is expanded only by about 11% compared with the one of native apoflavodoxin, as demonstrated by fluorescence correlation spectroscopy experiments (30). The molten globule is thus relatively compact. Far-UV CD reports that the molten globule contains helices (34). Fluorescence anisotropy of the tryptophans of the molten globule equals a value of 0.08, whereas fluorescence anisotropy of unfolded flavodoxin in GdnHCl is 0.05 (34). Thus, in the molten globule of flavodoxin, its three tryptophans are relatively immobilized.
The H/D exchange data presented in this study reveal that the flavodoxin molten globule must be almost entirely structured. Rates of backbone amide exchange could be determined for 68 of its residues, which are spread along its sequence (Fig. 4a). No exchange rates could be determined for the remaining residues, mainly because these residues exchange relatively fast in flavodoxin.3 The parts of the molten globule for which exchange rates are determined cannot be random coil, because in that situation, exchange of the corresponding backbone amides would be completed during the 10-s dead time of the interrupted H/D exchange experiment (see “Experimental Procedures”), and thus no exchange curves of these residues would be obtained.
Chemical shift changes of the NMR resonances of unfolded apoflavodoxin upon decreasing GdnHCl concentration to 1.58 m show that structure formation in virtually all parts of the unfolded protein precedes folding to the molten globule state (43). The chemical shift data show that only residues Ile21–Val36 behave as a random coil at 1.58 m GdnHCl. However, the data of Fig. 4a show that in the absence of denaturant, also several of the residues that reside in the region comprising residues Ile21–Val36 of the molten globule are protected against exchange. Thus, the corresponding region of the flavodoxin molten globule cannot be random coil in absence of denaturant but is structured instead. Indeed, the H/D exchange data show that the molten globule of flavodoxin is almost entirely structured.
Residues Leu110–Val125 Form the Stable Core of the Off-pathway Molten Globule of Flavodoxin
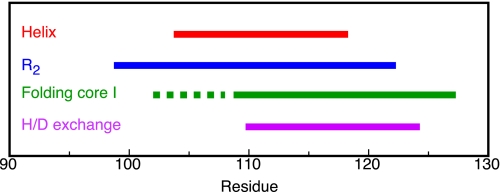
The wealth of information obtained until now for residues 99–127 of flavodoxin, including the results obtained in the study presented here, is summarized in Fig. 6. In unfolded flavodoxin residues Glu104–Lys118 transiently form an α-helix (44). In addition, restricted flexibility on the (sub)nanosecond time scale has been observed in unfolded flavodoxin for residues Gln99–Ala122 (44). Midpoints of denaturant-induced protein unfolding, as determined at the residue level by NMR spectroscopy, show that part of the ordered core of the off-pathway molten globule is formed by residues Ala109–Ser127 (43). The H/D exchange data presented here show that residues Leu110-Val125 of the molten globule have the highest protection factors against exchange. In addition, in a previous study, detection by NMR spectroscopy of native state H/D exchange in the presence of small amounts of a denaturant identified five unfolding clusters of residues within native apoflavodoxin (25). Four of these clusters unfold subglobally in a cooperative manner. The resulting conformations are partially unfolded forms (PUFs) of the protein. Both PUF1 and PUF2 are unfolding excursions that start from native apoflavodoxin but do not continue to the unfolded state and do not reside on the productive folding route. In contrast, both PUF3 and PUF4 probably are PUFs of the off-pathway folding intermediate (25). In PUF4, only the backbone amides of residues Leu110–Val125 are inaccessible to water (Fig. 5c).
FIGURE 6.
Overview of experimental data obtained for residues Gln99–Ser127 of the flavodoxin molten globule. Residues Glu104–Lys118 of unfolded flavodoxin in 3.4 m GdnHCl are colored red, because these residues transiently form an α-helix (44). Residues Gln99–Ala122 of unfolded flavodoxin in 3.4 m GdnHCl are colored blue, because transverse 1H-15N relaxation rates show that these residues have restricted flexibility on the (sub)nanosecond time scale (44). Residues Ala109–Ser127 are colored green, because they belong to the group of residues that have the largest midpoints of folding toward the molten globule (i.e. 2.64 ± 0.05 m GdnHCl) and belong to its ordered core (43). Unfortunately, no midpoints of unfolding could be determined for residues that reside in the dashed region. Residues Leu110–Val125 of the molten globule of Tyr44-flavodoxin are colored purple, because these residues have the highest protection factors against H/D exchange.
Comparison of b and c of Fig. 5 shows that the residues of the molten globule of flavodoxin that are protected most against H/D exchange coincide with those that are inaccessible to water in PUF4. Consequently, now conclusive proof is obtained that PUF4 indeed is an unfolding excursion of the flavodoxin off-pathway intermediate. The collection of data obtained implies that the residues that are protected against exchange in PUF4 are helical.
Three transiently formed helices, including the one formed by residues Glu104–Lys118, are equally populated for about 10% of the time in unfolded flavodoxin in 3.4 m GdnHCl (44). Consequently, in unfolded flavodoxin, these helices are equally stable against unfolding and thus are equally protected against H/D exchange. The H/D exchange data presented here show that one of the helices of the off-pathway molten globule (i.e. the region comprising residues Leu110–Val125; Figs. 4b and 5b) is better protected against exchange than the others. The enhanced protection against exchange suggests that this helix is better buried in the flavodoxin molten globule compared with the other helices mentioned. Hydrophobic interactions of this helix with the other ordered parts of the molten globule, although loose in nature, cause context-dependent stabilization of this helix against unfolding. Most likely, in the flavodoxin molten globule, the ordered regions detected in unfolded flavodoxin surround residues Leu110–Val125. Residues Leu110–Val125 thus form the stable core of the helical molten globule of the α-β parallel protein flavodoxin. Non-native docking of helices in the flavodoxin molten globule prevents formation of the parallel β-sheet of native flavodoxin. Hence, to produce native α-β parallel protein molecules, the off-pathway species needs to unfold.
Y. J. Bollen and C. P. M. van Mierlo, unpublished observations.
- H/D exchange
- hydrogen/deuterium exchange
- GdnHCl
- guanidine HCl
- HSQC
- heteronuclear single quantum coherence
- PUF
- partially unfolded form.
REFERENCES
- 1.Baldwin R. L. (1994) Nature 369, 183–184 [DOI] [PubMed] [Google Scholar]
- 2.Dinner A. R., Sali A., Smith L. J., Dobson C. M., Karplus M. (2000) Trends Biochem. Sci. 25, 331–339 [DOI] [PubMed] [Google Scholar]
- 3.Mayor U., Guydosh N. R., Johnson C. M., Grossmann J. G., Sato S., Jas G. S., Freund S. M., Alonso D. O., Daggett V., Fersht A. R. (2003) Nature 421, 863–867 [DOI] [PubMed] [Google Scholar]
- 4.Chiti F., Dobson C. M. (2006) Annu. Rev. Biochem. 75, 333–366 [DOI] [PubMed] [Google Scholar]
- 5.Ohgushi M., Wada A. (1983) FEBS Lett. 164, 21–24 [DOI] [PubMed] [Google Scholar]
- 6.Ptitsyn O. B. (1995) Adv. Protein Chem. 47, 83–229 [DOI] [PubMed] [Google Scholar]
- 7.Arai M., Kuwajima K. (2000) Adv. Protein Chem. 53, 209–282 [DOI] [PubMed] [Google Scholar]
- 8.Redfield C. (2004) Methods 34, 121–132 [DOI] [PubMed] [Google Scholar]
- 9.Jahn T. R., Radford S. E. (2008) Arch. Biochem. Biophys. 469, 100–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arai M., Kuwajima K. (1996) Fold Des. 1, 275–287 [DOI] [PubMed] [Google Scholar]
- 11.Forge V., Wijesinha R. T., Balbach J., Brew K., Robinson C. V., Redfield C., Dobson C. M. (1999) J. Mol. Biol. 288, 673–688 [DOI] [PubMed] [Google Scholar]
- 12.Arai M., Ito K., Inobe T., Nakao M., Maki K., Kamagata K., Kihara H., Amemiya Y., Kuwajima K. (2002) J. Mol. Biol. 321, 121–132 [DOI] [PubMed] [Google Scholar]
- 13.Hughson F. M., Wright P. E., Baldwin R. L. (1990) Science 249, 1544–1548 [DOI] [PubMed] [Google Scholar]
- 14.Jennings P. A., Wright P. E. (1993) Science 262, 892–896 [DOI] [PubMed] [Google Scholar]
- 15.Raschke T. M., Marqusee S. (1997) Nat. Struct. Biol. 4, 298–304 [DOI] [PubMed] [Google Scholar]
- 16.Kato H., Feng H., Bai Y. (2007) J. Mol. Biol. 365, 870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spence G. R., Capaldi A. P., Radford S. E. (2004) J. Mol. Biol. 341, 215–226 [DOI] [PubMed] [Google Scholar]
- 18.Bollen Y. J., Sánchez I. E., van Mierlo C. P. (2004) Biochemistry 43, 10475–10489 [DOI] [PubMed] [Google Scholar]
- 19.Dobson C. M. (2003) Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 20.Dill K. A., Chan H. S. (1997) Nat. Struct. Biol. 4, 10–19 [DOI] [PubMed] [Google Scholar]
- 21.Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. (1995) Proteins 21, 167–195 [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Recio J., Genzor C. G., Sancho J. (2001) Biochemistry 40, 15234–15245 [DOI] [PubMed] [Google Scholar]
- 23.Melo E. P., Chen L., Cabral J. M., Fojan P., Petersen S. B., Otzen D. E. (2003) Biochemistry 42, 7611–7617 [DOI] [PubMed] [Google Scholar]
- 24.Bollen Y. J., Nabuurs S. M., van Berkel W. J., van Mierlo C. P. (2005) J. Biol. Chem. 280, 7836–7844 [DOI] [PubMed] [Google Scholar]
- 25.Bollen Y. J., Kamphuis M. B., van Mierlo C. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4095–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollen Y. J., van Mierlo C. P. (2005) Biophys. Chem. 114, 181–189 [DOI] [PubMed] [Google Scholar]
- 27.Steensma E., van Mierlo C. P. (1998) J. Mol. Biol. 282, 653–666 [DOI] [PubMed] [Google Scholar]
- 28.Steensma E., Nijman M. J., Bollen Y. J., de Jager P. A., van den Berg W. A., van Dongen W. M., van Mierlo C. P. (1998) Protein Sci. 7, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Mierlo C. P., van den Oever J. M., Steensma E. (2000) Protein Sci. 9, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engel R., Westphal A. H., Huberts D. H., Nabuurs S. M., Lindhoud S., Visser A. J., van Mierlo C. P. (2008) J. Biol. Chem. 283, 27383–27394 [DOI] [PubMed] [Google Scholar]
- 31.Kathuria S. V., Day I. J., Wallace L. A., Matthews C. R. (2008) J. Mol. Biol. 382, 467–484 [DOI] [PubMed] [Google Scholar]
- 32.Otzen D. E., Giehm L., Baptista R. P., Kristensen S. R., Melo E. P., Pedersen S. (2007) Biochim. Biophys. Acta. 1774, 323–333 [DOI] [PubMed] [Google Scholar]
- 33.Lorenz T., Reinstein J. (2008) J. Mol. Biol. 381, 443–455 [DOI] [PubMed] [Google Scholar]
- 34.Nabuurs S. M., Westphal A. H., aan den Toorn M., Lindhoud S., van Mierlo C. P. (2009) J. Am. Chem. Soc. 131, 8290–8295 [DOI] [PubMed] [Google Scholar]
- 35.Engel M. F., Visser A. J., van Mierlo C. P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11316–11321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura C., Dyson H. J., Wright P. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4765–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steensma E., Heering H. A., Hagen W. R., Van Mierlo C. P. (1996) Eur. J. Biochem. 235, 167–172 [DOI] [PubMed] [Google Scholar]
- 38.van Mierlo C. P., van Dongen W. M., Vergeldt F., van Berkel W. J., Steensma E. (1998) Protein Sci. 7, 2331–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hvidt A., Nielsen S. O. (1966) Adv. Protein Chem. 21, 287–386 [DOI] [PubMed] [Google Scholar]
- 40.Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura C., Dyson H. J., Wright P. E. (2008) J. Mol. Biol. 378, 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamasaki K., Yamasaki T., Kanaya S., Oobatake M. (2003) Biopolymers 69, 176–188 [DOI] [PubMed] [Google Scholar]
- 43.Nabuurs S. M., Westphal A. H., van Mierlo C. P. (2009) J. Am. Chem. Soc. 131, 2739–2746 [DOI] [PubMed] [Google Scholar]
- 44.Nabuurs S. M., Westphal A. H., van Mierlo C. P. (2008) J. Am. Chem. Soc. 130, 16914–16920 [DOI] [PubMed] [Google Scholar]
- 45.Alagaratnam S., van Pouderoyen G., Pijning T., Dijkstra B. W., Cavazzini D., Rossi G. L., Van Dongen W. M., van Mierlo C. P., van Berkel W. J., Canters G. W. (2005) Protein Sci. 14, 2284–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]