Abstract
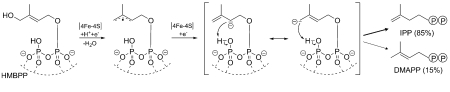
Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) represent the two central intermediates in the biosynthesis of isoprenoids. The recently discovered deoxyxylulose 5-phosphate pathway generates a mixture of IPP and DMAPP in its final step by reductive dehydroxylation of 1-hydroxy-2-methyl-2-butenyl 4-diphosphate. This conversion is catalyzed by IspH protein comprising a central iron-sulfur cluster as electron transfer cofactor in the active site. The five crystal structures of IspH in complex with substrate, converted substrate, products and PPi reported in this article provide unique insights into the mechanism of this enzyme. While IspH protein crystallizes with substrate bound to a [4Fe-4S] cluster, crystals of IspH in complex with IPP, DMAPP or inorganic pyrophosphate feature [3Fe-4S] clusters. The IspH:substrate complex reveals a hairpin conformation of the ligand with the C(1) hydroxyl group coordinated to the unique site in a [4Fe-4S] cluster of aconitase type. The resulting alkoxide complex is coupled to a hydrogen-bonding network, which serves as proton reservoir via a Thr167 proton relay. Prolonged x-ray irradiation leads to cleavage of the C(1)-O bond (initiated by reducing photo electrons). The data suggest a reaction mechanism involving a combination of Lewis-acid activation and proton coupled electron transfer. The resulting allyl radical intermediate can acquire a second electron via the iron-sulfur cluster. The reaction may be terminated by the transfer of a proton from the β-phosphate of the substrate to C(1) (affording DMAPP) or C(3) (affording IPP).
Keywords: iron-sulfur protein, LytB protein, nonmevalonate pathway, terpene biosynthesis, isoprenoid biosynthesis
Biosynthetic precursors of isoprenoids are generated via two independent pathways (1, 2). In the more recently discovered nonmevalonate pathway, the terminal intermediate, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate (HMBPPFig. 1), is reductively converted into a mixture of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) by the action of a single, monomeric iron-sulfur protein specified by the ispH gene (initially designated lytB) (3, 4). This pathway is crucial for most pathogenic bacteria as well as for Plasmodium falciparum (the parasite causing malaria) and plant plastids, yet it is absent in animals, and accordingly IspH has received considerable attention as potential drug target. Previously reported x-ray structures of IspH protein from Escherichia coli (5) and Aquifex aeolicus (6) show that the single peptide chain folds into three closely similar domains, which are arranged in a clover-like shape with pseudo-C3 symmetry. At its center of mass, the protein features an iron-sulfur cluster which is coordinated by the cysteine residues 12, 96, and 197 (one from each of the three folding domains, amino acid residue numbers refer to E. coli). Although it is mostly assumed that IspH protein requires a [4Fe-4S] cluster for catalytic activity (7, 8), all published structures so far showed [3Fe-4S] clusters (5, 6). Notably, the E. coli protein crystallized in a closed conformation with the iron-sulfur cluster located inside a solvent-inaccessible central cavity that also contained a pyrophosphate or malonate ion, depending on crystallization conditions (5). On the other hand, the A. aeolicus enzyme crystallized in an open conformation where the iron-sulfur cluster is solvent-accessible (6).
Fig. 1.
Reaction catalyzed by IspH. Suggested sequence of electron and proton transfer during the reduction of HMBPP to IPP/DMAPP.
Ever since the discovery of IspH protein, its reaction mechanism has been the subject of intense investigation (5, 9, 10). It is generally agreed that at least one intermediate must be an allyl derivative (Fig. 1), which might be either radicalic or anionic. Here we report x-ray structures of IspH protein from E. coli in complex with; (i) substrate (HMBPP), (ii) converted substrate (CON), (iii) Isopentenyl diphosphate (IPP), (iv) DMAPP, and (v) inorganic pyrophosphate. Implications of the crystallographic data on the reaction mechanism are discussed.
Results and Discussion
IspH Protein with Bound Substrate HMBPP (Fig. 2B).
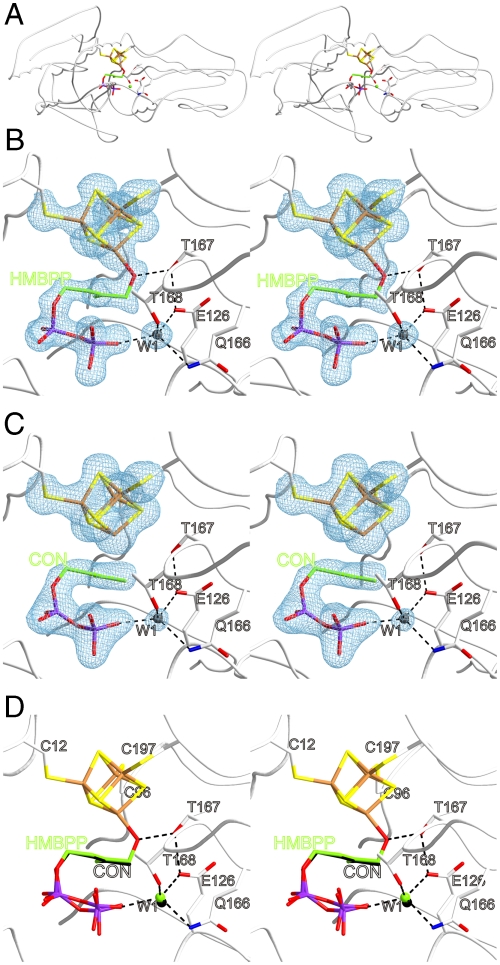
Recombinant E. coli IspH protein (comprising an N-terminal His6 fusion tag) was purified and subsequently crystallized in the presence of the substrate, HMBPP under anaerobic conditions (5) as described in the method section. The protein samples had high catalytic activities, in the range of 40 μmol mg-1 min-1, and were used as isolated. A diffraction dataset to 1.7 Å resolution was obtained at the Swiss Light Source (SLS) synchrotron radiation facility. The crystal structure was determined by molecular replacement using the published structure of E. coli IspH protein (5) as starting model (Rfree 0.269; Fig. 2A and B, Table S1). Interestingly, it only was possible to collect a full dataset of the intact HMBPP ligand bound to IspH when exposed to a significantly short irradiation time (4 min). The crystal structure of the complex reveals a closed conformation of IspH with the position of the protein backbone being virtually identical to that previously reported for E. coli IspH protein in complex with inorganic pyrophosphate (rmsd, 0.3 Å) (5). Inside the central cavity, the electron density of the bound substrate is well defined. The pyrophosphate moiety assumes the same position as the inorganic pyrophosphate ion and is embedded in a polar environment comprising the side chains of histidines 41, 74, and 124, serines 225 and 269, threonine 168, asparagine 227 and glutamine 166. The overall conformation of the bound substrate is hairpin-shaped resembling the recently published modeled ligand (5). A single fixed water molecule (subsequently designated W1) is well-defined inside the densely packed active site cavity where it is hydrogen bonded to the β-phosphate moiety as well as to Glu126Oϵ, Gln166Nϵ and Thr168Oγ which all are strictly conserved (Fig. 2B). Concomitantly, the substrate oxygen at C(1) is placed in hydrogen-bonding distance to Thr167Oγ (2.7 Å) resulting in the formation of a cyclic, 17-membered hydrogen bonded system, which includes substrate, W1, Glu126Oϵ-H and Thr167Oγ-H (Fig. 3). This arrangement positions the C(1)-O axis orthogonal to the plane of the three allylic carbon atoms with its oxygen in close proximity to Glu126Oϵ- (3.4 Å).
Fig. 2.
(A) Stereoview of the active site of IspH with bound HMBPP (Green) including coordinated water at position W1 (Gray). (B) Close-up view of the active site with the covalently bound substrate HMBPP and a converted substrate (C) interacting with the [4Fe-4S] cluster. The hydrogen-bonding networks at the IspH active sites including protein, ligand and the W1 water molecule are indicated by dashed black lines. Electron densities represented in blue are contoured at 1.0σ with 2F0-Fc coefficients; ligands have been omitted for the electron densities calculations. (D) Overlay of substrate (Green) and converted substrate (Black) bound to IspH active site.
Fig. 3.
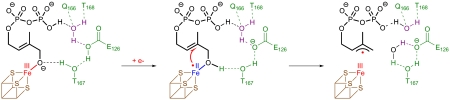
Proposed reaction sequence initiating HMBPP reduction as it was derived from crystallographic data. Schematic representation of bound substrate and the 17 member cyclic hydrogen-bonding system including substrate, T167, E126 and W1 depicts the anticipated role of the individual amino acids as proton relay and proton source.
The pivotal element in this crystal structure is the central iron-sulfur cluster, which unambiguously reveals a fourth iron atom. Interestingly, the fourth iron atom features a typical Fe-O bond distance of 2.0 Å to the oxygen at C(1). Formulation of an alkoxide group complexed at a localized Fe(III) site (Fig. 3) is in agreement with recently published Mössbauer data (7, 11) and takes into account the strong acidification of the hydroxyl functionality upon binding to the ferric iron center. The three allylic carbon atoms of the bound substrate are sandwiched between the pyrophosphate moiety and the apical iron center featuring Fe-C distances of 2.8–3.0 Å, which is longer than observed for organometallic iron allyl complexes (2.0–2.1 Å) (12), yet well below the sum of the van der Waals radii (3.6 Å).
IspH Protein in Complex with Converted Substrate (Fig. 2C).
Prior to synchrotron measurements, crystals of the IspH:HMBPP complex were tested for their diffraction quality by our in-house CuKa x-ray rotating anode. This data acquisition was carried out with a 30 s radiation exposure time. Thereafter appropriate candidates were stored in liquid nitrogen and data collection was then performed two weeks later at the SLS synchrotron. Surprisingly, the 2FOFC-electron density revealed that the conformation of the pyrophosphate ester is similar to that of the IspH:HMBPP complex (Fig. 2D). However, the data lack electron density of the alkoxide group at C(1) of the substrate performing a covalent bond with the iron-sulfur cluster and hence the apical iron lost its tetragonal form. Thus, the coordination of the apical iron atom with the substrate is no longer observed. It’s worth noting that the Fe-C distances between the apical iron atom and the allylic moiety of the converted substrate (2.6–2.7 Å) are even shorter than the ones observed for bound HMBPP. Therefore, an interaction of the olefinic π-system with the transition metal’s d-orbitals is likely. Interestingly, the electron density inside the active site cavity reveals only the water molecule at the W1 position, suggesting that the water molecule generated by the reaction is structurally disordered. We anticipate that conversion of HMBPP is induced by reducing photo electrons generated by x-ray exposure. This is confirmed by a crystal structure of IspH:HMBPP complex using CuKa radiation for long time diffraction data acquisition (24 h), which also features the converted substrate (Fig. S1). These data reveal that in the presence of reducing agents, amorphously frozen crystalline IspH protein still exhibits catalytic activity even at a temperature of 100 K, which yet is not sufficient to complete the hydrolytic reaction within 4 min. However the nature of the bound converted substrate cannot be determined unequivocally from the crystallographic data, since the electron density is compatible with an allyl radical, an allyl anion, as well as both of the reaction products.
IspH Protein in Complex with Reaction Products (Fig. 4A and 4B).
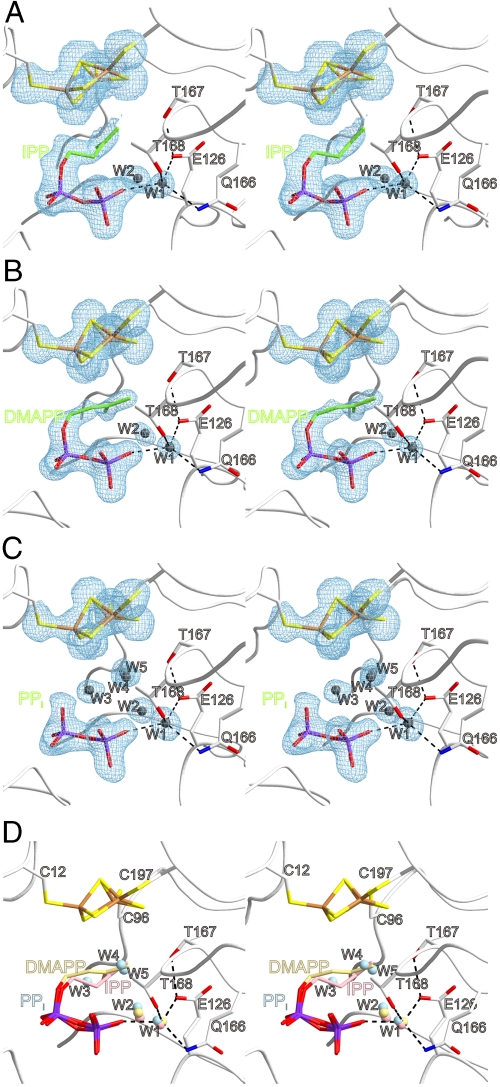
Crystals of IspH protein in complex with either of the products IPP or DMAPP were obtained by cocrystallization and diffracted to 2.0 and 1.7 Å resolution, respectively. Although the two product complexes crystallized in different space groups (C2 in case of the IspH:IPP complex, P212121 in case of the IspH:DMAPP complex; Fig. S2) the structures were fairly equal. While all protein and ligand conformations in the five crystal structures described in this article are of close resemblance (Fig. S2 and Fig. S3) each structure of the product complexes comprises a [3Fe-4S] cluster, as opposed to the [4Fe-4S] clusters in the IspH:HMBPP and IspH:CON complex. This discrepancy might be explained by a ligand induced directed crystallization from a heterogeneous mixture of purified IspH protein containing either [3Fe-4S] or [4Fe-4S] clusters. The differences in cluster composition complicate the comparison of binding modes of substrate and products. We anticipate that due to the lack of the hydrophilic hydroxyl moiety in both products, binding to the more lipophilic [3Fe-4S] cluster is favored over the more polar cluster in the active enzyme. Moreover, the enlarged central cavity provides space for binding a second water molecule besides W1 (Fig. 4A and B).
Fig. 4.
Active site of IspH with bound ligands (A) IPP, (B) DMAPP and (C) PPi. Defined water molecules are represented in gray. Note, that all crystal structures display an [3Fe-4S]-cluster. Color coding and orientation of protein and ligands are identical to Figure 2. (D) Structural superposition of products IPP and DMAPP and PPi.
IspH Protein Bound to Inorganic Pyrophosphate (Fig. 4C).
In an earlier report (5), inorganic pyrophosphate was serendipitously found inside the IspH active site under crystallization conditions without added pyrophosphate. We have now determined the structure of the unique crystal form that was obtained with PEG as precipitating agent and with 50 mM pyrophosphate added (Fig. 4C and D). The structure is virtually identical with the one reported earlier. The iron-sulfur cluster shows the [3Fe-4S] stoichiometry and the W1 water molecule. Interestingly, the active site cavity is furthermore densely packed with four additional water molecules. This result indicates, that the larger [4Fe-4S] cluster present in the active enzyme may prevent binding of pyrophosphate including its enlarged hydration sphere.
Structural Influence on Stability of the Iron-Sulfur Cluster.
The iron-sulfur cluster stoichiometries observed crystallographically with different ligands are highly reproducible. Notably, crystals obtained with IPP from two different enzyme batches always showed [3Fe-4S] clusters, and crystals from the same respective enzyme batches grown with the substrate, HMBPP, consistently showed [4Fe-4S] clusters. These observations prompted us to study the influence of the substrate, HMBPP, and one of the products, DMAPP on the stability of IspH. Whereas IspH protein is deactivated rapidly by molecular oxygen, we found that oxygen is tolerated for at least 1 d in the presence of HMBPP but in the absence of electron donors. Remarkably, IspH retained 95% activity in the presence of oxygen and HMBPP. On the other hand, DMAPP failed to protect IspH. Similar to IspH treated with oxygen in the absence of additives, only 5–7% activity was found. The stabilizing effect of the substrate appears to be directly related to the status of the iron-sulfur cluster as shown by rapid absorption changes at a wavelength of 410 nanometer (nm) (Fig. S4). Thus, it can be concluded that IspH is more stable in its closed conformation with a substrate molecule bound to the cluster, compared to its open conformation in solution.
Reaction Mechanism.
The crystal structures reported in this article lead to unique insights into the catalytic mechanism of IspH protein. From the structural data we draw the following conclusions:
(i) the central cavity is confirmed as the active site.
(ii) in line with earlier proposals, the IspH:HMBPP complex structure reveals complexation of the oxygen at C(1) with the apical iron atom of the [4Fe-4S] cluster (Fig. 2B). Since the substrate is not converted, the iron-sulfur cluster in this complex is assumed to be in the oxidized state.
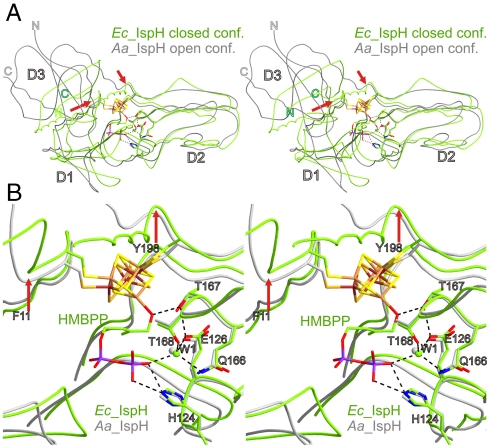
(iii) the oxygen at C(1) participates in a network of hydrogen bonds involving Thr167Oγ, Glu126Oϵ, the W1 water molecule and the β-phosphate residue of the substrate. Remarkably, the structural superposition of closed and opened IspH from E. coli and A. aeolicus (6) reveal similar conformations of all amino acids involved in the hydrogen-bonding network (Fig. 5). In contrast, amino acids coordinating the pyrophosphate side chains of bound ligands are structurally rearranged between open and closed conformation, except His124, which might play a crucial role for substrate docking. Thus, the major structural rearrangement of the domain D3 between the open and closed conformation does not allosterically affect the previously described hydrogen-bonding network, essential for catalysis (Fig. 5). Based on typical pKa values ([Fe(OH2)6]3+∶2.4, Glu: 4-6,
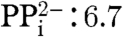 ) this oxygen is best considered as an alkoxide moiety coordinated to the apical ferric iron.
) this oxygen is best considered as an alkoxide moiety coordinated to the apical ferric iron.(iv) IspH protein requires an electron donor for catalytic turnover, but none was present in our experiments. However, when amorphously frozen IspH crystals are exposed to x-ray irradiations, even at cryogenic temperature, photo generated electrons can induce the cleavage of the carbon oxygen bond at C(1) after transfer to the bound substrate via the iron-sulfur cluster embedded in IspH.
(v) upon one electron reduction of the [4Fe-4S] cluster, the pKa of the iron-coordinated oxygen should increase markedly (pKa of [Fe(OH2)6]2+∶6.8) and hence should attract a proton within the hydrogen-bonding network. While Thr167Oγ may act as a proton relay Glu126Oϵ should serve as the ultimate proton donor inside the network; notably, the replacement of Glu126 by Gln or Asp results in catalytically inactive protein (5).
(vi) injection of one electron into the allylic system, possibly in conjunction with the transfer of a second proton to the hydroxyl group at C(1), results in cleavage of a carbon oxygen bond affording an allyl radical and a water molecule (Fig. 1). There is precedent for the facilitation of heterolytic dehydration reactions in nonactivated substrates by generation of a free radical state, e.g. by ribonucleotide reductase (13) and hydroxyglutarate dehydrogenase (14). For the transfer of the second proton, W1 and Glu126Oϵ may act as proton relay (distance OC(1)-Glu126Oϵ∶3.4), while the substrate’s pyrophosphate moiety could serve as proton donor (Fig. 3).
(vii) subsequent injection of a second electron converts the allyl radical into a strongly basic allyl anion, which might be stabilized by interaction with the apical iron center of the iron-sulfur cluster. Reprotonation has to occur from the diphosphate side (HSi-side), since the opposite face is blocked by the [4Fe- 4S] cluster. Proton transfer to either C(1) (affording DMAPP) or C(3) (affording IPP) may take place by abstraction of a proton either from the adjacent β-phosphate moiety or the released water molecule (Fig. 1). As discussed earlier (5), the substrate topology is consistent with the experimentally observed stereochemistry of IPP formation, since one side of the substrate is blocked by the iron-sulfur cluster.
(viii) reduction of HMBPP to IPP and DMAPP contributes two negative charges (two electrons transmitted via the iron-sulfur cluster) to the active site cavity, possibly inducing release of the product which is negatively charged as well.
Fig. 5.
Stereoview of the structural superposition of IspH protein from E. coli (Green) and A. aeolicus (Gray) comparing closed (E. coli) and open (A. aeolicus) conformations. Structural superposition was performed with a matching rate of 78% of Cα-atoms of the domains D2 with an rms deviation of 1.4 Å. (A) Overview of the orientation of the domains D1, D2, and D3; red arrows point to the hinge motive allowing the induced-fit of domain D3 upon substrate binding. (B) Close-up view and hydrogen-bonding network at the active site (black dashed lines, only drawn for HMBPP bound to IspH). Note, that amino acids involved in the hydrogen bond network are conserved over species and occupy identical locations within the crystal structures of open and closed conformation of E. coli and A. aeolicus.
The nature of the allylic intermediate as well as the character of its interaction with the apical iron center could not be deduced experimentally so far. Based on the structural information available from this study, the reaction sequence of the IspH catalyzed reductive dehydroxylation should be accessible using theoretical methods. Since IspH protein is a putative target for the treatment of malaria, tuberculosis and infections by a broad range of Gram-negative bacteria, such detailed knowledge of the reaction mechanism will facilitate the design and development of IspH inhibitors for antiinfective therapy.
Methods
Cell Culturing and Protein Purification.
The ispH gene was amplified from E. coli K12 genomic DNA by PCR and then ligated into the pQE30 Plasmid. The E126Q mutant was prepared as reported earlier (5) using mismatch PCR (15). The recombinant ispH gene was expressed under aerobic conditions in an E. coli XL-1 strain harboring the additional pACYCiscS-fdx plasmid (15) to ensure assembly of a functional iron-sulfur cluster using Terrific Broth (12 g peptone, 24 g yeast extract, 4 mL glycerol, 12 g KH2PO4 and 62 g K2HPO4 in 1 L | water)supplemented with ampicillin (180 mg/L), chloramphenicol (25 mg/L), 1 mM cysteine and ferric ammonium citrate (35 mg/L). Cells were grown over night at 30 °C, harvested, washed with degassed saline in a glove box, and stored anaerobically at -20° C. The glove box contained an atmosphere of 95% nitrogen and 5% hydrogen. Residual oxygen was removed by a palladium catalyst. Frozen cell paste was resuspended in 150 mL anaerobic lysis buffer (100 mM Tris/HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole, 5 mM dithionite). Prior to flushing the French Press with anaerobic lysis buffer, the inlet and outlet of the French Press were set under argon atmosphere. The suspension was passed through the French Press and was centrifuged subsequently. Centrifuge bottles contained cell lysate with 5 mM dithionite and an argon gas phase. The resulting crude extract showed intense brown color; the protein concentration was about 30 mg/mL. The solution was placed on a column of Ni2+ Chelating Sepharose Fast Flow that was then developed with 150 mM imidazole. Fractions were combined, and the solution was passed through a column of Sepharose Q that was then developed with 300 mM KCl yielding protein with an approximate purity of 99%. All buffers and saline were boiled for 20 min to get rid of oxygen. Dithionite only was added to the lysis buffer. Fractions were combined and dialyzed against 50 mM Tris/HCl, pH 8.0 over night.
Enzyme Assay.
All assays were carried out under strictly anaerobic conditions. Reaction mixtures contained 50 mM Hepes/NaOH, pH 7.5, 3 mM dithionite, 400 μM methyl viologen, 3 mM HMBPP, and IspH protein. The reaction was monitored photometrically at 310 nm and 30 °C.
Crystallization and Structure Determination.
All manipulations during crystallization were carried out under anaerobic conditions in a glove box under an atmosphere of 95% N2 and 5% H2. Crystallization plates were put into the glove box at least one week prior to usage. Buffers for crystallization trials were degassed under vacuum. Crystals were grown by using the sitting drop vapour diffusion method at 20 °C. IspH protein concentration used for crystallization was 35 mg/mL in 50 mM Tris/HCl (pH 8.0). Crystals were obtained in drops containing 4 μL of protein solution and 4 μL of reservoir solution (100mM BIS-Tris/HCl, pH 6.5, 200 mM lithium sulfate and 25% PEG3350). The final pH value was 6.8. IspH crystals in complex with ligands were prepared by cocrystallization (the concentrations of the ligands were about 50 mM). Crystals grew to a final size of 400 × 300 × 500 μm3 within 10 d. Crystals were soaked for 15 min in 2 M lithium sulfate, pH 6.5, and were subsequently shock-frozen in a stream of nitrogen gas at 100 K (Oxford Cryo Systems). Native datasets were collected using either synchrotron radiation at the X06SA-beamline, SLS, Villigen, Switzerland or an in-house CuKα rotating anode. Collected data were processed using the program package XDS (16). IspH protein crystallized in the orthorombic space group P212121 with cell parameters of about a = 71 Å, b = 81 Å and c = 112 Å, with 46% solvent content. For more details and crystal parameters of space group C2 see Table S1. Crystal structure analysis was performed by molecular replacement using coordinates of IspH from E. coli deposited at the Protein Data Bank (PDB ID: 3DNF) (5). The anisotropy of diffraction was corrected by an overall anisotropic temperature factor by comparing observed and calculated structure amplitudes using the program Crystallography and NMR system, (CNS) (17). Electron density was improved by averaging and back transforming the reflections 10 times over the twofold noncrystallographic symmetry axis using the program package MAIN (18). Conventional crystallographic rigid body, positional, and temperature factor refinements were carried out with CNS. The model was completed using the interactive three-dimensional graphic program MAIN.
The atomic coordinates for IspH in complex with the various ligands have been deposited with the Protein Data Bank, Research Collaboratory for Structural Bioinformatics at Rutgers University (IspH:HMBPP: PDB ID 3KE8; IspH:Int.: PDB ID 3KE9; IspH:IPP: PDB ID 3KEM; IspH:DMAPP: PDB ID 3KEF; IspH: PP: PDB ID 3KEL).
Supplementary Material
Acknowledgments.
We are grateful to our technician Katrin Gärtner for cloning of IspH mutants and to the staffs of PXII at the Paul Scherrer Institute, Swiss Light Source, Villigen, Switzerland, in particular Clemens Schulze-Briese, for help during data collection. Special thanks to Patrick Schreiner, Stephan Krapp, Mario Moertl, Jark Boettcher, and Claus Kuhn, Proteros Biostructures GmbH, Martinsried, Germany, for enabling several synchrotron trips and for collecting datasets. We thank the Hans-Fischer Gesellschaft and the Stifterverband für die Deutsche Wissenschaft (Projekt-Nr. 11047: Forschungsdozentur Molekulare Katalyse) for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913045107/DCSupplemental.
References
- 1.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae, and higher plants. Nat Prod Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 2.Rohdich F, Hecht S, Bacher A, Eisenreich W. Deoxyxylulose phosphate pathway of isoprenoid biosynthesis. Discovery and function of ispDEFGH genes and their cognate enzymes. Pure Appl Chem. 2003;75:393–405. [Google Scholar]
- 3.Rohdich F, et al. Studies on the nonmevalonate terpene biosynthetic pathway: Metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altincicek B, et al. LytB protein catalyzes the terminal step of the 2-C-methyl–erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett. 2002;532:437–440. doi: 10.1016/s0014-5793(02)03726-2. [DOI] [PubMed] [Google Scholar]
- 5.Gräwert T, et al. Structure of Active IspH Enzyme from Escherichia coli Provides Mechanistic Insights into Substrate Reduction. Angewandte Chemie International Edition. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]
- 6.Rekittke I, et al. Structure of (E)-4-Hydroxy-3-methyl-but-2-enyl Diphosphate Reductase, the Terminal Enzyme of the Non-Mevalonate Pathway. J Am Chem Soc. 2008;130:17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seemann M, et al. Isoprenoid Biosynthesis via the MEP Pathway: In Vivo Mössbauer Spectroscopy Identifies a [4Fe-4S]2+ Center with Unusual Coordination Sphere in the LytB Protein. J Am Chem Soc. 2009;131:13184–13185. doi: 10.1021/ja9012408. [DOI] [PubMed] [Google Scholar]
- 8.Röhrich RC, et al. Reconstitution of an apicoplast-localized electron transfer pathway involved in the isoprenoid biosynthesis of Plasmodium falciparum. FEBS Lett. 2005;579:6433–6438. doi: 10.1016/j.febslet.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Rohdich F, et al. The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: Studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc Natl Acad Sci USA. 2003;100:1586–1591. doi: 10.1073/pnas.0337742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Liu P. IspH Protein of the Deoxyxylulose Phosphate Pathway: Mechanistic Studies with C1-Deuterium-Labeled Substrate and Fluorinated Analogue. Angew Chem Int Ed. 2008;47:9722–9725. doi: 10.1002/anie.200803452. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Chu L, Sanakis Y, Liu P. Revisiting the IspH Catalytic System in the Deoxyxylulose Phosphate Pathway: Achieving High Activity. J Am Chem Soc. 2009;131:9931–9933. doi: 10.1021/ja903778d. [DOI] [PubMed] [Google Scholar]
- 12.Bolm C, Legros J, Le Paih J, Zani L. Iron-Catalyzed Reactions in Organic Synthesis. Chem Rev. 2004;104:6217–6254. doi: 10.1021/cr040664h. [DOI] [PubMed] [Google Scholar]
- 13.Ramos MJ, Fernandes PA. Computational Enzymatic Catalysis. Acc Chem Res. 2008;41:689–698. doi: 10.1021/ar7001045. [DOI] [PubMed] [Google Scholar]
- 14.Buckel W, Martins BM, Messerschmidt A, Golding BT. Radical-mediated dehydration reactions in anaerobic bacteria. Biol Chem. 2005;386:951–959. doi: 10.1515/BC.2005.111. [DOI] [PubMed] [Google Scholar]
- 15.Gräwert T, et al. IspH Protein of Escherichia coli: Studies on Iron-Sulfur Cluster Implementation and Catalysis. J Am Chem Soc. 2004;126:12847–12855. doi: 10.1021/ja0471727. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 17.Brünger A, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;1:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Turk D. Technische Universitaet Muenchen; 1992. Improvement of a programm for molecular graphics and manipulation of electron densities and its application for protein structure determination. Thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.