Abstract
Major histocompatibility complex (MHC) genetics dictate adaptive cellular immune responses, making robust MHC genotyping methods essential for studies of infectious disease, vaccine development, and transplantation. Nonhuman primates provide essential preclinical models for these areas of biomedical research. Unfortunately, given the unparalleled complexity of macaque MHCs, existing methodologies are inadequate for MHC typing of these critical animal models. Here, we demonstrate pyrosequencing of cDNA-PCR amplicons as a general approach to determine comprehensive MHC class I genotypes in nonhuman primates. More than 500 unique MHC class I sequences were resolved by sequence-based typing of 92 rhesus, cynomolgus, and pig-tailed macaques. We identified an average of 22 distinct MHC class I cDNA sequences in each macaque, nearly half of which have not been reported previously. The remarkable sensitivity of this approach in macaques demonstrates that pyrosequencing is viable for ultra-high throughput MHC genotyping of primates, including humans.
INTRODUCTION
Major histocompatibility complex (MHC) gene products determine the repertoire of T-cell responses that an individual can generate against pathogens and foreign tissues1,2. The genes encoding MHC class I sequences are among the most polymorphic in vertebrate genomes3. Therefore, comprehensive MHC genotyping methods are an important foundation for the study of T-cell responses.
Rhesus (Macaca mulatta), cynomolgus (M. fascicularis), and pig-tailed (M. nemestrina) macaque monkeys provide essential preclinical models for infectious disease, vaccine, biodefense, and transplantation research4–9. Unfortunately, the utility of macaque models for immunological research has been hindered by the unprecedented complexity of their MHC. While human leukocyte antigen (HLA) haplotypes contain only three classical class I genes (HLA-A, -B, and -C), macaque class I loci have undergone a complex series of segmental duplications such that gene content varies between macaque MHC haplotypes10. Genomic sequencing of the MHC region suggests that rhesus and cynomolgus macaques have at least 22 functional class I genes transcribed at varying levels11–14. Furthermore, MHC class I allelic polymorphisms are largely species-specific, with geographically isolated subpopulations of the same species rarely sharing MHC class I sequences15–19. More than 900 macaque MHC class I sequences are currently known, but many more remain to be characterized. Robust genotyping assays are available for less than 5% of these sequences20.
The development of an ultra-high throughput platform for comprehensive MHC class I genotyping of macaques is urgently needed to maximize the utility of these animals as research models. Here we describe the adaptation of massively parallel pyrosequencing of cDNA-PCR amplicons for MHC genotyping of rhesus, cynomolgus, and pig-tailed macaques. This technology reveals that the number of MHC class I transcripts in each macaque is higher than previously recognized, underscores the number of novel MHC class I sequences yet to be characterized, and provides a feasible approach for complete MHC class I genotyping of all macaques used in biomedical research.
RESULTS
Macaque MHC genotyping by pyrosequencing
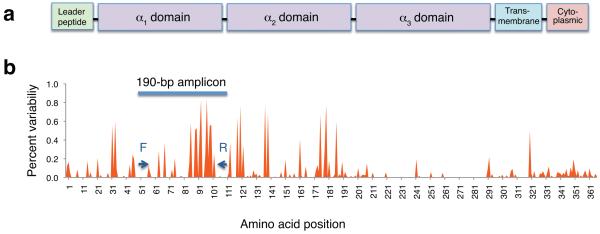
We designed a universal 190 base pair (bp) cDNA-PCR amplicon with primers based on highly conserved sequences within macaque MHC class IA and IB loci (Fig. 1). This amplicon spans the first of two highly polymorphic peptide binding domains encoded by class I loci1. Diagnostic polymorphisms within this amplicon allow for unambiguous resolution of 175 of 418 (42%) rhesus macaque class I sequences currently available in the Immuno Polymorphism Database21. The vast majority of MHC sequences that cannot be uniquely resolved are closely related variants that can be assigned to distinct class I lineages.
Figure 1. Polymorphic variation of known Mamu class I gene products.
(a) Domain structure of macaque class I genes. Exon two corresponds to the α1 domain. (b) Distribution of amino acid variability for Mamu class I gene products. We aligned predicted amino acid sequences of 418 previously described Mamu-A and Mamu-B alleles and plotted the frequency of differences from consensus for each amino acid residue. Arrows indicate locations of the PCR primers used in this study in highly conserved domains flanking the peptide-binding domain encoded by exon two.
We performed pyrosequencing of amplicons from 48 cynomolgus, pig-tailed, Indian-origin and Chinese-origin rhesus macaques in a single pilot run on a Genome Sequencer FLX (GS FLX) instrument. We subdivided these amplicons into four pools, each containing products from 12 animals that were distinguished by 10 bp Multiplex Identifier (MID) tags, molecular barcodes incorporated during the primary PCR (Supplementary Note online). We acquired nearly 500,000 high quality sequence reads containing a total of just over 100 million high quality bases. These data translated into an average of 9,315 reads per animal (range = 7,538–10,769 reads) for the Indian rhesus macaque amplicon pool.
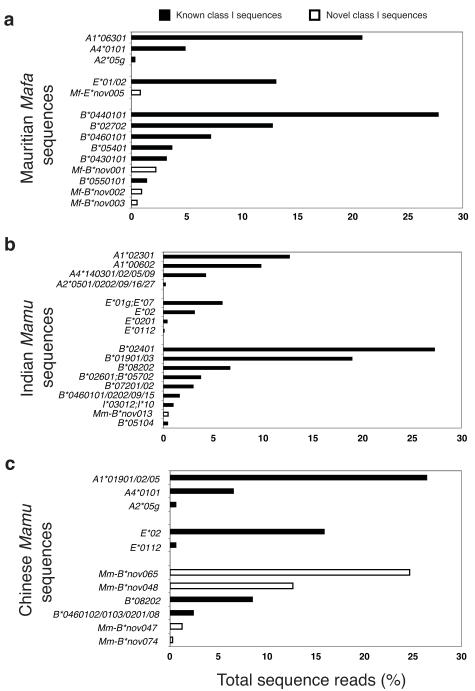
In order to evaluate the detection of known macaque class I alleles and test the sensitivity of the GS FLX pyrosequencing approach, we first examined four Mauritian cynomolgus macaques that are homozygous for well-characterized MHC haplotypes22. This geographically isolated population has extremely limited MHC diversity due to its recent expansion from a small founder population. We observed all Mafa-A and Mafa-B sequences previously described for the most frequent Mauritian M1 haplotype, with transcript levels ranging from 27.8% of total class I sequence reads for Mafa-B*0440101 down to 1.4% for Mafa-B*0550101 (Fig. 2a). In addition, we detected five novel sequences not previously observed by cloning and Sanger sequencing (transcript levels between 0.3–2.2% of total sequence reads). We obtained comparable results for the remaining three MHC homozygous Mauritian cynomolgus macaques, as well as eight heterozygous animals (Supplementary Figs. 1 and 2 online). Each of the Mauritian MHC haplotypes carries an average of seven transcribed Mafa-B sequences plus two or three classical Mafa-A and nonclassical Mafa-E class I sequences.
Figure 2. MHC class I transcript abundance profiles.
The frequency of each class I sequence is indicated as a percentage of the total MHC class I sequence reads that we evaluated for each animal. Open bars indicate MHC class I sequences that have not been described previously. Group specific designations such as Mafa-A2*05g indicate the large Mafa-A2*05-like family of sequences which differ by a few nucleotide substitutions outside exon two. (a) Mauritian cynomolgus macaque that is homozygous for the M1 haplotype22. (b) Indian rhesus macaque that is homozygous for the B24 haplotype17. (c) Chinese rhesus macaque that is homozygous for a novel Mamu-B haplotype and expresses several abundant Mamu-B sequences that have not been described previously.
We obtained analogous results from rhesus macaques (Supplementary Figs. 1 and 3 online). For example, one Indian-origin rhesus macaque (Fig. 2b) is homozygous for a common Mamu-B haplotype that we detected in nine unrelated animals (Supplementary Fig. 3 online). Together with the abundant transcripts for Mamu-B*02401 and Mamu-B*01901, we detected seven additional Mamu-B-like sequences that had not previously been associated with this haplotype at relatively low transcript levels (0.4–6.7% of total class I sequence reads)17. In contrast to the comparatively well-characterized class I sequences of Indian-origin rhesus macaques, in a homozygous Chinese-origin rhesus macaque (Fig. 2c) four of six Mamu-B-like sequences have not been reported previously; two of these represent the predominant Mamu-B transcripts expressed by this animal. The prevalence of novel sequences is even more pronounced for pig-tailed macaques where only limited class I allele discovery efforts have been described to date. Of the 136 distinct MHC class I sequences observed in 12 pig-tailed macaques, we detected over 100 novel MHC class I transcripts (Supplementary Figs. 1 and 4 online).
The success of our pilot study prompted us to examine whether we could maximize the efficiency of GS FLX genotyping for large cohorts by reducing the depth of sequence coverage. In a follow-up study, we pyrosequenced four amplicon pools containing 12 rhesus macaques each in 1/16 regions of a 70×75 PicoTiterPlate. This reduced the sequencing depth by an order of magnitude, to ~800 sequence reads per animal. Even with this reduced depth of coverage, we identified an average of 20.5 distinct MHC class I sequences per animal, as compared to 24.3 sequences per animal in our pilot study. This modest reduction in sensitivity notwithstanding, GS FLX analysis still provides considerably more comprehensive genotyping than existing methods15–20. The MHC class I sequences detected for these additional 48 macaques, as well as their relative transcript levels, are included (Supplementary Figs. 1 and 3 online).
Accuracy of pyrosequencing-based MHC genotyping of macaques
Sequence-based genotyping methods may be confounded by errors that accumulate due to polymerase misincorporations or sequencing artifacts. To diminish the number of sequence artifacts evaluated manually for each animal, we added a simple filtering step, requiring a minimum of five (pilot study) or two (follow-up study) identical reads in order for a sequence to be included in the downstream BLASTN analysis (Supplementary Note online). Greater than 98.3% of the resulting filtered reads were consistent with known or novel MHC class I sequences by BLASTN analysis (Table 1). With the filter step, we reduced the overall error rate of these data to <1.7% of the sequence reads evaluated subsequently, for both the representative animals illustrated in Fig. 2 and the full cohort (detailed analysis available in Supplementary Fig. 5 online). Excluding this low level of artifacts entails straightforward, manual editing, accomplished by intra- and inter-animal sequence comparison. Thus, the error rate in GS FLX pyrosequencing is acceptably low. We applied this multi-step analysis process to all of the MHC class I genotyping data presented here.
Table 1. Analysis of sequence artifacts.
| Unfiltered Error Rate (Fig. 2 Animals) |
Filtered Error Rate (Fig. 2 Animals) |
Filtered Error Rate (Whole Cohort) |
||||
|---|---|---|---|---|---|---|
| # Reads | (%) | # Reads | (%) | # Reads | (%) | |
| Total High Quality Reads | 21,950 | 21,950 | 484,985 | |||
| Failed Assembly | NA | 1,548 | (7.1) | 73,114 | (15.1) | |
| Passed Assembly | NA | 20,402 | (92.9) | 411,871 | (84.9) | |
| Total Reads for Error Analysis | 21,950 | 20,402 | 411,871 | |||
| MHC Class I | 20,497 | (93.4) | 20,050 | (98.3) | 406,274 | (98.5) |
| Insertions/Deletions | 762 | (3.5) | 300 | (1.5) | 2,914 | (0.8) |
| Single Base Substitutions | 321 | (1.5) | 22 | (0.1) | 565 | (0.1) |
| Multiple Base Substitutions | 124 | (0.6) | 30 | (0.1) | 2,118 | (0.5) |
| Short Reads (<180bp) | 246 | (1.1) | 0 | (0) | 0 | (0) |
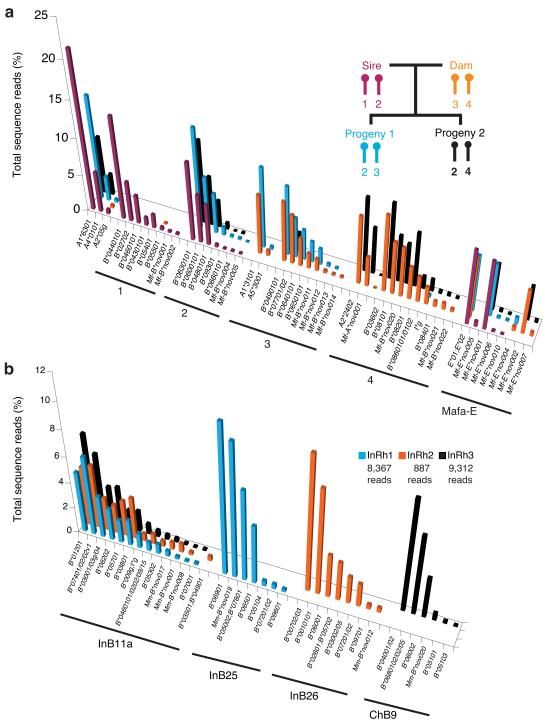
To exclude the possibility that novel sequences detected at low levels represented experimental artifacts, we examined the distribution of MHC class I sequences in pedigreed cynomolgus macaques. These novel class I sequences should not be inherited if they resulted from random errors during reverse transcription or PCR. Each progeny inherits the same haplotype from the sire while the haplotypes of the dam segregate between her offspring (Fig. 3a). The relative abundance of each MHC transcript is remarkably consistent on the haplotypes shared among the offspring and their parents. Importantly, we detected even those alleles that are present in as little as 0.2% of the total class I transcripts for these shared haplotypes.
Figure 3. Shared MHC class I transcript abundance profiles.
(a) The four haplotypes present in a breeding group of cynomolgus macaques are labeled 1–4. Both progeny inherited haplotype two from the sire while haplotypes three and four of the dam segregated between the offspring. (b) These three Indian rhesus macaques share the Mamu-B11a haplotype for which a complete genomic sequence has been published. Note the similarity of transcript profiles even though the values for InRh2 are based on an order of magnitude fewer sequence reads. Interestingly, InRh3 appears to have inherited a Mamu-B haplotype commonly found in Chinese rhesus macaques (ChB9; Supplementary Fig. 7 online) and is therefore likely to be of hybrid origin.
As a second approach to examine the accuracy of this genotyping method, we analyzed Indian rhesus macaques that share the B11a haplotype11,17. This haplotype is of special interest since this represents the only complete macaque genomic sequence currently available for this exceptionally complex region12. The B11a haplotype carries 19 Mamu-B-like loci that have the potential to encode at least 14 functional gene products. Previous cDNA cloning and Sanger sequencing identified transcripts for only eight of these loci previously11,17. However, with the increased sensitivity of GS FLX analysis we identified mRNA transcripts from at least 13 of the loci predicted by genomic sequencing (Fig. 3b). Between six and 13 Mamu-B sequences are transcribed from each of the haplotypes carried by these three animals (Fig. 3b). As with the cynomolgus macaque breeding group described above, the relative transcript abundance of class I sequences detected from the shared B11a haplotype was very similar despite the order of magnitude difference in depth of sequencing. Furthermore, we consistently observed similar class I transcript profiles for other ancestral haplotypes shared by unrelated animals, suggesting that GS FLX analysis provides at least a semi-quantitative representation of the relative class I transcript levels within an individual. We illustrate transcript profiles for additional shared haplotypes in Supplementary Fig. 6 online, further demonstrating the reproducibility of this technique.
Identification of high frequency Mamu class I sequences
Overall, we generated comprehensive MHC class I genotypes and expression profiles for 68 Indian- and Chinese-origin rhesus macaques obtained from four independent sources. These results allow us to begin to identify class I sequences that are relatively frequent in rhesus macaques. Of the 287 distinct class I sequences detected within our rhesus macaque cohort, there were 33 distinct Mamu-A, -B and -E sequences present in at least 10% of this cohort and expressed at relatively high transcript levels (ε4% of the total sequences per animal) (Table 2). These high-frequency alleles may represent high priority targets for additional functional immune characterization.
Table 2. Common rhesus macaque class I sequences that are highly expressed.
| MHC Class I Sequence | Frequency Observeda (%) |
Relative Transcript Abundanceb (Ave. % Total Sequence Reads) |
Putative Geographic Origin |
|---|---|---|---|
|
Mamu-A | |||
| A4*140301/02/05/09 | 63 | 4.3 | Both |
| A1*0040101/0102/0201/0202 | 34 | 12.1 | Both |
| A1*00801c | 13 | 16.0 | Indian |
| A3*1303/11 | 13 | 5.3 | Indian |
| A3*1308;A4*0102/A4*0201/02/03 | 13 | 4.8 | Chinese |
| A1*02301 | 10 | 17.0 | Indian |
|
Mamu-B | |||
| B*00702/03 | 28 | 14.7 | Both |
| B*03002/05 | 25 | 3.6 | Both |
| B*03001/0301/0302/0303/04 | 24 | 4.2 | Both |
| B*08202 | 22 | 3.7 | Indian |
| B*0010101 c | 21 | 9.7 | Both |
| B*02401 | 19 | 12.8 | Both |
| B*00501/02 | 18 | 4.8 | Both |
| B*04001/02 | 18 | 9.4 | Both |
| B*07401/02/02v1 | 18 | 5.0 | Indian |
| B*0440101/0102/02/03/04/05 | 18 | 3.9 | Both |
| B*01901/03 | 16 | 9.9 | Both |
| B*0680102/02/05 | 15 | 4.2 | Chinese |
| B*04101 | 13 | 6.6 | Both |
| B*05002;B*07801 | 13 | 7.4 | Both |
| B*01201 | 12 | 5.9 | Indian |
| B*03101/02 | 10 | 3.8 | Both |
| B*04301 | 10 | 6.3 | Both |
| B*06502 | 10 | 5.9 | Chinese |
| B*06503 | 10 | 8.9 | Chinese |
| B*06701/02 | 10 | 4.0 | Chinese |
|
Mamu-E | |||
| E*010101/0102/10/11/14/E*07/070102 | 59 | 9.5 | Both |
| E*030101/E*08 | 29 | 6.8 | Both |
| E*02 | 25 | 5.7 | Both |
| E*0113 | 19 | 5.5 | Both |
| E*05 | 19 | 8.4 | Chinese |
| E*0104/E*06 | 10 | 7.6 | Indian |
Percentage of animals that express specific class I sequences in the combined cohort (32 Indian- and 36 Chinese-origin rhesus macaques).
Transcript abundance is given as a percentage of sequence reads for a specific class I sequence relative to all class I sequences detected in an animal. Sequences listed were detected in 10% or more of the cohort at an abundance of at least 4% of total sequence reads when averaged across all macaques expressing this sequence in the cohort.
Mamu-A1*00801 and Mamu-B*0010101 (previously known as Mamu-A*08 and Mamu-B*01, respectively) are the only class I sequences shown here whose population frequencies have been determined by PCR-SSP assays20.
Using this genotype data, we also inferred the gene content of MHC haplotypes (Supplementary Figs. 3 and 7 online) and considerably extended the number of MHC class I sequences associated with previously described Mamu-A and Mamu-B haplotypes of Indian- and Chinese-origin rhesus macaques11,16,17. Surprisingly, all but six of 64 haplotypes observed in our Indian rhesus macaques could be accounted for by twelve previously described Indian-origin Mamu-B haplotypes (Supplementary Figs. 3 and 7 online). Consistent with the greater genetic diversity expected for Chinese-origin rhesus macaques, less than 1/3 of the 72 Mamu-B haplotypes in our cohort reflected previously reported configurations17,18. However, we did infer at least eight new Mamu-B haplotypes in these macaques, based on the sharing of five or more identical class I sequences between two or more animals (Supplementary Figs. 3 and 7 online).
DISCUSSION
These data prove that massively parallel pyrosequencing can provide comprehensive and cost effective MHC class I genotyping. We applied this technology to macaques, which have the most complex MHC genetics of any primate species described to date and have frustrated genotyping efforts for more than a decade. Comprehensive MHC genotyping has the potential to revolutionize the use of macaques in infectious disease and transplantation research and to guide functional immunology studies. Retrospective genotyping of macaques previously used in pathogenesis research may provide a more complete understanding of MHC restriction in cellular immune responses that are important in protective immunity and resistance to infectious diseases6,23,24. Pre-screening of macaques used in vaccine trials could balance these MHC sequences between experimental groups and reduce complications from over-representation of specific sequences that influence the quality of the cellular immune response25. This technology could also rapidly identify the most common MHC class I sequences in every macaque population used in biomedical research, enabling the selection of animals predicted to share T-cell responses or prioritizing sequences for functional characterization.
There are straightforward ways to improve upon the results obtained here. We designed the 190 bp amplicon to span the most polymorphic region of MHC class I molecules (Fig. 1) while retaining compatibility with current sequencing technology. Longer amplicons would allow for unique discrimination of more alleles and allelic variants, with the ultimate goal of full-length transcript sequencing to unambiguously determine the exact complement of class I sequences in an individual. We have performed preliminary studies with a 367 bp amplicon that utilizes an alternative reverse primer located in exon three. This longer amplicon provides improved resolution between closely related class I alleles and overcomes concerns about sequence artifacts resulting from contamination with genomic DNA as the longer amplicon spans an intron (data not shown). Pyrosequencing technology is rapidly improving and will soon allow for read lengths up to 500 bp. With this advance in mind, we have designed a new amplicon that spans 477 bp between conserved sequences in exons two and four of macaque class I genes. Genotyping with this longer amplicon will allow unambiguous resolution of 3/4 of the rhesus macaque class I sequences currently available in the Immuno Polymorphism Database21. Additionally, data from overlapping amplicons could be assembled to provide full-length MHC class I sequences. In silico studies with representative Indian rhesus macaques suggest that full-length class I sequences can be reconstructed from three overlapping amplicons once pyrosequencing read length of at least 400 bp can be achieved. Together, these approaches will allow for the novel sequence fragments identified by genotyping to be resolved into full-length MHC class I transcript sequences.
Pyrosequencing may also be used to dramatically improve upon existing technologies for genotyping other highly polymorphic loci. Obvious candidates include MHC class II, killer immunoglobulin receptor or T-cell receptor transcripts. This approach may also accelerate HLA class I genotyping of humans. Since there are only three HLA class I genes per chromosome, each transcribed at roughly equal levels, genotyping can be achieved with far fewer sequence reads than in macaques. Based on the yield from our macaque studies, HLA class I genotypes for thousands of individuals could be generated in a single GS FLX instrument run. Such ultra-high throughput typing may be valuable for tissue donor registry programs (http://bioinformatics.nmdp.org) as well as genetic epidemiology and whole genome association studies36.
Supplementary Material
ACKNOWLEDGEMENTS
Fresh blood or frozen peripheral blood mononuclear cell samples from various macaques were graciously provided by L. Picker (Oregon National Primate Research Center), P. Johnson (New England Primate Research Center), D. Read (Battelle Biomedical Research Center), N. Miller (US National Institute of Allergy and Infectious Diseases), J. Hoxie (University of Pennsylvania), J. Mankowski (Johns Hopkins University), T. Andrus (Charles River Biomedical Research Foundation, Inc.), and I. Lussier (Alpha Genesis, Inc.). L. Hetrick, A. Lane, E. Vlach and J. Thimmapuram provided outstanding emPCR, pyrosequencing and informatics support at the University of Illinois at Urbana-Champaign. We thank D. Watkins and R. DeMars for insightful comments on this manuscript. This work was supported by US National Institute of Allergy and Infectious Diseases contract number HHSN266200400088C/N01-AI-40088. Some support was also provided by a subcontract from the Battelle Biomedical Research Center under US National Institute of Allergy and Infectious Diseases contract N01-A1-30061. This publication was made possible in part by grant numbers P51 RR000167 and P40 RR019995 from the US National Center for Research Resources, a component of the US National Institutes of Health to the Wisconsin National Primate Research Center, University of Wisconsin-Madison. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01.
Appendix
METHODS
Macaque samples
We examined samples from 92 macaques obtained from nine different institutions (Supplementary Note online). Indian-origin and Chinese-origin rhesus macaques were represented by 32 and 36 samples, respectively, while 12 samples each came from cynomolgus and pig-tailed macaques. All animals were cared for according to the regulations and guidelines of the Institutional Care and Use Committees at their respective institutions.
Primary cDNA-PCR and pooling strategy
We converted total cellular RNAs to cDNA using a Superscript™III First-Strand Synthesis System (Invitrogen). We generated primary cDNA-PCR amplicons spanning 190 bp of exon two of macaque class I sequences with high-fidelity Phusion™ polymerase (New England Biolabs). Each PCR primer we utilized contained one of 12 distinct 10 bp MID tags along with adaptor sequences for 454 Sequencing™ (Supplementary Note online). After purification, we normalized primary amplicons to equimolar concentrations and pooled groups of 12 animals for GS FLX analysis.
Emulsion PCR and pyrosequencing
We performed the emulsion PCR and pyrosequencing steps with Genome Sequencer FLX instruments (Roche/454 Life Sciences) using GS FLX protocols according to the manufacturer’s specifications (454 Life Sciences)27,28 at the 454 Sequencing Center (Branford, CT) and the University of Illinois at Urbana-Champaign High-Throughput Sequencing Center (Supplementary Note online). We sequenced each amplicon pool of twelve animals in 1/4 of a 70×75 PicoTiterPlate for the pilot study while we utilized 1/16 plate regions for each of four pools in the follow-up experiment.
Data analysis
After image processing and base calling with GS FLX software (454 Life Sciences), we binned high quality sequence reads by their respective MID tags and assembled the reads into contigs with 100% identity for each animal using SeqMan Pro Version 8.0.2 (DNASTAR). We performed BLASTN analyses for the resulting contigs against a custom in-house database of macaque MHC class I sequences (Supplementary Note online). To normalize transcript abundance levels between animals, we divided the number of sequence reads detected for each distinct class I sequence by the total number of sequences reads which formed contigs in each animal. We designated MHC class I sequences not previously deposited in GenBank with a species abbreviation and the locus to which they are most similar (Mf-B*nov001 is the first novel class IB-like sequence identified in cynomolgus macaques). We deposited novel MHC class I sequences identified in this study to GenBank under accession numbers GQ153320-GQ153527 (Supplementary Fig. 1 online). Finally, it is important to note that macaque class I nomenclature has been modified recently to include an extra “0” in the allele lineage designations in order to maintain consistency with human HLA nomenclature and cover ever-expanding allele lists (Mamu-A*01 is now Mamu-A1*001). Information concerning relationships to previous nomenclature and details for each sequence are available at the Immuno Polymorphism Database (www.ebi.ac.uk/ipd/mhc/nhp/nomenclature.html)21.
Footnotes
COMPETING INTEREST STATEMENT
The authors declare the following competing financial interests. P.B., N.L., C.L.T. and E.S. are employed by 454 Life Sciences and T.H. is employed by Roche Applied Sciences.
REFERENCES
- 1.Marsh SGE, Parham P, Barber LD. The HLA fact book. Academic Press; London: 2000. [Google Scholar]
- 2.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 3.Horton R, et al. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J. 2008;49:220–255. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haigwood N. Predictive value of primate models for AIDS. AIDS Rev. 2004;6:189–198. [PubMed] [Google Scholar]
- 6.Watkins DI, et al. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 2008;14:617–621. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson JL, Carrion R. Demand for nonhuman primate resources in the age of biodefense. ILAR J. 2005;46:15–22. doi: 10.1093/ilar.46.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale DA, Dhanireddy K, Bruno D, Kirk AD. Induction of transplantation tolerance in non-human primate preclinical models. Proc.Trans. R. Soc. B. 2005;360:1723–1737. doi: 10.1098/rstb.2005.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 10.Bontrop RE. Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum. Immunol. 2006;67:388–397. doi: 10.1016/j.humimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Otting N, et al. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2005;102:1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulski JKT, et al. Rhesus macaque class I duplicon structures, organization and evolution within the alpha block of the major histocompatibility complex. Mol. Biol. Evol. 2004;21:2079–2091. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe A, et al. A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility region. Genomics. 2007;89:402–412. doi: 10.1016/j.ygeno.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J. Immunol. 2005;175:5230–5239. doi: 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- 16.Otting N, et al. Mhc class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otting N, et al. A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics. 2008;60:507–514. doi: 10.1007/s00251-008-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karl JA, et al. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics. 2008;60:37–46. doi: 10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell KJ, et al. Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics. 2008;61:177–187. doi: 10.1007/s00251-008-0351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaizu M, et al. Molecular typing of major histocompatibility complexclass I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- 21.Robinson J, Waller MJ, Stoehr P, Marsh SGE. IPD-the immuno polymorphism database. Nuc. Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiseman RW, et al. Siman immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder PJR, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loffredo JT, Valentine LE, Watkins DI. In: HIV Molecular Immunology 2006/2007. Korber BTM, et al., editors. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, NM: 2007. pp. 29–51. [Google Scholar]
- 25.Loffredo JT, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawashima Y, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–646. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas RK, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat. Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler DA, et al. Complete genome sequence of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.