Abstract
Despite widespread use of antibiotics, few studies have measured their effects on the burden or diversity of bacteria in the mammalian intestine. We developed an oral antibiotic treatment protocol and characterized its effects on murine intestinal bacterial communities and immune cell homeostasis. Antibiotic administration resulted in a ten-fold reduction in the amount of intestinal bacteria present and sequencing of 16S rDNA segments revealed significant temporal and spatial effects on luminal and mucosal-associated communities including reductions in luminal Firmicutes and mucosal-associated Lactobacillus species, and persistence of bacteria belonging to the Bacteroidetes and Proteobacteria phyla. Concurrently, antibiotic administration resulted in reduced RELMβ production, and reduced production of IFNγ and IL-17A by mucosal CD4+ T lymphocytes. This comprehensive temporal and spatial metagenomic analyses will provide a resource and framework to test the influence of bacterial communities in murine models of human disease.
Introduction
The human intestine is colonized by 100 trillion microorganisms belonging to each of the three domains of life1. Of these, bacteria are the most abundant—the colon is home to approximately 1011 to 1014 bacterial cells per mL of stool2 and has a diversity of at least 1000 species3. Through recent advances in surveying unculturable intestinal communities using 16S ribosomal DNA (16S rDNA) sequencing4 and microarray-based methods, it has become clear that the major bacterial phyla in the mammalian intestine include Bacteroidetes (Bacteroidetes class), Firmicutes (Clostridia class), Proteobacteria (Alpha, Beta, Gamma, and Epsilon classes), Actinobacteria, and Fusobacteria5, 6. Some intestinal bacteria are mutualists that promote normal mammalian physiology including proper digestion7, metabolism8, epithelial cell function9, angiogenesis10, enteric nerve function11, and immune system development12. While bacterial communities promote normal immune homeostasis, patients with inflammatory bowel disease (IBD)13 and atopic allergies14 have altered intestinal communities, indicating that microbial communities might influence the pathogenesis of these diseases15, 16.
Antibiotic treatment has the adverse effect of altering intestinal microbial communities17 and antibiotic exposure is linked to increased risk of Clostridium difficile colitis18 and asthma16 in humans. In keeping with these findings, antibiotic treatment in animal model systems has identified complex pro-inflammatory and immunoregulatory roles for intestinal communities in modulating intestinal cytokine responses19–21, in altering resistance to enteric pathogens19, 22–25, in maintaining mucosal homeostasis26, 27, and in controlling allergic inflammation28. However, while the use of oral broad-spectrum antibiotics in humans and animal models is common, knowledge of how antibiotics modify intestinal bacterial communities is limited.
Previous studies using culture and fluorescence in situ hybridization based methods have provided some insights into antibiotic effects on a limited subset of intestinal bacteria23, 29. More recently, molecular based examination of antibiotic treatment in humans showed effects on intestinal communities, but was confounded by inter-individual variability30. Animal models have proven well-suited for metagenomic studies because they allow for control of host genetic background, feeding practices, and antibiotic administration. In one such study of bacterial recovery after antibiotic perturbation, single-antibiotic effects were characterized at the phylum level at a single time point and anatomic location31. Other groups have reported the general effects of oral antibiotic treatment on intestinal bacteria, including significant changes to lactobacilli, enterococci/group D streptococci, Firmicutes, Bacteroidales, or segmented filamentous bacteria, depending on the specific antibiotic and treatment protocol22–25. However, the interpretation of these studies is limited as in-depth temporal and spatial analysis of the effects of antibiotic treatment on microbial communities was not undertaken.
Here, we characterized the effects of two different antibiotic treatment protocols on murine intestinal bacterial communities and immune cell homeostasis. We found that published protocols involving administration of multiple antibiotics (ampicillin, gentamicin, metronidazole, neomycin, and vancomycin) in drinking water altered intestinal microbiota, but were complicated by the development of severe dehydration in the host. For this reason, we developed an oral gavage-based protocol that facilitated controlled antibiotic dosing without the development of dehydration. Using DNA bar-coding and pyrosequencing of 16S rDNA gene segments, we examined temporal effects of antibiotics on bacterial communities and found significant reductions in the frequency of bacteria belonging to the Firmicutes phylum and persistence of the Bacteroidetes and Proteobacteria phyla over time. Additionally, we performed spatial analysis of luminal and mucosal-associated bacterial communities following antibiotic treatment and found significant effects in the cecum, proximal colon, and distal colon including reductions in mucosal-associated Lactobacillus species. Finally, we examined the effects of oral antibiotic treatment on mucosal immune cell homeostasis and found that antibiotic administration by gavage resulted in reduced production of RELMβ, as well as reduced IFNγ and IL-17A production by mucosal CD4+ T lymphocytes.
Results
Administration of antibiotics in drinking water alters bacterial communities but results in severe dehydration
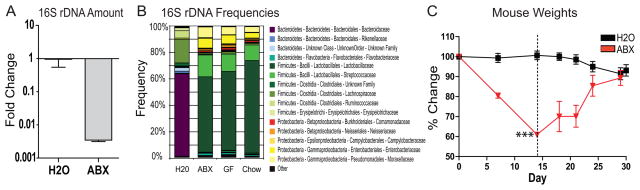
Antibiotic treatment has been used extensively to probe the effects of microbial signals in disease, including murine models of intestinal inflammation19, 20, 26, 27 and allergy28, 32. However, to date there has been limited analysis of how oral antibiotics specifically influence intestinal bacterial communities. We sought to characterize the effects of published antibiotic treatment protocols on intestinal bacterial communities through analysis of bacterial 16S rDNA compositions. Following published protocols, conventionally reared animals were given access to autoclaved water or autoclaved water containing ampicillin, gentamicin, metronidazole, neomycin, and vancomycin19, 20, 23, 26–28. Stool pellets were collected at day 10 post-treatment initiation and total DNA was extracted for quantitative analysis of 16S ribosomal DNA (16S rDNA) genes by real-time RT-PCR. We observed a greater than two log reduction in bacterial 16S rDNA after ten days of antibiotic treatment as compared to control-treated animals (Figure 1A).
Figure 1.
Antibiotic delivery in drinking water modulates intestinal communities but results in animal dehydration. A) 16S rDNA gene copies as quantified by real-time RT-PCR from stool pellets collected from naïve (H2O; N=5) or antibiotic-treated (ABX; N=5) animals (± s.e.m). B) Family level phylogenetic classification of 16S rDNA frequencies in stool pellets collected from naïve animals (H2O; N=5), antibiotic-treated animals (ABX; N=5), germ-free animals (GF; N=3), or autoclaved animal food (Chow; N=3). C) Weights of animals fed unsupplemented (H2O; N=4) or antibiotic supplemented (ABX; N=4) water (***, p<0.001; ± s.e.m). Antibiotic-treated animals were switched to unsupplemented water at day 14 (dashed line).
To examine the effects of antibiotics on the composition of bacterial communities in stool pellets, we used 454/Roche pyrosequencing of 16S rDNA segments and determined taxonomic assignments for each sequence using RDP Classifer. Antibiotic administration resulted in reduced frequencies of bacteria belonging to the Bacteroidetes phylum (Bacteroidaceae family) and Firmicutes phylum (Lachnospiraceae family). Antibiotic administration also resulted in increased frequencies of other members of the Firmicutes phylum (Leuconostocaceae and Streptococcaceae families) as well as members of the Proteobacteria phylum (Enterobacteriaceae and Moraxellaceae families) (Figure 1B, H2O vs. ABX).
We next investigated the origin of the bacterial 16S rDNA present in stool pellets after antibiotic treatment. To determine whether these sequences represented a living community that persisted after antibiotic treatment, we analyzed stool pellets from germ-free animals and found bacterial community sequences similar to those present in stool pellets from antibiotic-treated animals (Figure 1B). We hypothesized that antibiotic treatment reduced intestinal bacteria to levels at which residual DNA in autoclaved food became the major contributor of 16S rDNA sequences. To test this, DNA was extracted from sterile mouse chow and 16S communities were shown to be indistinguishable from those found in stool pellets from antibiotic-treated or germ-free animals (Figure 1B) indicating that stool pellets from these animals were contaminated with bacterial 16S rDNA normally present in autoclaved food.
We further sought to investigate the effects of antibiotics on intestinal immune homeostasis. Unfortunately, we found that animals fed antibiotics in drinking water rapidly showed signs of dehydration including weight loss, decreased skin turgor, and hard stools while animals fed autoclaved water showed no signs of dehydration (Figure 1C). Similar antibiotic treatment protocols without gentamicin or with added sweetener also resulted in dehydration (data not shown). These data reveal an adverse effect of antibiotic treatment that confounds the interpretation of the effects of antibiotics on immune cell homeostasis33.
Development of a new, broad-spectrum antibiotic treatment regimen that prevents animal dehydration
We developed an alternate protocol in which animals were orally gavaged with autoclaved water or autoclaved water containing ampicillin, gentamicin, metronidazole, neomycin, and vancomycin once daily for ten days. Daily weights and stool pellet samples were taken and animals were sacrificed on day 10 for histologic, microbiologic, and immunologic analyses. Animals treated with antibiotics by oral gavage maintained their body weight (Figure 2A) and displayed no signs of dehydration. Treated animals also developed loose stools (data not shown) as seen with antibiotic treatment in human patients34.
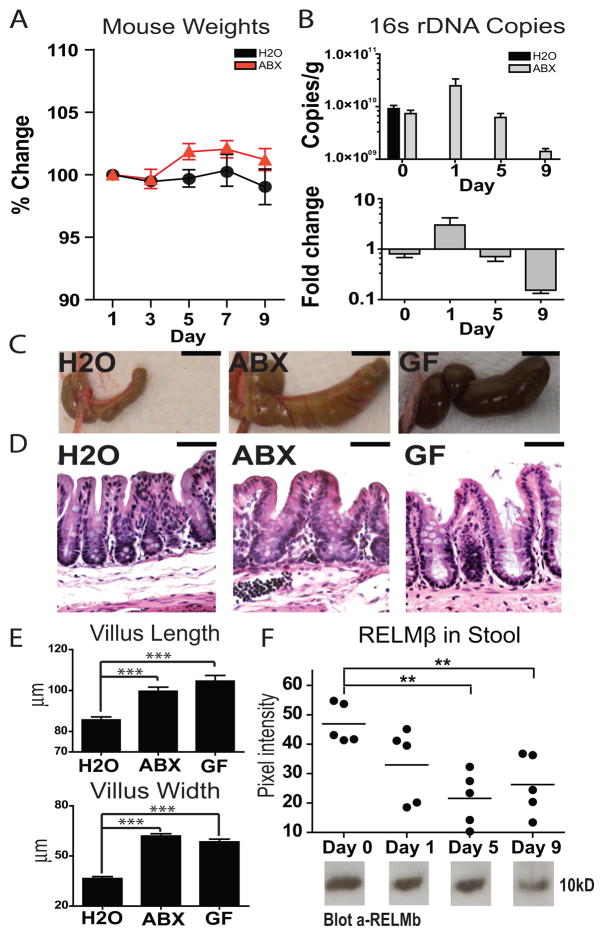
Figure 2.
Antibiotic administration by gavage mimics molecular, anatomic, histologic, and immunologic characteristics of reduced microbial stimulation without animal dehydration. A) Weights of animals gavaged with unsupplemented (H2O; N=4) or antibiotic-supplemented (ABX; N=5) water (± s.e.m). B) 16S rDNA gene copies as quantified from stool pellets collected before (day 0) or over the course of antibiotic treatment (days 1, 5, 9) (N=5; ± s.e.m). C) Cecal images from control-treated (H2O), day 10 antibiotic-treated (ABX), or germ-free animals (GF) (bar, 1 cm). D) Photomicrographs of H&E stained cecal sections from control-treated (H2O), day 10 antibiotic-treated (ABX), or germ-free animals (GF) showing expansion of the lamina propria and enterocyte hyperplasia in ceca from antibiotic-treated and germ-free animals (bar, 50 μm). E) Quantification of cecal villus length and width from control-treated (H2O; N=4), day 10 antibiotic-treated (ABX; N=5), or germ-free animals (GF; N=3) (***, p<0.001; ± s.e.m). F) Quantification of RELMβ protein in stool pellets by western blot over the course of antibiotic treatment (N=5) (**, p 0.01; ± s.e.m).
To examine the effects of this treatment regimen on intestinal bacteria, 16S rDNA copy number was quantified in stool pellet samples using real-time RT-PCR. We observed an initial increase in 16S rDNA copies in stool pellets from antibiotic treated animals at day one post treatment initiation suggesting a flushing of bacterial DNA into the stool, likely representing dead bacteria (Figure 2B, day 1). After nine days, we observed a ten-fold reduction in total 16S rDNA in stool pellets of treated animals (Figure 2B) consistent with a reduction in total bacterial load.
We next undertook gross anatomic and histologic analyses of antibiotic-treated animals. Treated animals displayed characteristic enlarged ceca, similar to those seen in germ-free animals (Figure 2C)35, as well as expansion of the lamina propria and enterocyte hyperplasia resulting in increased intestinal villus width and length (Figure 2D and E). Further, examination of RELMβ, a protein secreted from intestinal goblet cells in response to intestinal colonization36, showed a trend towards reduction at day one that reached statistical significance by day five (p=0.001) and day nine (p=0.005) post-treatment initiation (Figure 2F). Thus, antibiotic treatment by oral gavage resulted in bacterial depletion without dehydration, and this was accompanied by anatomic, histologic, and immunologic changes characteristic of reduced microbial stimulation.
Antibiotic treatment results in two phases of bacterial community restructuring
The temporal effects of antibiotic treatment on bacterial communities were examined using pyrosequencing of bacterial 16S rDNA gene segments. We analyzed 49,994 sequences from 144 samples representing stool pellet, luminal, and mucosal-associated bacterial communities originating from the cecum, proximal large intestine, and distal large intestine of nine animals (four naïve and five antibiotic-treated). To characterize the global effects of antibiotic treatment on the full intestinal microbiome, bacterial communities were compared in control-treated or antibiotic-treated animals by quantifying similarities based on phylogenetic distances using UniFrac as described previously37–39. UniFrac distances were calculated in two different ways, using only presence/absence information (unweighted) or taking into account the abundance of each bacterial lineage (weighted) and Principal Coordinate Analysis was used to cluster communities along orthogonal axes of maximal variance.
In unweighted UniFrac analysis, the first coordinate separated the stool pellet, luminal, and mucosal-associated samples on the basis of antibiotic treatment, and explained 26.8% of the variance (Figure 3A). Weighted UniFrac analysis also separated samples on the basis of antibiotic treatment, with 75.4% of the variance represented by the first principal coordinate (Figure 3B). These findings indicated that antibiotic treatment resulted in new luminal and mucosal-associated bacterial communities that were distinct from those seen in control-treated animals both in the proportions of the different groups, and in the types of bacteria present. Treated samples grouped tightly in our weighted analysis with respect to the second coordinate axis (Figure 3B, 14.1% of group variance) indicating that intestinal communities in treated animals were more similar than those existing prior to treatment.
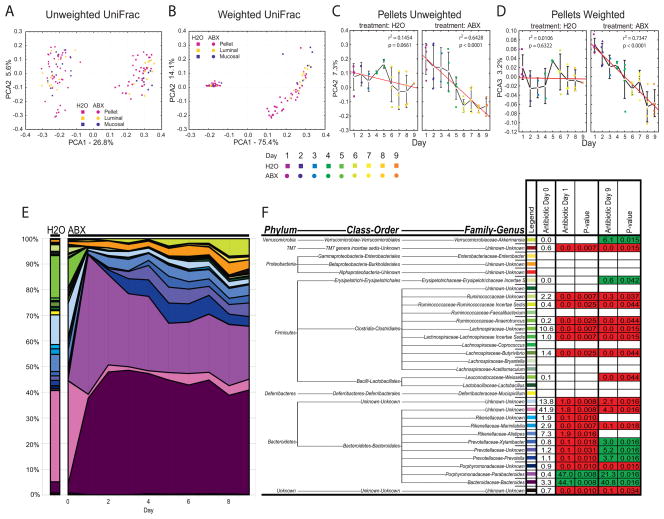
Figure 3.
Global restructuring of intestinal bacterial communities over time. A) Unweighted or B) weighted UniFrac analysis of stool pellet, luminal content, and mucosal-associated samples from control-treated or antibiotic-treated animals. C) Unweighted or D) weighted UniFrac analysis of stool pellet samples from control-treated (H2O; N=4) or antibiotic-treated (ABX; N=5) animals from day 1 to day 9 post treatment initiation. Colored symbols represent treatment day, white squares represent means, red line represents linear regression analysis (± s.e.m, significance determined by regression analysis). E) Genus level phylogenetic classification of 16S rDNA frequencies in stool pellets collected from control-treated (H2O; N=4) or antibiotic-treated (ABX; N=5) animals from day 0 to day 9. F) Average frequency of bacterial groups before (Antibiotic Day 0) or during (Antibiotic Day 1, 9) antibiotic treatment. Mann-Whitney P-values of changes in group frequency with antibiotic treatment. Frequency reductions upon antibiotic treatment in red, increases in green, non-significant changes blank.
To examine the temporal effects of antibiotic treatment analysis of stool pellets was utilized. Stool pellet analysis is both non-invasive and representative of luminal community composition5, allowing for longitudinal analyses. We found that samples from day zero and day one had distinct graphical locations in both unweighted and weighted UniFrac analyses (Figure 3A and B). This finding indicated an initial shift in community structure that occurred quickly after initiation of antibiotic treatment. Furthermore, unweighted and weighted UniFrac analyses revealed that the second and third coordinates, respectively, separated longitudinal pellet samples on the basis of treatment day (Figure 3C and D) indicating a second, more gradual change in community structure over time. This trend reached statistical significance in antibiotic-treated (p<0.0001, unweighted and weighted) but not control-treated animals. In summary, UniFrac analysis of bacterial communities from control-treated or antibiotic-treated animals revealed a dramatic shift in community structure that occurred quickly after antibiotic initiation, followed by a more gradual alteration in community structure over time.
Antibiotic treatment causes reductions in Firmicutes with the persistence of Bacteroidetes and Proteobacteria phyla over time
Phylogenetic placements allow the identification of bacterial taxa affected by antibiotic treatment. Temporal analysis of stool pellet samples revealed a significant shift in community composition on day one post-antibiotic treatment initiation (Figure 3E, F) that was not observed in control-treated animals (data not shown). The timing of this shift is consistent with the flushing of 16S rDNA into stool observed during quantitative analysis (Figure 2B, day 1) and the graphical separation between day zero and day one samples from treated animals in UniFrac analyses (Figure 3A, B). These findings indicate that even short antibiotic courses can result in dramatic alterations to intestinal bacterial communities. Temporal analysis of stool pellet samples over the course of treatment further revealed a gradual change in bacterial proportions over time (Figure 3E) that was consistent with the gradual alteration in community structure observed by UniFrac (Figure 3C, D).
After nine days of antibiotic treatment, there were several statistically significant differences between stool pellets from antibiotic-treated and control-treated animals. These included reductions in the Porphyromonadaceae family, Marinilabilia genus, Bacteroidales order, Bacteroidetes phylum, Weissella genus, Butyrivibrio genus, Lachnospiraceae Incertae Sedis genus, Lachnospiraceae family, Anaerotruncus genus, Ruminococcaceae Incertae Sedis genus, Ruminococcaceae family, and TM7 genera incertae sedis (p values listed in Figure 3F; groups significantly reduced shaded in red). Upon antibiotic treatment, there were increases in the frequency of bacteria belonging to the Bacteroides genus, Parabacteroides genus, Prevotella genus, Prevotellaceae family, Xylanibacter genus, Erysipelotrichaceae Incertae S, and Akkermansia genus (p values listed in Figure 3F; groups significantly increased shaded in green). We did not detect any statistically significant differences in bacterial frequencies over the course of treatment in stool pellets from control-treated animals (data not shown). In summary, we found that antibiotic treatment caused significant changes in stool pellet bacterial communities and that these changes occurred in at least two phases, one that occurred immediately after initiating treatment and one that progressed over the course of treatment.
Antibiotic treatment results in significant alterations to luminal and mucosal-associated bacterial communities along the length of the colon
Bacterial communities are distinct along the length of the intestinal tract5, and between luminal and mucosal sites40. Therefore, we analyzed the spatial effects of antibiotic treatment on luminal bacterial communities longitudinally along the colon (cecum, proximal colon, distal colon) (Figure 4A). In the lumen, antibiotic treatment was associated with lower frequencies of bacteria belonging to the Porphyromonadaceae family, Alistipes genus, Marinilabilia genus, Rikenellaceae family, Bacteroidales order, Bacteroidetes phylum, Mucispirillum genus, Butyrivibrio genus, Coprococcus genus, Lachnospiraceae Incertae Sedis, Lachnospiraceae family, Anaerotruncus genus, Faecalibacterium genus, Ruminococcaceae Incertae Sedis, Ruminococcaceae family and Clostridiales order (p values listed in Figure 4B; groups significantly reduced shaded in red). Antibiotic treatment also resulted in significant increases in the frequency of bacteria belonging to the Bacteroides genus, Parabacteroides genus, Prevotella genus, Xylanibacter genus, Burkholderiales order, Enterobacter genus, and Akkermansia genus (p values listed in Figure 4B; groups significantly increased shaded in green).
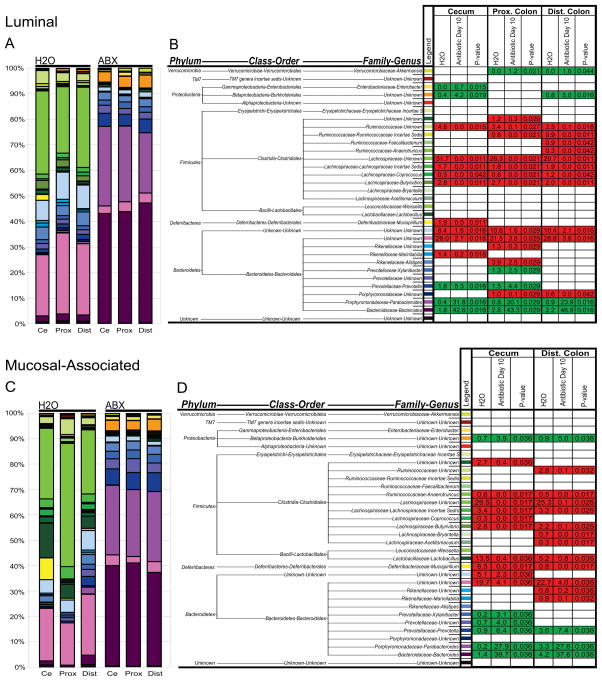
Figure 4.
Antibiotics modify luminal and mucosal-associated bacterial communities along the length of the colon. A) Genus level phylogenetic classification of 16S rDNA frequencies in luminal samples collected from control-treated (H2O; N=4) or day 10 antibiotic-treated (ABX; N=5) animals from the cecum (Ce), proximal colon (Prox), or distal colon (Dist). B) Average frequency of bacterial groups in samples from control-treated (H2O; N=4) or antibiotic-treated (Antibiotic Day 10; N=5) animals. C) Genus level phylogenetic classification of 16S rDNA frequencies in mucosal-associated samples collected from control-treated (H2O; N=4) or day 10 antibiotic-treated (ABX; N=5) animals from the cecum (Ce), proximal colon (Prox), or distal colon (Dist). D) Average frequency of bacterial groups in mucosal-associated samples from control-treated (H2O; N=4) or antibiotic-treated (Antibiotic Day 10; N=5) animals. Mann-Whitney P-values of changes in group frequency with antibiotic treatment. Frequency reductions upon antibiotic treatment in red, increases in green, non-significant changes blank.
Comparison of naïve mucosal-associated bacterial communities to naïve luminal communities revealed statistically higher frequencies of bacteria belonging to the Mucispirillum genus (p=0.012) and Lactobacillus genus (p=0.001), and lower frequencies of the Alistipes genus (p=0.017) (Figure 4C vs. 4A). Upon antibiotic treatment, there were significant reductions in the frequency of mucosal-associated bacteria belonging to the Marinilabilia genus, Rikenellaceae family, Bacteroidales order, Bacteroidetes phylum, Mucispirillum genus, Lactobacillus genus, Acetitomaculum genus, Bryantella genus, Butyrivibrio genus, Coprococcus genus, Lachnospiraceae Incertae Sedis, Lachnospiraceae family, Anaerotruncus genus, Ruminococcaceae family, and the Clostridiales order as compared to samples from control-treated animals (p values listed in Figure 4D; groups significantly reduced shaded in red). Upon antibiotic treatment, there were also significant increases in the frequency of mucosal-associated bacteria belonging to the Bacteroides genus, Parabacteroides genus, Prevotella genus, Prevotellaceae family, Xylanibacter genus, and the Burkholderiales order (p values listed in Figure 4D; groups significantly increased shaded in green). We were unable to amplify enough sequences from proximal colon mucosal-associated samples for statistical analysis. In summary, phylogenetic analyses revealed significant changes to luminal and mucosal-associated bacterial communities following antibiotic treatment.
Antibiotic treatment results in altered cytokine production by mucosal CD4+ T lymphocytes
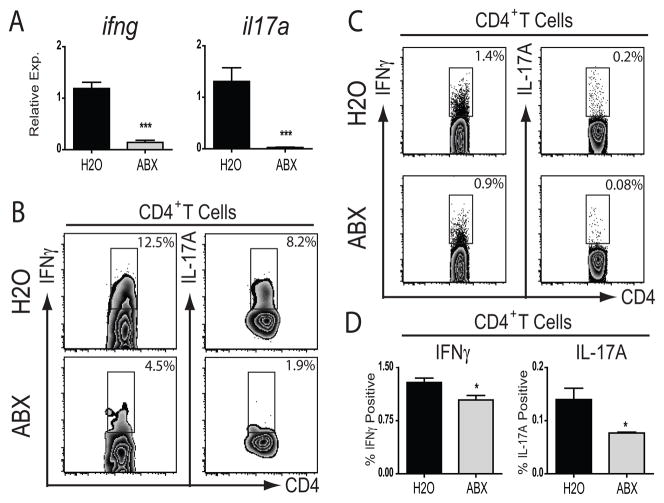
Microbial signals are known to influence many facets of immune function19–21, 48–50. We therefore sought to examine whether administration of antibiotics influenced intestinal immune cell homeostasis. Animals were treated with antibiotics by gavage for ten days, total RNA was isolated from sections of small intestine, and cytokine transcripts were quantified using real-time RT-PCR. Transcript levels of ifng and il17a were significantly reduced in the small intestine of antibiotic-treated animals as compared to controls (Figure 5A). Flow cytometric analysis revealed a reduced frequency of CD4+ T lymphocyte expression of IFNγ or IL-17A in the small intestinal lamina propria (Figure 5B). In addition, in the mesenteric lymph nodes which drain the small and large intestine, the frequency of CD4+ T lymphocytes that expressed IFNγ or IL-17A was significantly lower following antibiotic treatment (Figure 5C, D). Collectively, these data support a role for microbial signals in the maintenance of normal intestinal effector T lymphocyte populations.
Figure 5.
Antibiotic treatment alters mucosal CD4+ T lymphocyte homeostasis. A) Cytokine mRNA expression on day 10 as assessed by real-time RT-PCR of small intestine samples from control-treated (H2O; N=4) or antibiotic-treated (ABX; N=5) animals (***, p<0.001; ± s.e.m). B) Expression of IFNγ or IL-17A by CD4+ T lymphocytes in the small intestinal lamina propria of control-treated (H2O) or day 10 antibiotic-treated (ABX) animals as analyzed by flow cytometry. C) Expression of IFNγ or IL-17A by CD4+ T lymphocytes in the mesenteric lymph nodes of control-treated (H2O) or day 10 antibiotic-treated (ABX) animals analyzed by flow cytometry. D) Statistical analysis of mesenteric lymph node CD4+ T lymphocyte cytokine expression from control-treated (H2O; N=4) or antibiotic-treated (ABX; N=5) animals (*, p<0.05; ± s.e.m).
Discussion
We have developed an antibiotic treatment regimen that significantly alters intestinal bacterial communities and immune cell homeostasis without animal dehydration. We employed deep sequencing to quantify temporal and spatial antibiotic effects on intestinal bacterial communities and show significant changes in several bacterial groups that correlate with altered cytokine production by CD4+ T lymphocytes in intestinal-associated lymphoid tissues.
We initially treated animals with antibiotics in drinking water in a similar manner to previously published protocols19, 20, 23, 26–28. This treatment protocol reduced intestinal bacteria to the extent that DNA from food composed the majority of recovered 16S rDNA sequences. However, this protocol was associated with animal dehydration that may complicate the interpretation of subsequent immunologic studies33. These findings also highlight the presence of sterile microbial-derived signals and emphasize the importance of controlling for DNA contamination from food in future immunologic and metagenomic studies. As an alternative, we chose acute administration of broad-spectrum antibiotics by gavage, while allowing animal access to untreated drinking water; more closely mimicking antibiotic administration in humans. Animals treated by gavage did not show signs of dehydration and developed loose stools similar to known consequences of antibiotic treatment in patients34. Antibiotic treatment by oral gavage reduced absolute bacterial numbers and mimicked anatomic, histologic, and immunologic characteristics of reduced bacterial stimulation.
We carried out a temporal and spatial analysis of antibiotic effects on bacterial communities using pyrosequencing of bacterial 16S rDNA gene segments. UniFrac analysis revealed that samples from antibiotic treated animals grouped separately from, and clustered more tightly than, samples from control-treated animals indicating that antibiotic treatment promoted a novel community structure that was more similar between antibiotic-treated as compared to control-treated animals. Day one stool pellet samples from treated animals grouped closely with later time points consistent with an initial, fast phase of community alteration. This finding was emphasized in temporal analysis of stool pellet bacterial frequencies during treatment and suggests that even short exposure to oral antibiotics creates distinct intestinal bacterial communities. UniFrac and frequency analyses of stool pellet communities over the course of antibiotic treatment further revealed progressive alterations in community structure, indicating that intestinal bacterial communities are continuously modified over the course of antibiotic treatment.
The influence of antibiotic treatment by oral-gavage on stool bacterial communities was noticeably different from that observed during antibiotic administration in animal drinking water. For example, sequences from the Bacteroidetes group represented a relatively small proportion of sequences recovered from mice treated continuously with antibiotics while a higher proportion of Bacteroidetes sequences were recovered from animals treated with antibiotics by gavage. These differences likely arise due to differences in the duration of treatment (four weeks vs. ten days), in the method of antibiotic administration (continuous vs. gavage), in animal hydration state (physiologic vs. non-physiologic), and in the relative contribution that 16S rDNA from food makes to measured bacterial frequencies (higher with more efficient microbial depletion). This finding highlights the importance of controlling for 16S rDNA in food in future metagenomic studies.
Previous studies have shown that antibiotic-treated animals develop allergic responses28 and antibiotic use in children is associated with increased risk of developing asthma later in life16. In addition, early colonization with Bacteroides fragilis has been implicated as a risk factor for asthma development in humans41. We therefore examined the effects of antibiotic treatment on members of the Bacteroidies phylum. We found that members of the Bacteroidies phylum, namely the Bacteroidales order, represented 60–70% of intestinal bacteria in control-treated animals. After one day, these bacteria represented a combined frequency of greater than 95% in samples from antibiotic-treated but not control-treated animals, and subsequently stabilized at a frequency of approximately 90% in all sampled compartments. Outgrowths of the Bacteroidies phylum were not observed in previous metagenomic studies that examined recovery of intestinal microbiota after antibiotic exposure31, likely representing differences in antibiotic type and administration and highlighting the utility of temporal analyses in metagenomic studies. It is intriguing to speculate that rapid increases in Bacteroidies frequency after antibiotic initiation may be responsible for associations between early antibiotic exposure in humans and increased susceptibility to allergic inflammation.
We also detected significant reductions in the frequency of bacteria belonging to the Firmicute phylum in samples from antibiotic-treated as compared to control-treated animals. This bacterial phylum has been shown to be present in an increased frequency in obese patients as compared to healthy individuals42 and may provide the host an enhanced ability to extract energy from otherwise indigestible dietary polysaccharides43. As such, antibiotic treatment may result in a reduced capacity for energy harvest by the host which has implications for prolonged antibiotic treatment in patients. In addition, these findings highlight a possible role for antibiotics as a means of modulating intestinal bacterial communities in future treatments for obesity42.
Consistent with previous studies, we observed increased frequencies of Enterobacteriaceae, as well as other members of the Proteobacteria phyla, in samples from antibiotic-treated as compared to control-treated animals31. The Enterobacteriaceae include a number of nosocomial pathogens, with considerable antibiotic resistance, including Escherichia, Enterobacter, and Salmonella. Given that DNA transfer between intestinal bacteria plays a role in shaping bacterial communities3, 44 and in the development of pathogenic, antibiotic-resistant organisms45, Enterobacteriaceae that survive in an antibiotic exposed intestine may act as a clinically relevant reservoir that could seed subsequent infections with antibiotic resistant bacteria. Outgrowth of Clostridium difficile, a common cause of antibiotic-associated colitis that is thought to colonize the intestine when indigenous microbiota are disrupted by antibiotic use18, was not observed as a result of our antibiotic treatment protocol. However, this result is not unexpected as metronidazole and vancomycin, two antibiotics commonly used to treat C. difficile colitis in humans, were included in the antibiotic cocktails used in this study.
We extended our analysis to spatial examinations of antibiotic effects along the length of the intestine, and between luminal and mucosal-associated bacterial communities. We found that luminal and stool pellet bacterial communities from naïve animals were similar, suggesting that examination of stool pellets is an adequate, rough approximation of luminal communities5. However, we did find that naïve animals displayed significantly higher frequencies of Lactobacillus species in mucosal-associated as compared to luminal communities, suggesting that distinct bacterial communities at different anatomical locations may be relevant for mammalian physiology.
Upon antibiotic treatment, there were significant changes to both luminal and mucosal-associated bacterial communities including increases in the Bacteroidies and Proteobacter phyla, and reductions in the Firmicute phylum. In particular, significant reductions in the frequency of mucosal-associated Lactobacillus species were observed in antibiotic-treated as compared to control-treated animals. Lactobacillus species have been used as “probiotic” bacteria with mixed effectiveness in treating human diseases46, 47. It may therefore be useful to examine whether reductions in this potentially beneficial mucosal-associated bacterial group are responsible for changes in immune homeostasis observed following antibiotic treatment.
Finally, we examined the effects of acute antibiotic treatment on mucosal immune cell homeostasis. We found that production of IFNγ and IL-17A by mucosal CD4+ T lymphocytes was reduced in antibiotic-treated as compared to control-treated animals. We have previously shown that intestinal microbes negatively regulate IL-17A in the large intestine as germ-free and antibiotic-treated animals display increased Th17 cell populations20. Other reports indicate that germ-free and antibiotic treated mice display lower frequencies of Th17 cells in the lamina propria of the small intestine as compared to conventionally reared animals48–50 indicating that Th17 cell differentiation by microbial signals may be dependent on anatomic location. Other potential reasons for reported differences in Th17 cell responses in germ-free animals include diet, non-live microbial exposure, indigenous viral stimulation, or other facility-specific environmental signals. We now extend these observations by showing that antibiotic treatment reduces mucosal IFNγ and IL-17A production by CD4+ T lymphocytes in both the lamina propria of the small intestine and the mesenteric lymph nodes. This observation may provide mechanistic insights into the known susceptibility of animals treated with short courses of antibiotics to enteric bacterial infections19, 22–25.
In summary, we describe the development and characterization of a new antibiotic treatment protocol that modifies intestinal microbiota without animal dehydration. We utilize deep sequencing to show that antibiotic treatment causes significant temporal and spatial alterations in bacterial groups that have been implicated to have causative or therapeutic roles in human diseases. We further find that antibiotic treatment causes significant alterations in the expression of proinflammatory cytokines in gut-associated lymphoid tissues. It is hoped that these findings will provide a resource and framework for analysis and manipulation of intestinal microbial communities in murine models of human infection and disease.
Methods
Animals
Conventionally reared, six-eight week old female C57BL/6 mice were obtained from Charles River Laboratory and maintained in a specific pathogen–free facility. Animals were housed by litter and fed autoclaved LabDiet 5010 mouse chow (LabDiet, Richmond, IN) and autoclaved water. Germ-free animals were maintained in plastic isolator units and fed autoclaved LabDiet 5021 mouse chow (LabDiet, Richmond, IN) and autoclaved water. Isolators were checked weekly and consistently cultured negative for microbial contaminants. All experiments were approved by and performed following the guidelines of the University of Pennsylvania Institutional Animal Care and Use committee.
Antibiotic treatment
For continuous antibiotic treatment animals were provided with autoclaved drinking water, or autoclaved drinking water supplemented with ampicillin (1 mg/mL), gentamicin (1 mg/mL), metronidazole (1 mg/mL), neomycin (1 mg/mL), and vancomycin (0.5 mg/mL). For antibiotic treatment by oral gavage, animals had access to autoclaved food and water and were subjected to oral gavage daily for ten days with 200 μL of autoclaved water or autoclaved water supplemented with ampicillin (1 mg/mL), gentamicin (1 mg/mL), metronidazole (1 mg/mL), neomycin (1 mg/mL), and vancomycin (0.5 mg/mL).
16S rDNA sample acquisition and quantification of 16S rDNA
Stool pellet, luminal content, and washed mucosal-associated tissue samples were collected and total DNA was extracted using the QIAampR DNA Stool Mini Kit (stool pellet/luminal samples)(Qiagen, Inc., Valencia, CA) or the DNeasy Blood and Tissue Kit (tissue samples)(Qiagen, Inc., Valencia, CA).
Quantification of 16S rDNA was performed by real-time RT-PCR using degenerate bacterial 16S rDNA specific primers (5′ AGAGTTTGATCCTGGCTCAG-3′; forward), (5′-CTGCTGCCTYCCGTA-3′; reverse), (5′-FAM-TA+ACA+CATG+CA+AGTC+GA-BHQ1-3′; probe; + precedes the position of LNA base).
DNA manipulations
16S rRNA gene fragments were obtained as previously described53. Samples were amplified using 8 nucleotide-barcoded primer pairs (BSF8; 5′-AGAGTTTGATCCTGGCTCAG-3′), (BSR357; 5′-CTGCTGCCTYCCGTA-3′). 50 μL PCR reactions were carried out using the AmpliTaq System (Applied Biosystems, Inc., Foster City, CA) (Buffer II, 2 mM MgCl2, 200 μM each dNTP, 10 μM each primer, 0.1 mg/mL BSA, 100 ng (stool) or 500 ng (tissue) of template, 2.5 units polymerase). Cycle parameters: 1 minute 95° C, 20 cycles (stool pellet and luminal samples) or 25 cycles (tissue samples) of 30 secs at 95° C, 30 secs at 56° C, and 90 secs at 72° C, final extension at 72° C for 8 minutes. PCR products were gel purified using the QIAquick® Gel extraction kit (Qiagen, Inc., Valencia, CA). 100 ng of each amplicon was pooled and subjected to pyrosequencing.
Bioinformatic analysis
Sequence quality was accessed and samples with >100 sequences were carried through to subsequent anayses. Sequences were inserted into the 16S rRNA gene tree54 using parsimony insertion implemented in ARB. The tree constructed using ARB was used to carry out UniFrac analysis to compare global communty structure as described previously37–39. Taxonomic assignments for each sequence were obtained using RDP Classifer55.
Histology, stool protein extraction, and western blots
At necropsy, cecal tissue sections were removed and fixed in 4% paraformaldehyde (PFA) and paraffin-embedded. 5 μm sections were cut and stained with hematoxylin and eosin (H&E). Crypt morphology was quantified using “NIS Elements BR” imaging software (Nikon, Melville, NY).
Fecal protein isolation was performed as previously described51. Samples were equalized by protein content, analyzed by SDS-PAGE, and immunoblotted for RELMβ using a polyclonal rabbit anti-murine RELMβ antibody (PeproTech, Rocky Hill, NJ). Blots were visualized using ECL™ (Amersham, Piscataway, NJ) and band intensity was quantified using “UN-SCAN-IT” (Silk Scientific, Inc., Orem, UT).
Tissue isolation, RNA isolation, and real-time RT-PCR
At necropsy, 1 cm distal small intestine tissue sections were isolated and stored at 4° C in RNAlater™ (Qiagen, Inc., Valencia, CA). Tissue RNA was isolated by disruption (TissueLyzer; Qiagen, Inc., Valencia, CA) and TRIzol extraction. RNA was reverse transcribed into cDNA using Superscript Reverse Transcriptase (Invitrogen, Carlsbad, CA) and real-time RT-PCR was performed on cDNA samples using primers for ifng (Qiagen, Inc., Valencia, CA) or il17a using SYBR Green chemistry (Applied Biosystems, Inc., Foster, CA) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc., Foster, CA). The primer set for il17a (5′-TCCAGAAGGCCCTCAGACTA-3′; forward), (5′-TTCATTGCGGTGGAGAGTC-3′; reverse). Samples were normalized to naïve controls.
Cell isolation, stimulation, and flow cytometry
At necropsy, mLNs were harvested and single-cell suspensions were prepared. Lamina propria lymphocytes were isolated as previously described 52. Cells were stimulated by incubation for 4 h with 50 ng/mL PMA (Sigma-Aldrich) and 750 ng/mL ionomycin (Sigma-Aldrich, St. Louis, MI) in the presence of 10 μg/mL Brefeldin A (Sigma-Aldrich, St. Louis, MI), surface stained with fluorochrome-conjugated antibodies against CD4 and CD3, and fixed in 2% paraformaldehyde. Fixed cells were permeabilized with 0.5% saponin (Sigma-Aldrich, St. Louis, MI), stained intracellularly for IL-17A and IFNγ (eBioscience, Inc., San Diego, CA), acquired on a FACSCanto using FACSDiva software (BD Biosciences, Franklin Lakes, NJ), and analyzed with FlowJo software (Version 8.5; Tree Star, Inc., Ashland, OR).
Statistics
Cytokine and FACS analysis: Results represent the mean ± s.e.m. Statistical significance was determined by Student’s t test using Prism 4.0 (GraphPad Software). UniFrac analysis: Regression analysis was performed on day 0–9 stool pellet samples from naïve or antibiotic-treated animals. Pyrosequencing analysis: The frequencies of bacterial group compared using a Mann-Whitney test. Each mouse was treated as a biological replicate.
Acknowledgments
We thank J. Bisgaier for critical reading of this manuscript and members of the Artis and Bushman laboratories for helpful discussions. Research in the Artis lab is supported by the National Institutes of Health (NIH), AI61570 (DA), AI074878 (DA), T32 AI060516 Training in Bacteriology (DAH), T32 AI05528 Training in Immune System Development and Regulation (MCA), the Burroughs Wellcome Fund (Investigator in Pathogenesis of Infectious Disease Award to DA), the Crohn’s and Colitis Foundation of America’s William and Shelby Modell Family Foundation Research Award (DA), pilot grants from the University of Pennsylvania (University Research Foundation Award (DA), Veterinary Center for Infectious Diseases Pilot Grant (DA)), a National Institute of Diabetes and Digestive Kidney Diseases center grant (DK50306), and a NIH instrument grant (S10RR024525). This work was supported in part by the Penn Genome Frontiers Institute and by a grant from the Pennsylvania Department of Health (FDB, DA). The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Acronyms/Abbreviations
- Chow
mouse food
- ABX
antibiotic
- H2O
water
- RELMβ
Resistin Like Molecule Beta
- 16S rDNA
16S ribosomal DNA
References
- 1.Eckburg PB, Lepp PW, Relman DA. Archaea and their potential role in human disease. Infect Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Pace NR, Olsen GJ, Woese CR. Ribosomal RNA phylogeny and the primary lines of evolutionary descent. Cell. 1986;45:325–326. doi: 10.1016/0092-8674(86)90315-6. [DOI] [PubMed] [Google Scholar]
- 5.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 10.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husebye E, Hellstrom PM, Midtvedt T. Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex. Dig Dis Sci. 1994;39:946–956. doi: 10.1007/BF02087542. [DOI] [PubMed] [Google Scholar]
- 12.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 13.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 15.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 16.Marra F, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009;123:1003–1010. doi: 10.1542/peds.2008-1146. [DOI] [PubMed] [Google Scholar]
- 17.Hoban DJ. Antibiotics and collateral damage. Clin Cornerstone Suppl. 2003;3:S12–20. doi: 10.1016/s1098-3597(03)90025-1. [DOI] [PubMed] [Google Scholar]
- 18.De La Cochetiere MF, et al. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol. 2008;56:395–402. doi: 10.1007/s00248-007-9356-5. [DOI] [PubMed] [Google Scholar]
- 19.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaph C, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 22.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garner CD, et al. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SS, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 29.MILLER CP, BOHNHOFF M. Changes in the Mouse’s Enteric Microflora Associated with Enhanced Susceptibility to Salmonella Infection Following Streptomycin Treatment. J Infect Dis. 1963;113:59–66. doi: 10.1093/infdis/113.1.59. [DOI] [PubMed] [Google Scholar]
- 30.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guseinov TS, Guseinova ST. Effect of dehydration on morphogenesis of the lymphatic network and immune structures in the small intestine. Bull Exp Biol Med. 2008;145:755–757. doi: 10.1007/s10517-008-0187-2. [DOI] [PubMed] [Google Scholar]
- 34.Rohde CL, Bartolini V, Jones N. The use of probiotics in the prevention and treatment of antibiotic-associated diarrhea with special interest in Clostridium difficile-associated diarrhea. Nutr Clin Pract. 2009;24:33–40. doi: 10.1177/0884533608329297. [DOI] [PubMed] [Google Scholar]
- 35.Thompson GR, Trexler PC. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut. 1971;12:230–235. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ML, et al. Regulation of RELM/FIZZ isoform expression by Cdx2 in response to innate and adaptive immune stimulation in the intestine. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1074–83. doi: 10.1152/ajpgi.00442.2004. [DOI] [PubMed] [Google Scholar]
- 37.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vael C, Nelen V, Verhulst SL, Goossens H, Desager KN. Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm Med. 2008;8:19. doi: 10.1186/1471-2466-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 44.Grasselli E, et al. Evidence of horizontal gene transfer between human and animal commensal Escherichia coli strains identified by microarray. FEMS Immunol Med Microbiol. 2008;53:351–358. doi: 10.1111/j.1574-695X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 45.Karami N, et al. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J Antimicrob Chemother. 2007;60:1142–1145. doi: 10.1093/jac/dkm327. [DOI] [PubMed] [Google Scholar]
- 46.Betsi GI, Papadavid E, Falagas ME. Probiotics for the treatment or prevention of atopic dermatitis: a review of the evidence from randomized controlled trials. Am J Clin Dermatol. 2008;9:93–103. doi: 10.2165/00128071-200809020-00002. [DOI] [PubMed] [Google Scholar]
- 47.Kozuch PL, Hanauer SB. Treatment of inflammatory bowel disease: a review of medical therapy. World J Gastroenterol. 2008;14:354–377. doi: 10.3748/wjg.14.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 50.Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 51.Artis D, et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci U S A. 2004;101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 53.McKenna P, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]