Abstract
The differentiation of activated CD4+ T cells into the T helper type 1 (TH1) or TH2 fate is regulated by cytokines and the transcription factors T-bet and GATA-3. Whereas interleukin 12 (IL-12) produced by antigen-presenting cells initiates the TH1 fate, signals that initiate the TH2 fate are not completely characterized. Here we show that early GATA-3 expression, required for TH2 differentiation, was induced by T cell factor 1 (TCF-1) and its cofactor β-catenin, mainly from the proximal Gata3 promoter upstream of exon 1b. This activity was induced after T cell antigen receptor (TCR) stimulation and was independent of IL-4 receptor signaling through the transcription factor STAT6. Furthermore, TCF-1 blocked TH1 fate by negatively regulating interferon-γ (IFN-γ) expression independently of β-catenin. Thus, TCF-1 initiates TH2 differentiation of activated CD4+ T cells by promoting GATA-3 expression and suppressing IFN-γ expression.
After being activated, naive CD4+ T cells differentiate into helper T cell subsets to orchestrate effective immunity to microorganisms. T helper type 1 (TH1) cells mediate immunity against intracellular viruses and bacteria, whereas TH2 cells protect against parasites and mediate allergic responses1,2. The best-characterized feature of helper T cells is that TH1 cells produce interferon-γ (IFN-γ), whereas TH2 cells produce interleukin 4 (IL-4; A001262), IL-5 and IL-13 (refs. 3,4). Whereas activated CD4+ T cells differentiate into the TH1 lineage in response to IL-12 provided by antigen-presenting cells (APCs) and IFN-γ produced by natural killer cells2, similar signals that initiate TH2 differentiation remain incompletely characterized.
Once the TH2 response is initiated, IL-4 production by activated CD4+ T cells reinforces the TH2 fate5. However, the source of IL-4 at early stages of TH2 differentiation remains controversial. Cells of the innate immune system that produce IL-4 and help initiate TH2 differentiation3 have been found to be dispensable in some circumstances4,6,7, which suggests that IL-4 produced by antigen-activated CD4+ T cells is sufficient to initiate the TH2 response8. Thus, CD4+ T cells themselves have the potential to provide the initial IL-4 for TH2 differentiation. IL-4 expression is regulated by the transcription factor GATA-3 (refs. 9–12), which is expressed from two transcripts, called ‘Gata3-1a’ and ‘Gata3-1b’ here, that initiate from regulatory regions upstream of noncoding exons 1a and 1b, respectively13. Various combinations of transcription factors ‘downstream’ of the T cell antigen receptor (TCR) and the IL-4 receptor (IL-4R; A001263) seem to regulate GATA-3 expression during TH2 differentiation14. Notch-dependent signals have been shown to directly regulate GATA-3 expression and critically control helper T cell fate in vivo15–19. However, another report has suggested that Notch signals provide survival and proliferative advantages to committed helper T cells rather than direct commitment to the helper T cell fate20. Thus, other factors may induce GATA-3 expression, early after CD4+ T cell activation, in the absence of IL-4 in a transcription factor STAT6–independent way, but these remain to be fully described.
The transcription factor T cell factor 1 (TCF-1; A002260) represses gene expression in conjunction with Groucho21, and activates gene transcription by acting together with β-catenin (A000506) ‘downstream’ of several signaling pathways, including the canonical Wnt signaling pathway22–24. Several reports have demonstrated an essential function for TCF-1 and β-catenin at stages of T cell development that are regulated by the TCR or pre-TCR25,26. TCF-1 has high expression in thymocytes27 and in resting and activated human T cells28 and mouse T cells (data presented here), and β-catenin is stabilized ‘downstream’ of TCR signaling in activated T cells29–32. In addition, TCF reporter activity has been noted in activated T cells in vivo33. These data suggest that TCF-1 and β-catenin might participate in gene transcription in activated T cells without the requirement for Wnt-dependent activity. However, involvement of TCF-1 and β-catenin in the activation and differentiation of mature T cells has not been established before.
Here we demonstrate that TCF-1 and β-catenin regulated the differentiation of TCR-activated CD4+ T cells into the TH2 lineage. Initial GATA-3 expression in activated CD4+ T cells arose mainly from Gata3-1b transcripts. Gata3-1b transcription was induced ‘downstream’ of TCR signals, at early time points, and was independent of IL-4R signaling through STAT6. TCF-1 and β-catenin bound to the TCF-1-recognition site upstream of Gata3 exon 1b to positively regulate GATA-3 expression. Deletion of TCF-1 resulted in less Gata3-1b expression and IL-4 production. In contrast, enforced expression of stabilized β-catenin in activated CD4+ T cells resulted in ‘preferentially’ higher Gata3-1b expression and IL-4 production. Moreover, enforced expression of ICAT, a specific inhibitor of the interaction of TCF-1 and β-catenin, impaired Gata3-1b expression and IL-4 production, demonstrating a functional requirement for both TCF-1 and β-catenin. TCF-1 negatively regulated IFN-γ production and impaired the TH1 fate in TCR-activated CD4+ T cells. Thus, our data demonstrate that TCF-1 and β-catenin are responsible for driving TCR-activated T cells to a TH2 fate and do so by initiating GATA-3 expression and inhibiting IFN-γ production.
RESULTS
TCR induces GATA-3 from the Gata3-1b promoter
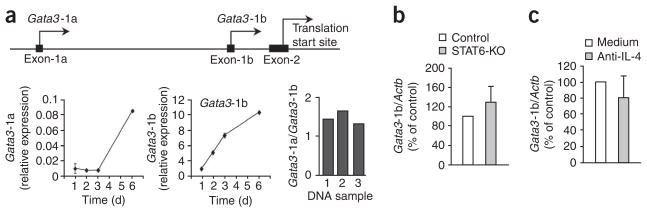
Gata3 is transcribed from two alternative promoters located 10 kilo-bases apart upstream of noncoding exon 1a and exon 1b13 (Fig. 1a). The resultant mRNA transcripts encode the same protein product but can be distinguished by the presence of promoter-specific sequences unique to exon 1a (Gata3-1a) or exon 1b (Gata3-1b). To assess GATA-3 expression, we activated CD4+ T cells with antibody to CD3 (anti-CD3) and anti-CD28 and studied the kinetics of expression of these two Gata3 transcripts. Unexpectedly, we found that Gata3-1b transcripts were 100-fold more abundant than Gata3-1a transcripts when normalized to a control gene (Fig. 1a). This difference was not due to different primer efficiencies, as verified by PCR amplification of Gata3-1a and Gata3-1b regions from three independent preparations of genomic DNA (Fig. 1a, right). Whereas the relative abundance of Gata3-1a transcripts remained low for the first 3 d and increased thereafter, the relative abundance of Gata3-1b transcripts increased within 24 h of CD4+ T cell activation (Fig. 1a). At all time points after TCR stimulation of CD4+ T cells, the relative abundance of Gata3-1b transcripts was much higher than that of Gata3-1a transcripts. Thus, Gata3-1b transcripts are the main contributors to GATA-3 expression in TCR-activated CD4+ T cells.
Figure 1.
Initial GATA-3 expression in TCR-activated CD4+ T cells is from Gata3-1b transcripts. (a) Real-time RT-PCR analysis of the abundance of Gata3-1a and Gata3-1b transcripts (left) in control CD4+ T cells stimulated with plate-bound anti-CD3 plus anti-CD28, assessed at various times after activation; results are presented relative to the abundance of Actb (n = 3 independent samples). Right, Gata3-1a PCR product relative to Gata3-1b PCR product (n = 3 independent samples). Top, approximately 10 kb of Gata3, showing two regulatory regions upstream of noncoding exon 1a and exon 1b and the translation start site in exon 2. (b) Real-time RT-PCR analysis of Gata3-1b mRNA expression in enriched, naive control or STAT6-deficient (STAT6-KO) CD25−CD4+ T cells stimulated for 3 d as described in a, presented relative to the abundance of mRNA in control cells, set as 100% (n = 4 independent samples). (c) Real-time RT-PCR analysis of Gata3-1b mRNA expression in CD4+ T cells stimulated for 3 d as described in a, in the presence (Anti-IL-4) or absence (Medium) of an IL-4-specific antibody, presented relative to the abundance of mRNA in control cells, set as 100% (n = 4 independent samples). Data are pooled from two independent experiments (error bars, s.e.m.).
To initiate TH2 differentiation, Gata3 must be induced in the absence of IL-4 signaling in a STAT6-independent way. To determine if Gata3-1b transcripts were sensitive to IL-4R signaling through STAT6, we activated control and STAT6-deficient CD4+ T cells34 with anti-CD3 and anti-CD28 in the absence of additional helper T cell–promoting agents. We found that the induction of Gata3-1b transcription in STAT6-deficient CD4+ T cells was similar to that in control CD4+ T cells (Fig. 1b), which suggested that transcription of Gata3-1b was STAT6 independent. In addition, antibody-mediated blockade of IL-4 did not diminish the abundance of Gata3-1b transcripts (Fig. 1c). Together these data demonstrate that early after activation of CD4+ T cells, GATA-3 is produced from Gata3-1b transcripts in a STAT6- and IL-4-independent way.
TCF-1 and β-catenin regulate Gata3-1b expression
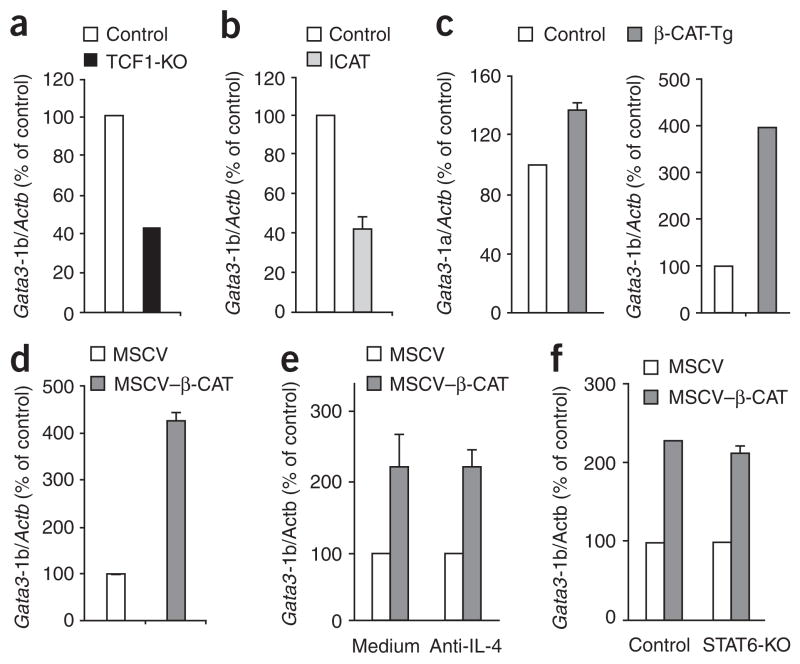
To study the transcriptional regulation of Gata3, we screened DNA sequences in the regulatory region upstream of Gata3 exon 1b. We found two closely placed binding sites for the transcription factor TCF-1 in the putative promoter region, upstream of Gata3 exon 1b. To determine if these sites functionally regulate expression of Gata3-1b, we quantified Gata3-1b mRNA in activated control and TCF-1-deficient CD4+ T cells. We purified CD25−CD4+ T cell populations that were enriched for naive cells from both TCF-1-deficient mice27 and control mice and confirmed that they contained similar proportions of CD62LhiCD44lo naive cells, which were also CD25−CD69−. TCF-1-deficient CD4+ T cells showed normal activation in response to APCs plus anti-CD3, as indicated by upregulation of CD25 and CD69 (Supplementary Fig. 1a). In addition, activated TCF-1-deficient CD4+ T cells proliferated and produced IL-2 similar to control CD4+ T cells (Supplementary Fig. 1b,c). Notably, we found that Gata3-1b expression was lower in activated TCF-1-deficient CD4+ T cells than in wild-type CD4+ T cells (Fig. 2a), which suggested that induction of Gata3-1b expression in CD4+ T cells after TCR stimulation required TCF-1.
Figure 2.
TCF-1 and β-catenin act together to regulate the production of GATA-3 from Gata3-1b transcripts. (a) Real-time RT-PCR analysis of Gata3-1b mRNA in control and TCF-1-deficient CD25−CD4+ T cells stimulated for 6 d with APCs plus anti-CD3; results are presented relative to the abundance of mRNA in activated control CD4+ T cells, set as 100% (n = 2 independent samples). (b) Real-time RT-PCR analysis of Gata3-1b mRNA in CD25−CD4+ T cells obtained from control and ICAT mice and stimulated for 6 d with APCs plus anti-CD3, presented as in a (n = 4 independent samples). (c) Real-time RT-PCR analysis of Gata3-1a and Gata3-1b mRNA in TCR-stimulated control and β-CAT-Tg CD25−CD4+ T cells, assessed as described in a (n = 6 independent samples). (d) Gata3-1b mRNA in C57BL/6 CD25−CD4+ T cells stimulated for 2 d with plate-bound anti-CD3 plus anti-CD28 and transduced with retrovirus expressing human CD8 alone (MSCV) or human CD8 and a stabilized form of β-catenin (MSCV–β-CAT), then sorted electronically as human CD8–positive cells at 24 h after infection (n = 3 independent samples). (e) Real-time RT-PCR analysis of Gata3-1b expression by C57BL/6 CD25−CD4+ T cells stimulated, transduced and sorted as described in d and cultured in the presence or absence of IL-4-specific antibody (n = 4 independent samples). (f) Real-time RT-PCR analysis of Gata3-1b expression by enriched naive control or STAT6-deficient CD25−CD4+ T cells stimulated, transduced and sorted as described in d (n = 4 independent samples). Data are pooled from two or three (b–d) independent experiments (error bars, s.e.m.).
As TCF-1 is not known to induce GATA-3 expression, it was important to further elucidate the mechanism by which TCF-1 participated in GATA-3 induction. Consequently, we sought to determine if induction of Gata3-1b by TCF-1 in activated CD4+ T cells required the cofactor β-catenin. ICAT is a 9-kilodalton protein that has been shown to bind to β-catenin and inhibit its interaction with TCF-1 and impair TCF-1- and β-catenin-dependent transactivation of target genes35. Mice with enforced expression of ICAT from the proximal promoter of the gene encoding the kinase Lck (called ‘ICAT mice’ here) have been generated and analyzed36. CD4+ T cells from ICAT mice and control mice showed similar activation, proliferation and IL-2 production (Supplementary Fig. 2). However, we found that activated ICAT CD4+ T cells also had lower expression of Gata3-1b than did control CD4+ T cells (Fig. 2b). These data show that deletion of TCF-1 or disruption of the TCF-1–β-catenin interaction by ICAT expression impairs GATA-3 expression. To determine if enforced expression of β-catenin enhanced Gata3-1b expression, we used CD4+ T cells from β-catenin-transgenic mice (β-CAT-Tg mice)37. We found that β-CAT-Tg CD4+ T cells showed normal activation and proliferation compared with that of control CD4+ T cells but produced less IL-2 (Supplementary Fig. 2). We also found that enforced expression of stabilized β-catenin ‘preferentially’ augmented Gata3-1b expression, without affecting Gata3-1a transcripts, in activated β-CAT-Tg CD4+ T cells (Fig. 2c). Additionally, we found that retroviral expression of β-catenin rapidly induced expression of Gata3-1b transcripts in activated CD4+ T cells (Fig. 2d). Finally, the abundance of total GATA-3 in the loss-of-function (TCF-1-deficient and ICAT) and gain-of-function (β-CAT-Tg) CD4+ T cells reflected the pattern noted for Gata3-1b (Supplementary Fig. 3). Thus, these data document involvement of TCF-1, acting with its cofactor β-catenin, in the regulation of Gata3-1b expression in TCR-activated CD4+ T cells.
Next we wanted to determine whether TCR stimulation of CD4+ T cells affected the expression and/or function, of TCF-1 or β-catenin. Resting (ex vivo) and TCR-activated CD4+ T cells express both TCF-1 and LEF (a TCF family protein that binds the same DNA sequence as TCF-1); however, the relative expression of TCF-1 was much higher than that of LEF when normalized to β-actin (Supplementary Fig. 4a). Ex vivo CD4+ T cells also expressed β-catenin, and this expression was slightly higher after TCR stimulation (Supplementary Fig. 4b). These data show that TCF-1 and β-catenin are expressed in resting and TCR-activated CD4+ T cells and that TCR signals lead to moderately greater β-catenin abundance, probably through protein stabilization. Expression of lacZ, which encodes β-galactosidase, was higher after TCR activation of CD4+ T cells from BAT-lacZ reporter mice (which express lacZ under control of a β-catenin-activated transgene; Supplementary Fig. 4c). These data suggest that TCF-1 and β-catenin function to induce TCR-dependent gene expression. Finally, activated CD4+ T cells had higher expression of Bambi, a TCF-1 target gene38, than did resting CD4+ T cells (Supplementary Fig. 4d). Thus, TCR-dependent signals induce TCF-1 and β-catenin transactivation activity and the expression of target genes in CD4+ T cells.
The paucity of IL-4 at the start of an infection suggests that TCR-driven GATA-3 expression must be independent of IL-4 signaling during the initial phase of TH2 differentiation. To test whether TCF-1- and β-catenin-induced Gata3-1b was independent of IL-4, we measured the abundance of Gata3-1b transcripts in vector-transduced and β-catenin-transduced CD4+ T cells treated with an IL-4-blocking antibody or without such antibody treatment. TCF-1 and β-catenin induced equivalent increases in Gata3-1b expression in the presence or absence of anti-IL-4 in the culture medium (Fig. 2e). To determine if TCF-1- and β-catenin-induced Gata3-1b expression required STAT6, we retrovirally transduced β-catenin into activated CD4+ T cells from STAT6-deficient and control mice and assayed the abundance of Gata3-1b transcripts relative to Actb (encoding β-actin). We found that there was greater relative abundance of Gata3-1b transcripts in β-catenin-transduced cells, which was not diminished in the absence of STAT6 (Fig. 2f). Thus, the TCF-1- and β-catenin-induced expression of GATA-3 from Gata3-1b transcripts was independent of IL-4, IL-4R and STAT6, which suggests involvement of these events in the initiation of TH2 differentiation.
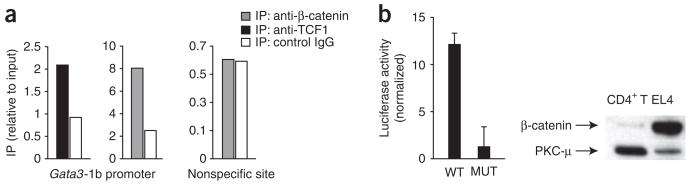
TCF and β-catenin bind Gata3-1b regulatory elements
To determine if TCF-1 directly regulates GATA-3 expression from the regulatory sites upstream of Gata3 exon 1b, we did chromatin-immunoprecipitation assays with DNA from activated CD4+ T cells and anti-TCF-1 and anti-β-catenin. We found that TCF-1 bound to the TCF-1-binding sites in the regulatory region upstream of Gata3-1b in activated CD4+ T cells (Fig. 3a). Likewise, β-catenin-specific antibodies immunoprecipitated the same DNA but not nonspecific DNA sequence that did not contain TCF-binding sites (Fig. 3a). These data show that TCF-1 and β-catenin bind to the TCF-1-binding sites in the Gata3-1b regulatory region. Using purified preparations of recombinant LEF protein39, we also found by electrophoretic mobility-shift assay that LEF specifically bound to oligonucleotides corresponding to the Gata3-1b promoter sequence containing the TCF-binding site but not to sequence with a mutated TCF-binding site (Supplementary Fig. 5). To further assess if the TCF-1-binding sites in the Gata3 regulatory region were functional, we generated a luciferase reporter plasmid containing Gata3-1b regulatory DNA sequences. We assayed reporter activity in the mouse T lymphoma cell line EL4, which has higher expression of β-catenin than that of resting CD4+ T cells (Fig. 3b). We found constitutive luciferase activity with the wild-type TCF-1 reporter; however, expression was lower when the TCF-1 site was mutated (Fig. 3b). These data show that TCF-1 and β-catenin bind to the identified binding sites upstream of Gata3 exon 1b and functionally activate transcription.
Figure 3.
TCF-1 and β-catenin bind TCF-1 sites upstream of Gata3 exon 1b. (a) Chromatin-immunoprecipitation assay of C57BL/6 CD4+ T cells stimulated for 16 h with plate-bound anti-CD3 plus anti-CD28 and then fixed, followed by immunoprecipitation (IP) of chromatin with anti-TCF, anti-β-catenin or nonspecific isotype-matched control antibody (IgG). DNA recovered from immunoprecipitates is presented relative to input DNA; DNA lacking a TCF-1-binding site serves as negative control (right). Data are representative of three independent experiments. (b) Luciferase activity of EL4 cells transfected with a luciferase reporter construct containing trimers of wild-type (WT) or mutated (MUT) TCF-1-binding sites from the Gata3-1b region, presented relative to that of cells transfected with a control pGL3-promoter (normalized to renilla). P = 0.0007. Right, immunoblot analysis of β-catenin in EL4 cells and primary mouse CD4+ T cells; protein kinase C-μ (PKC-μ) serves as a loading control. Data are pooled from three independent experiments with duplicate samples (error bars, s.e.m.).
TCF-1-induced GATA-3 leads to IL-4 production
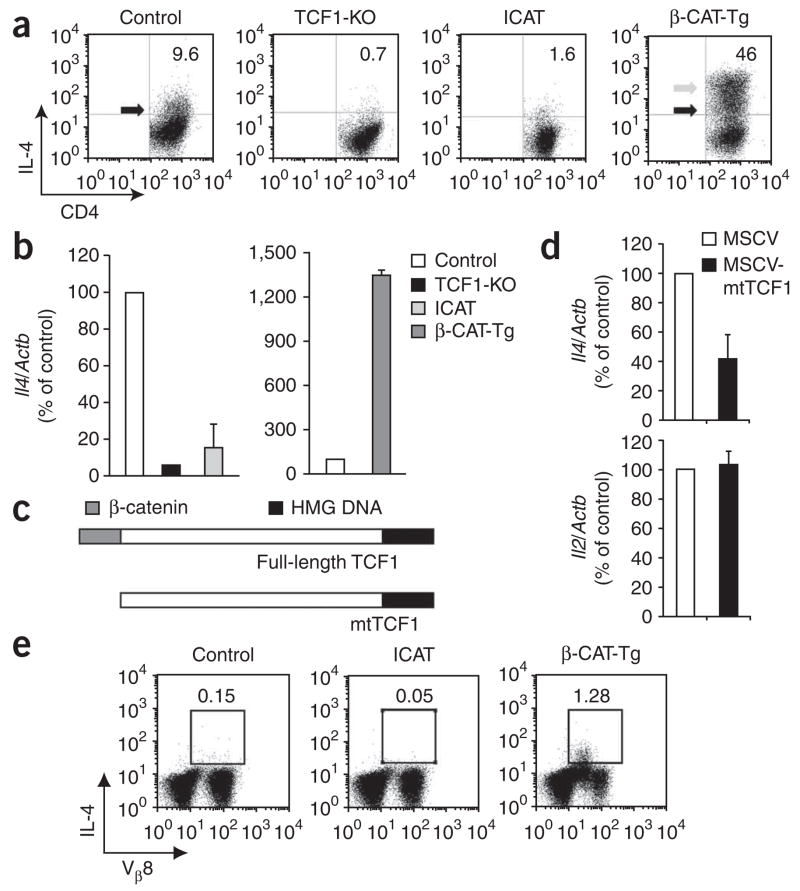
To determine if impaired GATA-3 expression affects IL-4 production, we assayed TCR-activated CD4+ T cells from TCF-1-deficient, ICAT and β-CAT-Tg mice. We found that tenfold more activated control CD4+ T cells than activated TCF-1-deficient or ICAT-expressing CD4+ T cells produced IL-4 (Fig. 4a). In contrast, fourfold more activated β-CAT-Tg CD4+ T cells than activated control CD4+ T cells produced IL-4 (Fig. 4a). In addition, activated β-CAT-Tg CD4+ T cells produced more IL-4 per cell (Fig. 4a, arrows). The changes in IL-4 production were not due to changes in IL-4R expression or the capacity of IL-4R to signal, as measured by IL-4-induced phosphorylation of STAT6, both of which were equivalent in TCF-1-deficient and control CD4+ T cells (Supplementary Fig. 6). Moreover, we found that the abundance of Il4 mRNA was also lower in TCR-activated TCF-1-deficient and ICAT T cells than in control CD4+ T cells (Fig. 4b). In contrast, β-CAT-Tg CD4+ T cells had more Il4 mRNA (Fig. 4b). These data show that TCF-1 and β-catenin regulate IL-4 production in activated CD4+ T cells in vitro.
Figure 4.
TCF-1 and β-catenin act together to promote IL-4 production and TH2 differentiation. (a) Flow cytometry of control, TCF-1-deficient (TCF1-KO), ICAT and β-CAT-Tg CD25− CD4+ T cells stimulated with APCs plus anti-CD3, then restimulated with phorbol 12-myristate 13-acetate and ionomycin on day 6 of culture and stained for intracellular IL-4 and surface CD4. Gray arrow indicates β-CAT-Tg cells with more IL-4 production; black arrows indicate control and β-CAT-Tg populations with similar IL-4 production. Numbers in plots indicate percent IL-4+CD4+ cells. Data are representative of six independent analyses. (b) Real-time RT-PCR analysis of Il4 mRNA in cells stimulated as described in a, presented relative to mRNA in control cells, set as 100%. Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.). (c) Structure of wild-type TCF-1 (Full-length TCF1) and mutant TCF-1 (mtTCF1); mtTCF1 lacks the amino-terminal β-catenin-binding domain but retains the Groucho- and DNA-binding domains. HMG, high-mobility group. (d) Real-time RT-PCR analysis of IL-4 and IL-2 mRNA in C57BL/6 CD25−CD4+ T cells stimulated for 1 d with plate-bound anti-CD3 plus anti-CD28 and infected with retrovirus expressing human CD8 alone (MSCV) or human CD8 and mtTCF1 (MSCV-mtTCF1) then sorted electronically as human CD8–positive cells at day 2 after infection; results are presented relative to the abundance of mRNA in control cells, set as 100%. Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.). (e) Flow cytometry of spleen and lymph node cells from control, ICAT and β-CAT-Tg mice injected intraperitoneally with SEB or PBS; 2 h later, cells were isolated and restimulated in vitro as described in a, then analyzed by intracellular staining of cells with anti-IL-4 and surface staining with anti-NK1.1 and antibody to the variable region Vβ8 of the TCR. Numbers above outlined areas indicate percent IL-4+Vβ8+ cells in the NK1.1− population. Data are representative of five independent experiments.
To further substantiate the finding that TCF-1 and β-catenin act together to enhance IL-4 production, we used a mutant form of TCF-1 lacking the β-catenin-binding domain (mtTCF1)39 (Fig. 4c). We found that expression of mtTCF1 in activated CD4+ T cells resulted specifically in less Il4 mRNA but not less Il2 mRNA (Fig. 4d). These data show that overexpression of a mutant TCF-1 that is unable to recruit β-catenin impairs IL-4 expression. Collectively these data show that TCF-1 and β-catenin act together to induce GATA-3 expression, which in turn leads to more IL-4 production by activated CD4+ T cells.
TCF-1 and β-catenin also regulate IL-4 production in vivo, as assessed after injection of staphylococcal enterotoxin B (SEB), which is an agonist for Vβ8+ T cells. We found approximately 8.5-fold more IL-4 producing T cells in SEB-injected β-CAT-Tg mice than in SEB-injected control mice (Fig. 4e). Conversely, the frequency of IL-4 producing T cells in SEB-injected ICAT mice was one-third that in SEB-injected control mice (Fig. 4e). In these conditions, IL-4 production by cells of the innate immune system was very low and not different in control versus mutant mice (Supplementary Fig. 7). These data show that TCF-1 and β-catenin act together to regulate IL-4 production in vivo and in vitro in antigen-activated CD4+ T cells.
TCF-1-induced IL-4 production is Notch independent
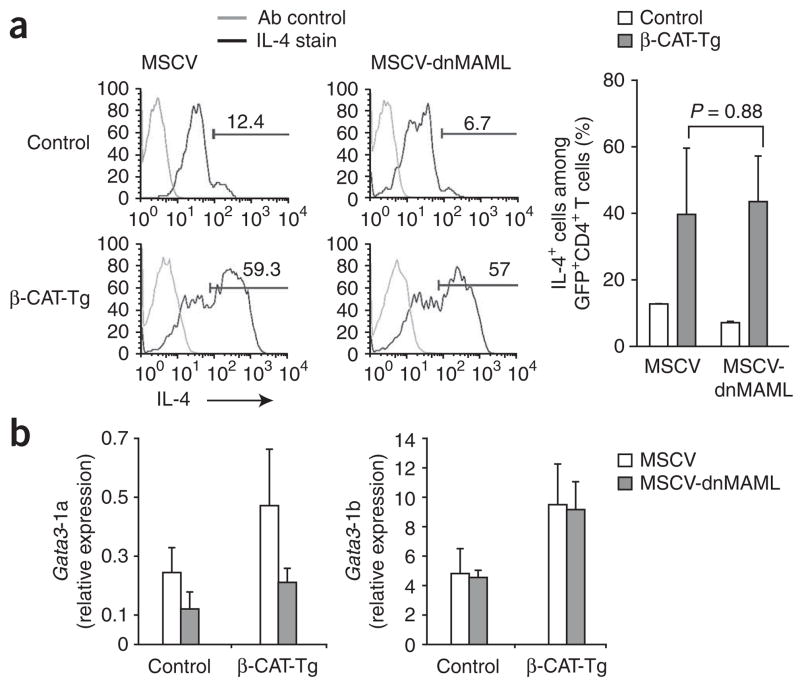
Notch-dependent transcription has been shown to regulate IL-4 production by inducing GATA-3 expression from the Gata3-1a promoter15,17. To determine whether the induction of IL-4 production by TCF-1 and β-catenin depends on Notch signaling, we blocked Notch activity with a dominant negative form of the transcriptional coactivator Mastermind-like (dnMAML) and assayed IL-4 production in the presence and absence of β-catenin. We transduced control and β-CAT-Tg CD4+ T cells with retroviral constructs coexpressing dnMAML and green fluorescent protein (GFP) and assigned scores to the frequency of IL-4-producing cells among GFP+ cells. As reported before17, expression of dnMAML resulted in a lower frequency of IL-4-producing cells among transduced control CD4+ T cells (Fig. 5). Notably, enforced expression of β-catenin resulted in a fivefold higher frequency of IL-4-producing cells, which was not diminished by blocking Notch signaling with dnMAML (Fig. 5a). Moreover, Gata3-1b transcripts were not lower in abundance in either control or β-catenin-expressing CD4+ T cells transduced with dnMAML (Fig. 5b). Thus, these data show that Gata3-1a and Gata3-1b are Notch-dependent and Notch-independent transcripts, respectively. We conclude that TCF-1 and β-catenin induce Gata3-1b expression and subsequent IL-4 production, which is independent of Notch signaling.
Figure 5.
TCF-1- and β-catenin-induced IL-4 production by CD4+ T cells is Notch independent. (a) IL-4 production in GFP-gated CD4+ T cells among control and β-CAT-Tg CD25−CD4+ T cells stimulated with plate-bound anti-CD3 plus anti-CD28 and infected with control retrovirus expressing GFP alone (MSCV) or retrovirus expressing dnMAML and GFP (MSCV-dnMAML), assayed by intracellular staining on day 6 after infection (left). Ab control, istoype-matched control antibody. Numbers above bracketed lines indicate percent IL-4+ cells. Right, frequency of IL-4-producing cells among GFP+CD4+ T cells. Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.). (b) Real-time RT-PCR analysis of Gata3-1a and Gata3-1b mRNA in GFP+CD4+ T cells sorted 6 d after retroviral infection as described in a, presented relative to Actb. Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.).
TCF-1 negatively regulates IFN-γ production
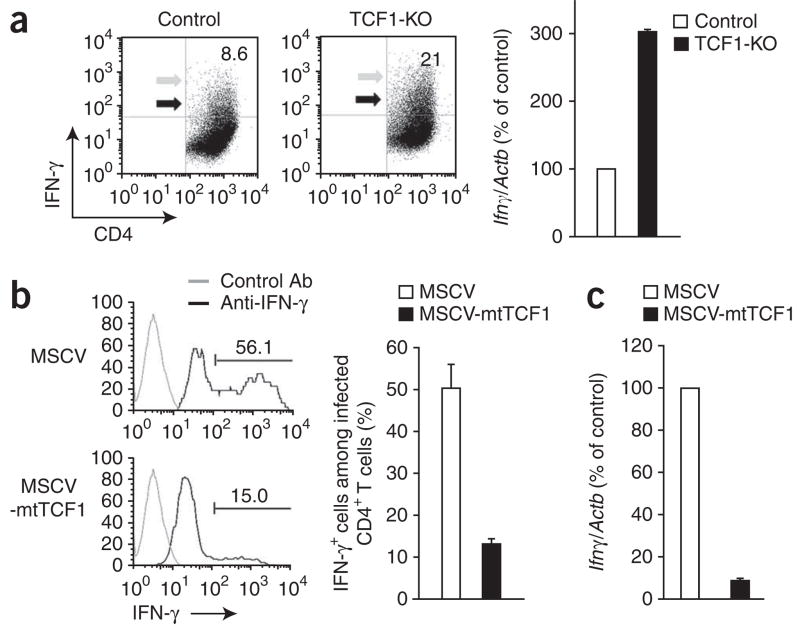
Signals that induce GATA-3 expression and the TH2 fate can also block IFN-γ expression and the TH1 fate2,3. Therefore, we assessed whether TCF-1 and/or β-catenin also regulate IFN-γ production and the TH1 fate. We found that activated TCF-1-deficient CD4+ T cells produced more IFN-γ than did wild-type control cells (Fig. 6a), which suggested that TCF-1 negatively regulates IFN-γ expression. On the basis of these data, we predicted that TCF-1 expression would be lower in TH1-differentiated CD4+ T cells than in undifferentiated cells. Indeed, within 3 d of the differentiation of CD4+ T cells toward the TH1 lineage, the abundance of TCF-1 mRNA decreased to one third that in control cells (Supplementary Fig. 8). Thus, TCF-1 is diminished after encounter with IL-12 and IFN-γ, cytokines that promote the TH1 fate. We also found that ICAT-expressing CD4+ T cells did not have more IFN-γ production (data not shown). Thus, we conclude that the repressive effect of TCF-1 on IFN-γ was not dependent on its interaction with β-catenin.
Figure 6.
TCF-1 negatively regulates IFN-γ production independently of β-catenin. (a) Flow cytometry of control and TCF-1-deficient CD4+ T cells stimulated for 6 d with APCs and anti-CD3 and assayed for IFN-γ production by intracellular staining (left). Gray arrows indicate TCF-1-deficient cells with higher IFN-γ production than that of control cells; black arrows indicate TCF-1-deficient cells with IFN-γ production similar to control cells. Numbers in plots indicate percent IFN-γ+CD4+ cells. Right, real-time RT-PCR analysis of Ifng mRNA, presented relative to that of TCR-activated control CD4+ T cells, set at 100%. Data are representative of six independent experiments (left) or two independent experiments with a total of four independent samples (right; error bars, s.e.m.). (b) IFN-γ production by C57BL/6 CD4+ T cells stimulated for 1 d with plate-bound anti-CD3 plus anti-CD28 and infected with retrovirus as described in Figure 4d, then cultured for 2 d in IL-2 and assessed by intracellular staining of human CD8–positive CD4+ T cells (left). Right, frequency of IFN-γ+ cells among human CD8–positive CD4+ T cells. P = 0.0006 (right). Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.). (c) Real-time RT-PCR analysis of Ifng mRNA in CD4+ T cells infected and sorted as described in b, presented relative to mRNA in TCR-activated control CD4+ T cells, set as 100%. Data are pooled from two independent experiments with a total of four independent samples (error bars, s.e.m.).
To further confirm that TCF-1 expression blocks IFN-γ independently of β-catenin, we transduced CD4+ T cells with control retro-virus or retrovirus expressing mtTCF1, the mutant TCF-1 that cannot interact with β-catenin (Fig. 6b) and assayed IFN-γ production. IFN-γ expression was lower in CD4+ T cells transduced with mtTCF1 (Fig. 6b,c). Together these data demonstrate that in the absence of TH1 cytokines, TCF-1 negatively regulates IFN-γ and thereby blocks the TH1 fate.
Deletion of TCF-1 impairs TH2 responses in vivo
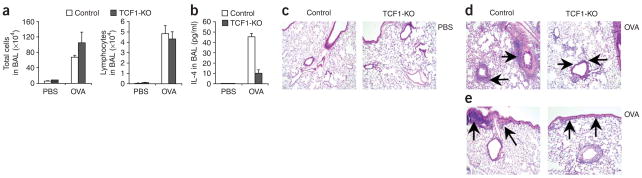
To determine if TCF-1-deficient mice had altered adaptive immune responses in vivo, we used a mouse model of allergic asthma. Nonimmunized control mice (injected with PBS) and TCF-1-deficient mice had similar cell counts in their bronchoalveolar lavage (BAL) fluid at baseline. As expected, after immunization and challenge with ovalbumin (OVA), control mice had significantly more total cells in the BAL fluid than did PBS-injected mice, as did immunized TCF-1-deficient mice (P = 0.001; Fig. 7a, left). In addition, analysis of differential cell counts showed that TCF-1-deficient mice and control mice had a similar number of lymphocytes in the BAL fluid (Fig. 7a, right). However, cytokine analysis showed that TCF-1-deficient mice had significantly less of the TH2 cytokine IL-4 in the BAL fluid than did control mice (P = 0.03; Fig. 7b). Furthermore, lung histology showed that OVA-immunized and OVA-challenged control mice had more inflammatory cell infiltration in the perivascular and peribronchial areas, involving medium and large airways (Fig. 7c–e). In contrast, immunized TCF-1-deficient mice had cell infiltration in the lung tissue (Fig. 7c–e), but it was mostly limited to the perivascular areas (Fig. 7d) with limited involvement of the peribronchial areas (Fig. 7e), particularly the large airways (Fig. 7e). These results demonstrate that after immunization, TCF-1-deficient mice attempt to mount an adaptive immune response to OVA allergen but with less production of the TH2 cytokine IL-4 and visibly less inflammation around the airways in the lung.
Figure 7.
TCF-1 regulates TH2 responses in vivo. Immune responses of control mice (PBS, n = 4; OVA, n = 9) and TCF-1-deficient mice (PBS, n = 5; OVA, n = 6) sensitized and challenged with PBS or OVA. (a) Total and differential cell counts in BAL fluid samples obtained 1 d after the final OVA challenge. (b) IL-4 concentration in BAL fluid. (c–e) Hematoxylin and eosin–stained lung sections from PBS-injected mice (c) or OVA-injected mice (d,e). Arrows indicate inflammation around blood vessels (d) or inflammation around large airways (e). Original magnification, × 10. Data are pooled from two independent experiments (a,b; error bars, s.e.m.) or are representative of two independent experiments with a total four to nine independent samples each (c–e).
DISCUSSION
We have demonstrated here that GATA-3 in CD4+ T cells was produced mainly from Gata3-1b transcripts early after TCR stimulation and in the absence of IL-4 and STAT6 signaling. We have shown that TCF-1 and β-catenin positively and directly regulated Gata3 expression by binding to TCF-1-binding sites in the Gata3 regulatory region upstream of exon 1b. Induction of GATA-3 by TCF-1 and β-catenin resulted in IL-4 production and initiation of the TH2 fate. In addition, TCF-1 inhibited IFN-γ production and TH1 differentiation independently of its interaction with β-catenin. Therefore, we propose that TCF-1 facilitates the initiation of TH2 differentiation by a dual mechanism: by inducing GATA-3 expression and by blocking IFN-γ production.
Naive CD4+ T cells have low expression of GATA-3, which is induced in response to TCR and IL-4R signaling9–12 from two alternative transcripts13,14. In this report we have demonstrated that Gata3-1b transcripts were the main isoform expressed at early time points after TCR activation of CD4+ T cells independently of IL-4R signaling through STAT6. In our experiments, the abundance of Gata3-1a transcripts was much lower in TCR-stimulated CD4+ T cells and was induced at late times after stimulation. Whereas GATA-3 is produced mainly from Gata3-1b transcripts in activated CD4+ T cells, both transcripts were expressed with different kinetics, and further analysis is needed to delineate the importance of alternative use of these transcripts in activated CD4+ T cells. Furthermore, Notch signaling regulates Gata3-1a transcripts15,17, whereas TCF-1 and β-catenin regulate Gata3-1b expression, during TH2 differentiation. Although further studies are needed to demonstrate independent regulation of GATA-3 expression by Notch and TCF-1 in vivo, these data show that both transcripts contribute to GATA-3 expression during TH2 differentiation. However, the timing and pattern of expression of Gata3-1b indicates it must initiate TH2 differentiation.
To demonstrate the importance of TCF-1 in mounting a TH2 response in vivo, we used an established mouse OVA model. We found that TCF-1-deficient mice were substantially impaired in mounting a TH2 response to OVA allergen in vivo, which indicates an important function for TCF-1 in TH2 responses in vivo. TCF-1 and β-catenin initiate the TH2 differentiation of TCR-stimulated CD4+ T cells, as indicated by the following observations. First, TCF-1 and β-catenin induced GATA-3 from Gata3-1b transcripts ‘downstream’ of TCR signals within 16–24 h. Second, TCF-1 and β-catenin induced GATA-3 independently of IL-4 signaling through STAT6. Third, TCF-1- and β-catenin-induced GATA-3 resulted in more IL-4 production. Thus, TCF-1-and β-catenin-induced GATA-3 expression meets the criteria for the initiation of TH2 differentiation in conditions of low IL-4. Finally, TCF-1 negatively regulated IFN-γ production and blocked TH1 differentiation. Therefore, downregulation of TCF-1 is essential for differentiation to the TH1 lineage. In agreement with that, we found that APC-derived IL-12 and IFN-γ downregulated TCF-1 expression in CD4+ T cells. Thus, we propose that TCF-1 regulates TH2 fate by two means: by inducing GATA-3 expression, which promotes IL-4 production, and by blocking IFN-γ production.
We have shown here and others have shown before that activated T cells express components that mediate canonical Wnt signals29,32. However, it remains unclear if TCR signals directly stabilize β-catenin or result in the induction of autocrine Wnt signaling, which then stabilizes β-catenin. Thus, the mechanism by which TCR signals lead to stabilization of β-catenin in activated CD4+ T cells requires further investigation. Pre-TCR signals stabilize β-catenin in thymocytes through phosphorylation of the kinase Erk40. By analogy with that, we speculate that Erk-dependent mechanisms may regulate inhibition of activity of the kinase GSK3β41 and TCR-induced stabilization of β-catenin. We have also found that TCR signals lead to higher expression of various Wnt receptor components (data not shown), which may enhance the responsiveness of activated T cells to Wnt ligands. Thus, β-catenin is stabilized directly or indirectly ‘downstream’ of the TCR in activated CD4+ T cells. In addition, β-catenin may also be stabilized by microbial byproducts, such as prostaglandin E2, that promote TH2 differentiation and have been shown to stabilize β-catenin in colon cancer cells42–46. Thus, stabilization of β-catenin by TCR signals as well as by TH2-promoting agents acts together with TCF-1 to regulate initial GATA-3 expression and initiate TH2 differentiation.
Finally, two lines of evidence suggest that IFN-γ production is negatively regulated by TCF-1. First, the higher frequency of activated TCF-1-deficient CD4+ T cells resulted in more production of IFN-γ. Second, expression of a mutant TCF-1 that cannot bind β-catenin repressed IFN-γ production in activated wild-type CD4+ T cells. However, at present there are no known, confirmed TCF-1-binding sites in the Ifng promoter. Thus, either the repression observed in our assays is indirect or TCF-1 acts directly through sequences distal from the promoter or through ‘cooperation’ with other factors that act on the proximal promoter. These and other possibilities will be an important focus of future studies in the field.
In conclusion, we have shown here that TCF-1 and β-catenin positively regulated Gata3-1b expression, which led to more IL-4 production in TCR-activated CD4+ T cells. These events occurred at early time points after CD4+ T cell activation, were cell intrinsic in nature and were independent of both IL-4 and Notch signaling. Together these data provide a model for the molecular basis of the initiation of the TH2 fate. We suggest that TCF-1 and β-catenin bind a response element in the Gata3 regulatory region upstream of exon 1b to initiate transcription of Gata3-1b mRNA. Higher GATA-3 expression promotes IL-4 production and initiates TH2 differentiation. Simultaneously, TCF-1 also inhibits IFN-γ production and TH1 differentiation independently of β-catenin. APC-derived IL-12 and IFN-γ inhibit TCF-1 expression and block GATA-3 expression, thereby promoting TH1 differentiation. Thus, TCF-1 initiates the TH2 differentiation of activated CD4+ T cells by a dual mechanism of activation of GATA-3 and repression of IFN-γ.
METHODS
Mice
The generation of β-CAT-Tg mice37 and ICAT mice36 has been described. TCF-1-deficient mice27 were provided by H. Clevers. BAT-lacZ mice47 and STAT6-deficient mice34 were from The Jackson Laboratory. Age-matched littermate control mice or C57BL/6 mice were used in all experiments. All mice were bred and maintained in the animal facility of the National Institute on Aging according to regulations of the National Institutes of Health and were in compliance with the guidelines of the animal resources facility of the National Institute on Aging, which operates under the regulatory requirements of the US Department of Agriculture and Association for Assessment and Accreditation of Laboratory Animal Care.
Antibodies and flow cytometry
Cells were collected, stained and analyzed on a FACSCalibur (Becton Dickinson). Dead cells were excluded by forward light scatter or by forward light scatter plus propidium iodide. All the data were acquired and are presented on a log scale. The following antibodies (all from BD Pharmingen) were used for staining: allophycocyanin-conjugated anti-CD4 (GK1.5), phycoerythrin (PE)-conjugated or peridinin chlorophyll protein (PerCP)–cyanine 5.5 (Cy5.5)–conjugated anti-CD8α (53-6.7), fluorescein isothiocyanate (FITC)-conjugated anti-CD69 (H1.2F3), PE–anti-CD25 (PC61), PE–anti-IL-4 (BVD4-1D11), FITC–anti-NK1.1 (PK136), FITC–anti-Vβ8.1.2 (MR5-2), PE–anti-CD44 (IM7), FITC–anti-CD62L (MEL-14), PE–FITC–anti-TCRβ (H57-597), PerCP-Cy5.5–anti-c-Kit (2B8), PE–anti-IL-4Rα (MIL4R-M1), FITC–anti-Gr-1 (RB6-8C5) and PE-conjugated antibody to phosphorylated STAT6 (pY641). Purified anti-CD3 (145-2C11), anti-CD28 (37.51), anti-IL-4 (11B11) and anti-IFN-γ (XMG1.2) were from BD Pharmingen. PerCP-Cy5.5–anti-CD11c (N418), PE–anti-IL-2 (JES6-5H4) and FITC–anti-IFN-γ (XMG1.2) were from eBioscience.
Labeling with the cytosolic dye CFSE
Cells were washed in PBS, incubated for 8 min at 20–25 °C at a density of 1 × 107 cells per ml with 0.25 μM CFSE (carboxyfluorescein diacetate succinimidyl ester; Molecular Probes) and washed in medium with serum before further culture.
In vivo CD4+ T cell stimulation
Control C57BL/6 mice, ICAT mice and β-CAT-Tg mice were injected intraperitoneally with 50 μg SEB (Sigma) or PBS (negative control). Then, 2 h after injection, spleen and lymph node cells were isolated and then restimulated for 4 h with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (1 μM; both from Calbiochem), with the addition of GolgiStop (BD Pharmingen) during the final 2 h. Cells were then collected and stained for intracellular cytokines as well as cell surface markers with the Cytoperm/Cytofix kit according to the manufacturer’s protocol (BD Pharmingen).
In vitro CD4+ T cell differentiation
CD25−CD4+ T cells were purified from spleen and lymph nodes by negative selection with the Mouse CD4+ T Cell Isolation kit (Miltenyi) in combination with biotin-conjugated anti-CD25 (7D4). The resulting populations were highly enriched for CD44loCD62Lhi naive cells, and the frequency of naive cells was similar in control mice and the various mutant mice. Splenic APCs were isolated by depletion of T cells by negative selection with the Mouse Pan T Cell Isolation kit (Miltenyi). CD4+ T cells were stimulated with plate-bound anti-CD3 (1 μg/ml; 145-2C11) plus anti-CD28 (1 μg/ml; 37.51) or with irradiated splenic APCs (2,000 rads) plus soluble anti-CD3 (1 μg/ml). On day 3 of culture, cells were diluted with medium, and IL-2 (10 ng/ml; R&D Systems) was added to cultures. On various days, cells were restimulated for 4 h with phorbol 12-myristate 13-acetate and ionomycin with the addition of GolgiStop during the final 2 h. Cells were then collected and stained for intracellular cytokines.
Retroviral infection
For retroviral infection, purified CD25−CD4+ T cells were activated for 2 d with plate-bound anti-CD3 and anti-CD28 and then incubated for a total of 3 h with culture supernatant containing mouse stem cell virus (MSCV)-based retrovirus (MSCV-β-CAT-huCD8, MSCV-mtTCF-huCD8 or MSCV-huCD8; MSCV-dnMAML-GFP or MSCV-GFP). Virus supernatant was then removed and cells were further cultured for 1 d in medium with IL-2 (10 ng/ml) and IL-7 (5 ng/ml; both from R&D Systems), and human CD8–positive or GFP+ and human CD8–negative or GFP− populations were electronically sorted on a MoFlo (Dako-Cytomation), followed by analysis of mRNA expression.
Quantitative (real-time) RT-PCR
Total mRNA was reversed transcribed with poly(dT) and Superscript III reverse transcriptase (Invitrogen). Then, cDNA was amplified by 40 cycles of real-time RT-PCR (Applied Biosystems) with annealing and extension at 60 °C (primer sequences, Supplementary Table 1).
Immunoblot analysis
Nuclear extracts were prepared by lysis of cells with 1.5 mM MgCl2 plus 10 mM KCl and pelleting of the nucleus, followed by lysis of the nucleus with 420 mM NaCl, 1.5 mM MgCl2 plus 25% (vol/vol) glycerol. For immunoblot analysis, nuclear extracts were resolved by 4–12% SDS-PAGE (Invitrogen) and then transferred to nitrocellulose membrane. Blots were incubated with monoclonal anti-β-catenin (14/Beta-Catenin; 610154; BD Pharmingen) or rabbit-anti-PKC-μ (C-20; sc-639; Santa Cruz) or rabbit-anti-SP1 (H-225; sc-14027; Santa Cruz), followed by horseradish peroxidase–conjugated anti–mouse IgG (sc-2005) or anti–rabbit IgG (sc-2004; both from Santa Cruz). Reactivity was visualized by enhanced chemiluminescence.
Chromatin-immunoprecipitation assay
CD4+ T cells were activated for 16 h with plate-bound anti-CD3 plus anti-CD28, and a kit from Upstate Cell Signaling was used for chromatin immunoprecipitation according to the manufacturer’s protocol. Chromatin was precipitated with 4 μg mouse anti-β-catenin (H-102; sc-7199), anti-TCF (H-118X; sc-13025X) or normal mouse IgG (sc-2027; all from Santa Cruz), followed by salmon sperm–coated protein A–agarose (Upstate Cell Signaling). Precipitates were analyzed by real-time RT-PCR for detection of Gata3-1b promoter with the following primer set: forward, 5′-GGGAAAGCAAGCAGAGACCA-3′; reverse, 5′-TTGCCTCCGAAC CAGCTTTC-3′. DNA immunoprecipitated with anti-TCF-1, anti-β-catenin or IgG is presented relative to input DNA according to the following formula: [(Gata3-1b DNA in immunoprecipitate)/(Gata3-1b DNA in total input)] × 4,000.
Reporter plasmid construction and reporter activity assays
The TCF-1-binding site in the Gata3 promoter (at a position −1.6 kilobases relative to the translation start site) and flanking sequences were cloned into pGL3 enhancer constructs to generate a luciferase reporter plasmid. Three copies of sequence with the TCF-1 site (underlined; 5′-GCGGGCGTCCGAATCAAAGCCCAGGTCCTC-3′) or sequence with a mutated TCF-1 site (underlined; 5′-GCGGGCGTCCGAATGAGATTCCAGGTCCTC-3′) were annealed, then were digested with SacI and XhoI, which generated a SacI overhang at the 5′ end and an XhoI overhang at the 3′ end, then sequences were ligated into the pGL3-Promoter luciferase vector (Promega) digested with SacI and XhoI. The identity of each clone was confirmed by DNA sequencing. Expression plasmid encoding renilla luciferase (pRL-TK) was from Promega. EL4 cells were seeded the day before transfection at a density of 2 × 105 cells per ml. Cells (2 × 106) were transiently cotransfected with 5 μg reporter construct and 25 ng pRL-TK (included as an internal control for normalization of data) using the Amaxa Cell Line Nucleofector kit L according to the manufacturer’s instructions (Amaxa). Transfected cells were collected 24 h after transfection and lysed, and luciferase activity was measured with a Dual-Luciferase Reporter Assay system (Promega).
Electrophoretic mobility-shift assay
The purification of LEF protein has been described39. The sequences of the oligonucleotides were as follows: wild-type probe, 5′-GCGGGCGTCCGAATCAAAGCCCAGGTCCTC-3′; mutant probe, 5′-GCGGGCGTCCGAATGAAATTCCAGGTCCTC-3′. [32P]-labeled oligonucleotide probes were incubated for 30 min at 4 °C with purified LEF protein and samples were resolved by PAGE. Gels were then dried and exposed to a phosphorimager plate.
Mouse model of allergen immunization (acute asthma model)
Mice were immunized intraperitoneally with 50 μg OVA (chicken egg albumin; Sigma-Aldrich) in 2 mg aluminum hydroxide gel on days 0 and 7. Mice were then challenged intranasally on 3 consecutive days (days 14, 15, and 16) with 50 μg OVA; 1 d after the final OVA challenge, mice were killed and lung inflammation was evaluated.
Evaluation of inflammatory cells in BAL fluid
BAL fluid samples were collected from anesthetized mice in 2 ml PBS (two successive volumes of 1 ml PBS) through the trachea as described48,49. After centrifugation and resuspension, total cell counts were determined with a hemocytometer. For differential cell counts of less than 1 × 105 cells, viable BAL cells were centrifuged onto slides with a Cytospin 3 apparatus (Thermo Shandon) and were stained with the Diff-Quik Stain set (Siemens) and cell types in a total of 300 cells were recorded. Supernatants of BAL samples were stored at −80 °C until use.
Histological evaluation of lung inflammation
Lung sections 5 μm in thickness were stained with hematoxylin and eosin after pressure fixation with 10% (vol/vol) formaldehyde (Ricca Chemical Company). Cell infiltration in lung tissue was analyzed.
IL-4 ELISA
IL-4 concentrations in BAL fluid were determined with a commercial ELISA kit (R&D Systems) according to the manufacturer’s protocols.
Statistics
Statistical significance was determined by the Student’s t-test.
Supplementary Material
Acknowledgments
We thank R. Wersto and team for cell sorting; the animal facility of the National Institute on Aging for maintaining animals; S. Luo and team for genotyping; H. Clevers (Hubrecht Institute) for TCF-1-deficient mice; W. Pear (University of Pennsylvania) for the dnMAML retroviral construct; and N. Taylor, A. Singer, R. Bosselut, R. Sen and A. Bhandoola for critically reading the manuscript. Supported by the Intramural Research Program of the National Institute on Aging of the National Institutes of Health and the Oak Ridge Institute for Science and Education’s Research Associates Program of the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS
Q.Y. and J.M.S. designed and orchestrated the study and wrote the manuscript; Q.Y., A.S., M.Z.H., T.M.S. and K.E.L. did all the in vivo and in vitro experiments; S.Y.O., H.-G.M. and Z.Z. contributed to the analysis of OVA-injected mice in vivo; H.D. did electrophoretic mobility-shift assays; and B.W. and M.L.W. provided reagents and advice.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A001262, A001263, A002260 and A000506.
Note: Supplementary information is available on the Nature Immunology website.
References
- 1.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 2.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 3.Corthay A. A three-cell model for activation of naive T helper cells. Scand J Immunol. 2006;64:93–96. doi: 10.1111/j.1365-3083.2006.01782.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown DR, et al. β2-microglobulin-dependent NK1.1+ T cells are not essential for T helper cell 2 immune responses. J Exp Med. 1996;184:1295–1304. doi: 10.1084/jem.184.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 6.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 7.von der Weid T, Beebe AM, Roopenian DC, Coffman RL. Early production of IL-4 and induction of Th2 responses in the lymph node originate from an MHC class I-independent CD4+NK1.1− T cell population. J Immunol. 1996;157:4421–4427. [PubMed] [Google Scholar]
- 8.Schmitz J, et al. Induction of interleukin 4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 10.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 11.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 13.Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- 14.Scheinman EJ, Avni O. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J Biol Chem. 2009;284:3037–3048. doi: 10.1074/jbc.M807302200. [DOI] [PubMed] [Google Scholar]
- 15.Amsen D, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amsen D, et al. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 17.Fang TC, et al. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minter LM, et al. Inhibitors of γ-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 19.Tanigaki K, et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 20.Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS ONE. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brantjes H, Barker N, van Es J, Clevers H. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol Chem. 2002;383:255–261. doi: 10.1515/BC.2002.027. [DOI] [PubMed] [Google Scholar]
- 22.Molenaar M, et al. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 23.Roose J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 24.Staal FJ, Burgering BM, van de Wetering M, Clevers HC. Tcf-1-mediated transcription in T lymphocytes: differential role for glycogen synthase kinase-3 in fibroblasts and T cells. Int Immunol. 1999;11:317–323. doi: 10.1093/intimm/11.3.317. [DOI] [PubMed] [Google Scholar]
- 25.Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 26.Staal FJ, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–1794. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verbeek S, et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 28.Willinger T, et al. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J Immunol. 2006;176:1439–1446. doi: 10.4049/jimmunol.176.3.1439. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Banerjee D, Huelsken J, Birchmeier W, Sen JM. Deletion of β-catenin impairs T cell development. Nat Immunol. 2003;4:1177–1182. doi: 10.1038/ni1008. [DOI] [PubMed] [Google Scholar]
- 30.Staal FJ, Clevers HC. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat Rev Immunol. 2005;5:21–30. doi: 10.1038/nri1529. [DOI] [PubMed] [Google Scholar]
- 31.Yu Q, Sen JM. β-catenin regulates positive selection of thymocytes but not lineage commitment. J Immunol. 2007;178:5028–5034. doi: 10.4049/jimmunol.178.8.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu B, Crampton SP, Hughes CC. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity. 2007;26:227–239. doi: 10.1016/j.immuni.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriyama A, et al. GFP transgenic mice reveal active canonical Wnt signal in neonatal brain and in adult liver and spleen. Genesis. 2007;45:90–100. doi: 10.1002/dvg.20268. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 35.Satoh K, et al. Anteriorization of neural fate by inhibitor of β-catenin and T cell factor (ICAT), a negative regulator of Wnt signaling. Proc Natl Acad Sci USA. 2004;101:8017–8021. doi: 10.1073/pnas.0401733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain MZ, Yu Q, Xu M, Sen JM. ICAT expression disrupts β-catenin-TCF interactions and impairs survival of thymocytes and activated mature T cells. Int Immunol. 2008;20:925–935. doi: 10.1093/intimm/dxn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulroy T, Xu Y, Sen JM. β-Catenin expression enhances generation of mature thymocytes. Int Immunol. 2003;15:1485–1494. doi: 10.1093/intimm/dxg146. [DOI] [PubMed] [Google Scholar]
- 38.Sekiya T, et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-β signaling, as a target of the β-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279:6840–6846. doi: 10.1074/jbc.M310876200. [DOI] [PubMed] [Google Scholar]
- 39.Atcha FA, et al. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352–8363. doi: 10.1128/MCB.02132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M, Sharma A, Wiest DL, Sen JM. Pre-TCR-induced β-catenin facilitates traversal through beta-selection. J Immunol. 2009;182:751–758. doi: 10.4049/jimmunol.182.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y, Shen S, Lino AC, Curotto de Lafaille MA, Lafaille JJ. Beta-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat Med. 2008;14:162–169. doi: 10.1038/nm1707. [DOI] [PubMed] [Google Scholar]
- 42.Kalinski P, Hilkens CM, Wierenga EA, Kapsenberg ML. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol Today. 1999;20:561–567. doi: 10.1016/s0167-5699(99)01547-9. [DOI] [PubMed] [Google Scholar]
- 43.Kalinski P, Vieira PL, Schuitemaker JH, de Jong EC, Kapsenberg ML. Prostaglandin E2 is a selective inducer of interleukin-12 p40 (IL-12p40) production and an inhibitor of bioactive IL-12p70 heterodimer. Blood. 2001;97:3466–3469. doi: 10.1182/blood.v97.11.3466. [DOI] [PubMed] [Google Scholar]
- 44.Woolard MD, et al. Francisella tularensis-infected macrophages release prostaglandin E2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol. 2007;178:2065–2074. doi: 10.4049/jimmunol.178.4.2065. [DOI] [PubMed] [Google Scholar]
- 45.Shao J, Jung C, Liu C, Sheng H. Prostaglandin E2 stimulates the β-catenin/T cell factor-dependent transcription in colon cancer. J Biol Chem. 2005;280:26565–26572. doi: 10.1074/jbc.M413056200. [DOI] [PubMed] [Google Scholar]
- 46.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-β-catenin signaling axis. Science. 2005;310:1504–1510. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 47.Maretto S, et al. Mapping Wnt/β-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng T, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.