Abstract
The herbicide glyphosate became widely used in the United States and other parts of the world after the commercialization of glyphosate-resistant crops. These crops have constitutive overexpression of a glyphosate-insensitive form of the herbicide target site gene, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS). Increased use of glyphosate over multiple years imposes selective genetic pressure on weed populations. We investigated recently discovered glyphosate-resistant Amaranthus palmeri populations from Georgia, in comparison with normally sensitive populations. EPSPS enzyme activity from resistant and susceptible plants was equally inhibited by glyphosate, which led us to use quantitative PCR to measure relative copy numbers of the EPSPS gene. Genomes of resistant plants contained from 5-fold to more than 160-fold more copies of the EPSPS gene than did genomes of susceptible plants. Quantitative RT-PCR on cDNA revealed that EPSPS expression was positively correlated with genomic EPSPS relative copy number. Immunoblot analyses showed that increased EPSPS protein level also correlated with EPSPS genomic copy number. EPSPS gene amplification was heritable, correlated with resistance in pseudo-F2 populations, and is proposed to be the molecular basis of glyphosate resistance. FISH revealed that EPSPS genes were present on every chromosome and, therefore, gene amplification was likely not caused by unequal chromosome crossing over. This occurrence of gene amplification as an herbicide resistance mechanism in a naturally occurring weed population is particularly significant because it could threaten the sustainable use of glyphosate-resistant crop technology.
Keywords: 5-enolpyruvylshikimate-3-phosphate synthase, herbicide resistance, mobile genetic element, evolution, Palmer amaranth
Global adoption of transgenic crops has been rapid, reaching 120 million ha in 2008. Approximately 85% of this area has been planted with herbicide-resistant crops, nearly all of which are glyphosate-resistant (1). Evolution of resistance to the widely used, nonselective herbicide glyphosate (N-[phosphonomethyl] glycine) in weedy species endangers the continued success of transgenic glyphosate-resistant crops and the sustainability of glyphosate as the world’s most important herbicide (2). Since commercialization of glyphosate-resistant cotton in the U.S. in 1997, some growers have relied exclusively on multiple glyphosate applications each season in a monoculture system to manage weeds including Amaranthus palmeri (Palmer amaranth) (3). A. palmeri is dioecious (4) and is an economically troublesome weed threatening the sustainability of cotton production in the southeastern United States (5), where glyphosate has been the principal tool for A. palmeri control since 1997. Unfortunately, glyphosate resistance has now evolved in A. palmeri populations within glyphosate-resistant cotton fields reported in Georgia (3), Tennessee (6), North Carolina (7), South Carolina (8), and Arkansas (9). In 2009, glyphosate-resistant A. palmeri was projected to occur on at least 250,000 ha of crop land (8).
The molecular target of glyphosate (10) is the chloroplast-targeted enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS, EC 2.5.1.19), a component of the shikimate pathway (11). In crop species, resistance to glyphosate has been conferred by expression of bacterial genes that metabolize glyphosate (12), overexpression of sensitive EPSPS, expression of glyphosate-resistant EPSPS from bacteria, and expression of glyphosate-resistant plant EPSPS containing one or more target-site mutations (13). After step-wise glyphosate selection, EPSPS gene amplification has occurred in plant cell lines, resulting in glyphosate resistance in cell culture (12).
Glyphosate resistance has been confirmed in 16 weed species as of 2009 (14). In weed species that have evolved glyphosate resistance, the resistance mechanisms thus far elucidated are reduced glyphosate translocation and/or target-site mutations in the EPSPS gene (15). Reduced glyphosate translocation is a common resistance mechanism in Conyza canadensis and Lolium rigidum and this mechanism provides a higher level of resistance (7- to 11-fold) than do known EPSPS mutations in weedy species (16). EPSPS mutations at Pro106 (using the maize mature EPSPS numbering system) confer glyphosate resistance in several glyphosate-resistant weed species, including Eleusine indica (17), L. rigidum (18), and L. multiflorum (19). The lower levels of resistance (2- to 3-fold) provided by the Pro106 mutations are sufficient for weeds to survive typical glyphosate application rates (18). To date, increased EPSPS expression has not been identified as a resistance mechanism in glyphosate-resistant weeds.
Crop yield loss due to A. palmeri is particularly problematic (20), in part because A. palmeri populations previously evolved herbicide resistance to photosystem II inhibitors, acetolactate synthase (ALS) inhibitors, and dinitroanilines (21). The first reported glyphosate-resistant A. palmeri population was 6- to 8-fold more resistant than a susceptible population (3), and the glyphosate resistance mechanism in this population was previously unknown but is not due to differences in absorption or translocation of glyphosate (3). The mechanism is also not due to a ploidy change (3), because glyphosate-resistant individuals had the reported A. palmeri genome size (22). Here, we use genetic and molecular analyses of EPSPS genes and proteins from glyphosate-resistant and -susceptible A. palmeri populations and demonstrate that amplification of the EPSPS gene is the glyphosate resistance mechanism.
Results
EPSPS cDNA Sequencing.
Target site mutations in the EPSPS gene confer 2- to 3-fold glyphosate resistance in several other weedy species (15). To determine whether a target site mutation was present in glyphosate-resistant A. palmeri, full-length cDNA of EPSPS was obtained by PCR from seven glyphosate-resistant (R) and two glyphosate-susceptible (S) A. palmeri plants collected from Georgia (United States). Sequence analysis did not reveal mutation in the R cDNA at the Pro106 residue known to confer glyphosate resistance in other weed species (Fig. S1). An SNP occurred in position 316 of all EPSPS fragments from R individuals (Fig. S1), resulting in a substitution of a lysine for arginine. Some plant species susceptible to glyphosate contain a lysine at this position, suggesting that this polymorphism is not conferring glyphosate resistance.
Effect of Glyphosate on EPSPS cDNA and Shikimate Levels.
Shikimate accumulates in plants when EPSPS is inhibited by glyphosate because shikimate-3-phosphate, a substrate in the reaction catalyzed by EPSPS, converts to shikimate and accumulates faster than it can be consumed in other metabolic pathways (11). Glyphosate R and S plants originating from Georgia populations were sampled for shikimate accumulation and RNA before and 8 h after treatment (HAT) with water or 0.4 kg ha−1 glyphosate. The S plants accumulated shikimate after glyphosate treatment, whereas R plants did not (Table 1). Using quantitative RT-PCR, EPSPS transcript abundance was measured relative to ALS (EC 4.1.3.18), a low-copy gene with known monogenic inheritance in Amaranthus species (23). Compared with S plants, R plants had, on average, 35-fold higher EPSPS expression relative to ALS (Table 1), and expression was unaffected by glyphosate treatment.
Table 1.
Expression of EPSPS cDNA in glyphosate-resistant and -susceptible A. palmeri is not affected by glyphosate treatment
| Biotype | Glyphosate | Shikimate 8 HAT (Δ ng shikimate μL−1) | EPSPS expression relative to ALS 8 HAT [2(ΔCt)] |
| Susceptible | − | 0.5 (0.3) | 0.8 (0.1) |
| Susceptible | + | 15.0 (1.8) | 0.8 (0.1) |
| Resistant | − | −0.9 (0.6) | 35.1 (4.7) |
| Resistant | + | −0.5 (0.3) | 35.0 (5.7) |
EPSPS cDNA was measured relative to ALS using quantitative PCR and expressed as 2^ΔCt (threshold cycle), where ΔCt = (Ct, ALS − Ct, EPSPS). The + glyphosate data were obtained 8 HAT with 0.4 kg ha−1 glyphosate, and the − glyphosate data were obtained 8 HAT with water. Means and standard errors (in parentheses) are from two experimental runs with four biologic replicates each.
EPSPS Gene Copy Number Correlates with Glyphosate Resistance.
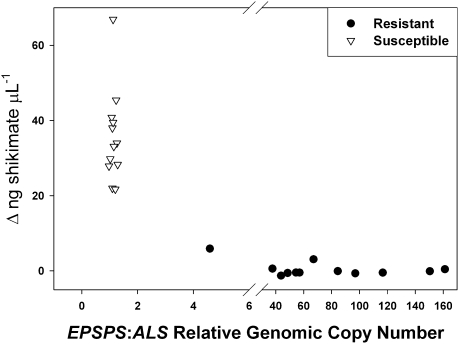
DNA blot hybridizations indicated an increase in EPSPS copy number in R relative to S plants (Fig. S2). We used quantitative PCR to more accurately measure relative genomic copy numbers of the EPSPS gene relative to ALS in R and S individuals. Genomic EPSPS copy numbers relative to ALS ranged from 1.0 to 1.3 (n = 12) for S plants, whereas relative copy numbers for R plants were much higher, varying from 5 to more than 160 (n = 12) (Fig. 1).
Fig. 1.
Increase in genomic copy number of EPSPS correlates with reduced shikimate accumulation in 12 individuals each of glyphosate-resistant (filled circles) and -susceptible (open triangles) A. palmeri plants. Increase in genomic copy number of EPSPS is relative to ALS as measured using quantitative PCR on genomic DNA. Shikimate accumulation was measured after incubation in 250 μM glyphosate in an in vivo leaf disk assay.
In a leaf disk assay using 250 μM glyphosate, all 12 S plants accumulated shikimate, an indication that EPSPS was inhibited, whereas 10 of 12 R plants did not accumulate shikimate, indicating that EPSPS was still functioning (Fig. 1). The R plant with the lowest relative EPSPS copy number accumulated a modest amount of shikimate, the R plant with a relative EPSPS copy number of 65 accumulated shikimate to levels only slightly above background, and both accumulated much less shikimate than the S plants (Fig. 1).
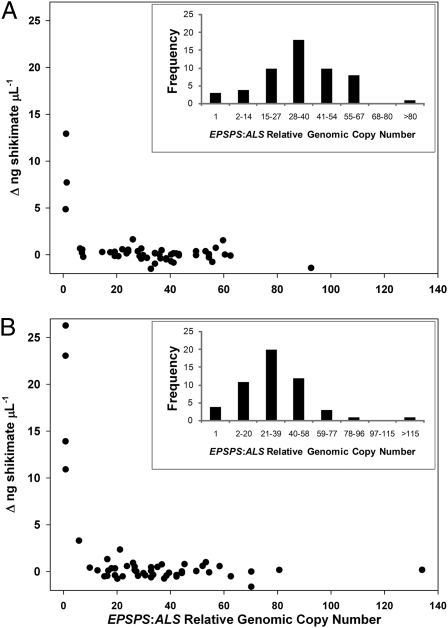
To determine whether the association between glyphosate resistance and increased EPSPS copy number was heritable, two pseudo-F2 populations were generated, one by hand-pollinating and one by open-pollinating F1 plants that were verified resistant by treatment with 0.4 kg ha−1 glyphosate. The F1 plants had a glyphosate R male parent and an S female parent. EPSPS relative copy number was determined for the parents of the hand-pollinated pseudo-F2 population, in which the F1 male parent had 18 relative EPSPS copies and the F1 female parent had 39 relative EPSPS copies. The pseudo-F2 populations segregated for both relative EPSPS copy number and glyphosate resistance, and these two traits were strongly associated (Fig. 2 A and B). Relative EPSPS copy number ranged from one to greater than the sum of copy numbers from both parents (Fig. 2A). Generally, pseudo-F2 individuals with increased copy number did not accumulate shikimate at 250 μM glyphosate, indicating that they were resistant to that glyphosate dose, although a few individuals with >20 relative copies accumulated shikimate at levels slightly higher than background after treatment with 250 μM glyphosate. All pseudo-F2 individuals with 1 relative EPSPS copy were distinguishable by high shikimate accumulation, indicating that they were susceptible to glyphosate and that the population was segregating for glyphosate resistance (Fig. 2 A and B).
Fig. 2.
EPSPS genomic copy number and glyphosate resistance cosegregate in pseudo-F2 A. palmeri populations. EPSPS copy number relative to ALS and accumulation of shikimate were determined as described in Materials and Methods. Insets: Relative copy number histograms in pseudo-F2 populations generated using (A) hand pollination (F1 male parent 18 relative EPSPS copies and F1 female parent 39 relative EPSPS copies) and (B) open pollination (parental relative copy number not measured).
EPSPS Transcript Abundance Correlates with EPSPS Genomic Copy Number.
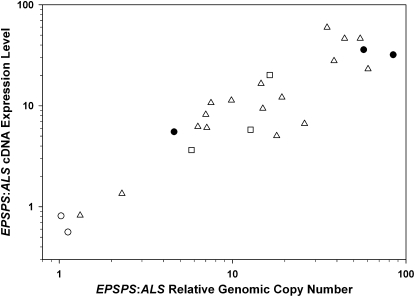
Selected individuals from pseudo-F2 and parental populations were measured for EPSPS transcript accumulation using quantitative RT-PCR. Plants with a relative EPSPS:ALS genomic copy number of 1:1 had a relative EPSPS:ALS transcript abundance of ≈1:1 (Fig. 3), whereas plants with increased relative EPSPS genomic copy number had increased EPSPS relative transcript abundance (Fig. 3). There was a strong correlation (r = 0.76, P < 0.0001) between relative EPSPS genomic copy number and transcript abundance (Fig. 3).
Fig. 3.
Increase in EPSPS genomic relative copy number is positively correlated with increase in EPSPS cDNA expression levels in selected A. palmeri glyphosate-resistant (filled circles), -susceptible (open circles), and pseudo-F2 individuals [pseudo-F2 hand pollinated (open triangles), open pollinated (open squares)]. Genomic copy numbers and expression levels are relative to ALS and were determined by quantitative PCR as described in Materials and Methods.
EPSPS Quantity and Activity Correlate with EPSPS Genomic Copy Number.
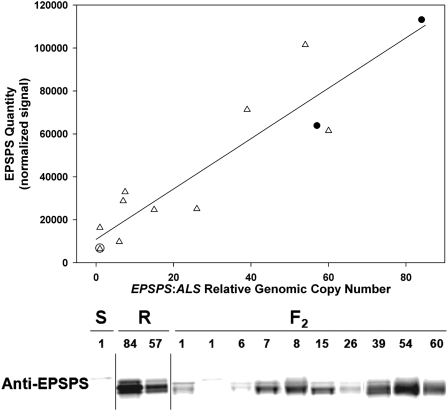
EPSPS protein quantity was measured with immunoblotting. The EPSPS signal in plants with increased EPSPS relative copy number rapidly saturated, preventing quantification relative to plants with lower copy number. Thus, we loaded half as much total soluble protein (TSP) for plants with >20 relative EPSPS copies as for plants with <20 relative copies. EPSPS signal intensity had a significant positive relationship (R2 = 0.85, P < 0.0001) with EPSPS genomic copy number in S, R, and pseudo-F2 plants (Fig. 4).
Fig. 4.
EPSPS protein levels in glyphosate-susceptible (S), glyphosate-resistant (R), and pseudo-F2 A. palmeri plants are correlated with relative EPSPS genomic copy number. Top: Regression of normalized EPSPS quantity on increase in relative EPSPS genomic copy number; open circles: S; filled circles: R; open triangles: F2. Bottom: Samples with <20 relative EPSPS copies had 30 μg TSP loaded per lane, and samples with >20 relative EPSPS copies had 15 μg TSP loaded per lane. Increase in relative EPSPS genomic copy number is indicated above each sample lane.
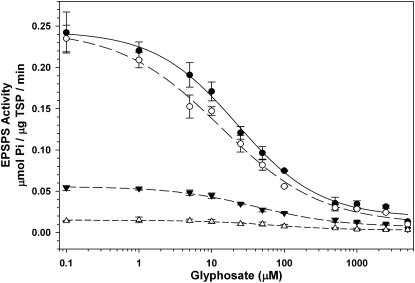
We conducted an EPSPS activity assay to compare the EPSPS activity in resistant pseudo-F2 plants with increased EPSPS genomic copy number relative to a susceptible pseudo-F2 plant with no increase in relative copy number. EPSPS activity was measured using phosphate released by EPSPS and was much lower in the susceptible pseudo-F2 plant than in resistant pseudo-F2 plants (Fig. 5), because the pseudo-F2 plant with 54 relative copies exhibited ≈20 times more EPSPS activity than the plant with 1 relative copy. The IC50 values (glyphosate dose that inhibited 50% of EPSPS activity) for three samples with greater than 1 relative EPSPS copy were lower but not statistically different (α = 0.05) from the IC50 for the sample with 1 relative copy (Fig. 5), indicating that EPSPS from plants with increased EPSPS copy number is as sensitive to glyphosate inhibition as EPSPS from plants lacking increased EPSPS copies.
Fig. 5.
Increased EPSPS enzyme activity is positively correlated with EPSPS relative genomic copy number in four pseudo-F2 A. palmeri plants. Glyphosate inhibition assays were normalized for TSP quantity. Data points are means and standard errors of three replications. Filled circles: 54 relative EPSPS copies, IC50 (glyphosate concentration that reduced enzyme activity by 50%) = 22 μM; open circles: 39 relative copies, IC50 = 15 μM; filled triangles: 8 relative copies, IC50 = 36 μM; open triangles: 1 relative copy, IC50 = 66 μM.
Distribution of the Amplified EPSPS Gene in the A. palmeri Genome.
We used FISH to determine the chromosomal locations and distributions of the amplified EPSPS genes. A 1-kb fragment from the EPSPS gene was used as the FISH probe. The reported chromosome number for A. palmeri is 2n = 34 (24), and we observed FISH signals dispersed throughout the genome in an R individual (Fig. 6). A uniform FISH signal pattern was also observed in most interphase nuclei (Fig. 6). These results suggest that the amplified EPSPS genes were randomly inserted into the A. palmeri genome. In contrast, we did not observe unambiguous FISH signals in the majority of metaphase or interphase cells prepared from an S individual. One or few putative FISH signal spots were observed in some interphase nuclei from the S individual. However, it was technically difficult to distinguish such putative FISH spots from background signals. The EPSPS copy number was not characterized in the R individual plant, but the average copy number in R plants (n = 12) was 77 (±14) (Fig. 1).
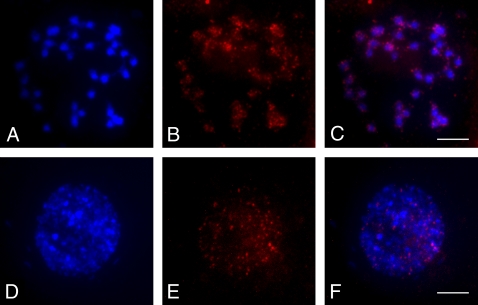
Fig. 6.
FISH mapping in an A. palmeri glyphosate-resistant individual. (A) Somatic metaphase chromosomes of glyphosate-resistant A. palmeri. (B) FISH signals from the EPSPS gene probe. (C) Merged image from A and B. Dispersed signals can be observed on every chromosome. (D) An interphase nucleus of glyphosate-resistant A. palmeri. (E) FISH signals from the EPSPS gene probe. (F) Merged image from D and E. (Scale bars, 5 μm.)
Discussion
We demonstrate that the recent evolution of glyphosate resistance in a weed population is due to EPSPS gene amplification and increased EPSPS expression. Increased expression of EPSPS as a molecular glyphosate resistance mechanism has been reported to endow relatively low-level glyphosate resistance in laboratory studies (25–27), but this report concerns a field weed population. The data reported here indicate that an EPSPS gene amplification in glyphosate-resistant A. palmeri from Georgia results in high levels of EPSPS expression and that this mechanism imparts high-level glyphosate resistance. The gene amplification is not due to genome duplication (i.e., ploidy change) (3). EPSPS is normally a low-copy gene in plants: rice has one EPSPS locus (28) and Arabidopsis has two loci (29).
It is unknown whether the EPSPS gene amplification existed within the A. palmeri population in Georgia before glyphosate selection pressure or whether EPSPS gene amplification occurred during a period of <7 years over which glyphosate was repeatedly applied. Interesting future questions include whether other loci are duplicated and whether this A. palmeri biotype carries a genetic trait that endows high levels of gene amplification without an increase in chromosome number. FISH analysis revealed that the amplified EPSPS genes were dispersed throughout the genome. Lack of large tandem arrays of the EPSPS gene suggests that the amplification is not due to unequal crossing-over or rolling circle replication–based mechanisms. The high number of copies and their location throughout the genome suggest that the amplification could have originated via a transposon- or RNA-mediated mechanism, followed by selection of a highly amplified individual from the population. Most transposons in plant genomes are inactive but may be activated by various conditions, including abiotic stress (30). Therefore, a testable hypothesis is that the original EPSPS locus was associated with a mobile genetic element that activated and amplified the EPSPS gene.
The most common glyphosate resistance mechanism selected in plant cell culture is increased EPSPS activity, typically due to gene amplification (12). There is evidence for enhanced EPSPS expression in glyphosate-resistant weeds, but no previous evidence for EPSPS gene amplification. Two- to threefold elevated EPSPS expression and enzyme activity were found in glyphosate-resistant L. rigidum, and EPSPS from glyphosate-resistant and -susceptible plants were equally sensitive to glyphosate (31). However, the elevated expression was not due to gene amplification because EPSPS gene copy number in L. rigidum was examined using DNA blot hybridizations and glyphosate-resistant lines did not have increased EPSPS gene copy number in comparison with glyphosate-susceptible lines. In glyphosate-resistant biotypes of C. canadensis and C. bonariensis, basal EPSPS mRNA levels were double the levels in susceptible biotypes, but the resistant biotypes also had reduced glyphosate translocation (32, 33).
In our studies of a segregating A. palmeri pseudo-F2 population, increasing EPSPS gene copy number correlated with increased EPSPS mRNA, increased EPSPS protein activity, and glyphosate resistance. The higher quantity of EPSPS in glyphosate-resistant pseudo-F2 plants was equally sensitive to glyphosate inhibition as EPSPS from glyphosate-susceptible pseudo-F2 plants, in contrast with E. indica, in which the IC50 for glyphosate-resistant lines with a Pro106 mutation was 5-fold higher than in S lines (17).
EPSPS protein levels and activity both increased as the number of EPSPS genomic copies increased. Therefore, the effect of additional EPSPS copies is additive, and additional copies confer higher levels of resistance. We measured the resistance phenotype with 250 μM glyphosate in an in vivo leaf disk assay, and this dose did not induce shikimate accumulation in most individuals with EPSPS gene amplification. This result should not be interpreted to indicate that plants with a 20-fold increase in copy number are as resistant as plants with a 60- or 100-fold increase in copy number. EPSPS activity can be reduced to nearly zero in plants with increased copy number, but the dose required to eliminate EPSPS activity increases with increasing copy number, indicating that additional EPSPS gene copies have an additive effect in conferring resistance.
The stability of EPSPS gene amplification in A. palmeri is unknown, because the extent of EPSPS gene amplification varied greatly in plants from the R field population. Additionally, one individual in an A. palmeri pseudo-F2 had a higher relative EPSPS copy number than the sum of the relative copy number from both parents, indicating that additional copies may be gained during recombination. Even if the EPSPS gene amplification is unstable during sexual recombination, apomixis may occur in A. palmeri (34), which could function to maintain the large amplification in the population. Further contributing to the dynamics of EPSPS copy number, its amplification and increased expression could have a fitness penalty in the resistant biotype in the absence of glyphosate selection (35).
Although not previously reported in naturally occurring plant populations, large gene amplifications that confer resistance to xenobiotic compounds have occurred in other organisms. Large tandem gene amplifications of metabolic genes confer insecticide resistance in Culex mosquitoes and Myzus aphids (36, 37). Organophosphate-resistant mosquitoes had ≈80-fold more copies of esterase genes than susceptible mosquitoes (37). Resistance to methotrexate in mammalian cancer cells is due to overproduction of the target enzyme, dihydrofolate reductase, from gene amplification (38). This adaptation occurred during step-wise selection with increasing methotrexate doses and resulted in gene amplification and overproduction of normal dihydrofolate reductase.
Our data demonstrate that glyphosate resistance in a Georgia A. palmeri population is due to many-fold amplification of the EPSPS gene on multiple chromosomes. This occurrence of gene amplification as an herbicide resistance mechanism was observed in a naturally occurring weed population. It remains to be seen whether the same mechanism exists in other glyphosate-resistant A. palmeri populations or in other glyphosate-resistant species. The occurrence of the EPSPS gene amplification in A. palmeri raises many questions about how the amplification occurred initially and has been subsequently maintained, including the frequency of other gene amplifications across the genome and the role of this process in the evolution of A. palmeri as an economically damaging weed with a history of multiple herbicide resistance traits.
Materials and Methods
Plant Material and Genetic Populations.
Seeds of R A. palmeri were collected from a field site in Macon County, Georgia (3), whereas seeds of a known S A. palmeri population were collected from the University of Georgia Ponder Farm Research Station. Seeds of R and S were germinated and transplanted into large pots for growth in a greenhouse. The resistance phenotype of each plant was confirmed using an in vivo leaf disk assay (39). Each plant was covered with pollination bags before flowering. R males were placed next to S females to create an F1 generation (S/R). Plants were shaken daily to ensure adequate pollination.
Seeds from the S female plants were stored at 4°C for 2 months, then germinated and grown to the four-leaf stage. These S/R F1 plants were sprayed with a low rate (0.4 kg ae ha−1) of formulated glyphosate (potassium salt, Roundup Weather Max, Monsanto) to select for heterozygous resistant progeny, because apomixis may occur in A. palmeri (34). One R F1 male was selected for hand crossing to one R F1 female to generate a hand-pollinated pseudo-F2 through half-sibling mating. Both parents of the hand-pollinated pseudo-F2 were sampled for DNA extraction (see below). Pollination bags were placed over female inflorescences before emergence, and pollen from the resistant male was applied by hand daily for 2 weeks. An open-pollinated pseudo-F2 population was generated by placing different R female and male half-siblings from the S/R F1 next to each other in the greenhouse. Seeds from female plants were stored at 4°C for 2 months.
EPSPS cDNA Sequencing.
The EPSPS sequence from A. tuberculatus (FJ869880) was obtained by 5′ and 3′ RACE (40) and used to design primers for A. palmeri EPSPS. The following primer sets were used to amplify overlapping fragments of the central, 5′, and 3′ regions, respectively, of the EPSPS gene from resistant and susceptible cDNA: EPSF1 (5′-ATGTTGGACGCTCTCAGAACTCTTGGT-3′) EPSR1 (5′-GTCATAAGTTTCAATGGCGGTGG-3′); EPSF5 (5′-GCCAAGAACACAAAGCGAAATTCAGAG-3′) × EPSR5 (5′-TCTTTACCAACAGGAAACAGACCACCAC-3′); and EPSF6 (5′-CAGGGAATCATCTGGAAGGAAACATTTG-3′) × EPSR6 (5′- CTATTAGTCTCAAATCAAAACCTTCGGCG-3′). PCRs contained 1 μL cDNA; 400 nM each of forward and reverse primers; 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 1.5 mM MgCl2; and 1 U of high-fidelity Taq polymerase (Invitrogen) with a 1× concentration of supplied buffer in a final volume of 25 μL. The thermoprofile included 5 min at 94°C followed by 30 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C, with a final extension of 10 min at 72°C. The EPSF1 × EPSR1 PCR product contained the Pro106 codon. Seven R individuals and two S individuals were used. The EPSF1 × EPSR1 PCR product was ligated into pGEM-T Easy plasmids (Promega). Plasmids were transformed into Escherichia coli cells, and transformed cells were cultured overnight in liquid LB media. Plasmids from six clones of each individual were isolated for Sanger sequencing using the M13F and M13R primers. EPSF5 × EPSR5 and EPSF6 × EPSR6 PCR products were isolated by gel electrophoresis for direct sequencing. Consensus sequences for each biotype were assembled using Lasergene v. 7.0 SeqMan (DNASTAR). Multiple sequence alignments of plant EPSPS, including selected accessions from GenBank, A. tuberculatus, and both A. palmeri biotypes were constructed using ClustalW2 (European Bioinformatics Institute).
Effect of Glyphosate on EPSPS cDNA and Shikimate Levels.
Seeds from the R and S populations were germinated in small pots and grown to the four-leaf stage. Five plants each of R and S were sampled for one 4-mm leaf disk for in vivo measurement of background absorbance in a leaf disk shikimate assay (39) and one leaf disk for RNA extraction (see below). Four plants each of R and S were then treated with 0.4 kg ae ha−1 glyphosate, and one plant of each was treated with water. At 8 HAT, all plants were again sampled for one leaf disk for shikimate measurement and one leaf disk for RNA extraction, to measure EPSPS cDNA expression level (see below). The 8 HAT leaf disk samples were taken from both sides of the midvein at the base of the leaf, and the 0 HAT leaf disk samples were taken distal to the 8 HAT location. The experiment was conducted twice.
EPSPS Gene Copy Number.
Seeds from the hand-pollinated and open-pollinated pseudo-F2 populations, along with R and S seeds, were germinated and grown in small pots. Fifty-four plants of each pseudo-F2 and 12 plants each of R and S were grown to the four-leaf stage. One leaf of each plant was used for an in vivo leaf disk shikimate accumulation assay (39) with glyphosate concentrations of 0 and 250 μM in 10 mM ammonium phosphate buffer. A shikimate standard curve was used to calculate the ng shikimate μL−1 accumulation above the background level. Each plant was assayed in duplicate. One leaf from each plant was sampled for genomic DNA extraction and one leaf for RNA extraction for subsequent measurement of genomic EPSPS copy number and EPSPS cDNA expression level (see below).
DNA and RNA Extraction and cDNA Synthesis.
Tissue samples were immediately frozen in liquid nitrogen, ground in a 1.5-mL microcentrifuge tube, and stored at −80°C. Genomic DNA was extracted using the Qiagen DNEasy Plant Mini Kit (Qiagen), quantified using a NanoDrop spectrophotometer (Thermo Scientific), and checked for quality by gel electrophoresis. DNA concentrations were adjusted to 1 ng μL−1 in sterile HPLC grade water. RNA was extracted using TRIzol reagent (Invitrogen), dissolved in sterile HPLC water, quantified using a NanoDrop spectrophotometer, and checked for quality and integrity by gel electrophoresis.
A. palmeri RNA (200 ng for time course treatments and 700 ng from each of 20 pseudo-F2 individuals) was used for cDNA synthesis with oligo-DT primers and the Verso cDNA kit (Thermo Scientific). This kit includes a DNase treatment. Final cDNA volume was 20 μL.
Quantitative PCR.
Quantitative real-time PCR was used to measure EPSPS genomic copy number relative to ALS and cDNA expression level of EPSPS relative to ALS. Primer efficiency curves were conducted for each primer set using a 1×, 1/5×, 1/25×, and 1/125× dilution series of resistant genomic DNA. The primer sets EPSF1 × EPSR8 (5′- TGAATTTCCTCCAGCAACGGCAA-3′) (195-bp product) and ALSF2 (5′-GCTGCTGAAGGCTACGCT-3′) × ALSR2 (5′- GCGGGACTGAGTCAAGAAGTG-3′) (118-bp product) were used for quantitative PCR on genomic DNA and cDNA. ALS primers were designed on the basis of conserved regions of published plant ALS gene sequence (41).
Triplicate genomic DNA templates (10 ng) or triplicate cDNA templates (1 μL) were amplified in a 25-μL reaction volume using Syber-Green master mix (Bio-Rad Laboratories) by the following thermoprofile on a MyiQ real-time PCR detection system (Bio-Rad): 95°C for 15 min, then 30 cycles of 95°C for 30 s and 60°C for 1 min. Real-time fluorescence data were captured during the amplification cycles. Melt-curve analysis was conducted by holding the samples at 95°C for 5 min, then reducing the temperature to 55°C for 5 min, followed by increasing the temperature by 0.5°C every 10 s to 95°C. Negative controls consisting of template with no primers and primers with no template were included. Threshold cycles (Ct) were calculated using iCycler iQ v. 3.1 (Bio-Rad). Melt-curve analysis of quantitative PCR products showed that no primer-dimers formed with either primer set. The melting peak for products of both primer sets was 86.0°C. Primer efficiency and slope were 100.2% and −3.318 for EPSPS and were 103.8% and −3.235 for ALS. No amplification products were observed in any controls lacking template.
Relative quantification using a modification of the 2-ΔΔCt method (42) was used to analyze data from the quantitative PCR experiments. The ALS gene was used as a low-copy control gene with known monogenic inheritance in other Amaranthus species (23). Relative quantification of EPSPS was calculated as ΔCt = (Ct, ALS – Ct, EPSPS). Increase in EPSPS copy number was expressed as 2ΔCt. Each individual sample was run in triplicate, and the average increase in EPSPS copy number and standard deviation were calculated for each sample. Results were expressed as fold increase in EPSPS copy number relative to ALS. The same relative quantification calculation was used for fold increase in EPSPS expression.
EPSPS Quantification and Activity Assay.
Young expanding leaf tissue was sampled from selected R, S, and pseudo-F2 plants for protein extraction and EPSPS quantification (SI Materials and Methods). A continuous assay for inorganic phosphate release (43) was conducted with a phosphate detection kit (Molecular Probes) to assay for EPSPS activity (SI Materials and Methods). Phosphate release above background level was measured for 10 min, and a slope was calculated to determine micromoles of phosphate released per microgram TSP per minute. Dose–response analysis in R was used to calculate the IC50, the glyphosate concentration that inhibited EPSPS activity by 50%, and to statistically compare IC50 values (44).
FISH Mapping of the EPSPS Gene.
FISH was conducted according to published protocols (45). The probe (1,044 bp) was synthesized using EPSF1 × EPSR1 primers from an R plant cDNA, cloned, sequenced, and then PCR amplified from the plasmid.
Supplementary Material
Acknowledgments
We thank Jacob Snelling and Rebecca Davidson for their assistance with initial EPSPS sequence analysis, and Dr. Stephen Powles for useful comments on manuscript drafts. This research was funded by Monsanto Company. B.B. was supported by the Turkish Scientific Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 955.
Data deposition: The sequences in this paper have been deposited in the GenBank database (accession nos. FJ861242, FJ861243, and FJ869880).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906649107/DCSupplemental.
References
- 1.James C. Global status of commercialized biotech/GM crops: 2008. 2008. Available at: www.isaaa.org/resources/publications/briefs/39/executivesummary/default.html. Accessed May 7, 2009.
- 2.Powles SB. Evolved glyphosate-resistant weeds around the world: Lessons to be learnt. Pest Manag Sci. 2008;64:360–365. doi: 10.1002/ps.1525. [DOI] [PubMed] [Google Scholar]
- 3.Culpepper AS, et al. Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 2006;54:620–626. [Google Scholar]
- 4.Grant WF. Cytogenetic studies in Amaranthus. I. Cytological aspects of sex determination in dioecious species. Can J Bot. 1959;37:413–417. [Google Scholar]
- 5.Webster TM. Weed survey—Southern states. Proc South Weed Sci Soc. 2005;58:291–306. [Google Scholar]
- 6.Steckel LE, Main CL, Ellis AT, Mueller TC. Palmer amaranth (Amaranthus palmeri) in Tennessee has low level glyphosate resistance. Weed Technol. 2008;22:119–123. [Google Scholar]
- 7.Culpepper AS, Whitaker JR, MacRae AW, York AC. Distribution of glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in Georgia and North Carolina during 2005 and 2006. J Cotton Sci. 2008;12:306–310. [Google Scholar]
- 8.Culpepper AS, York AC, Marshall MW. Glyphosate-resistant Palmer amaranth in the Southeast. Proc South Weed Sci Soc. 2009;62:371. [Google Scholar]
- 9.Norsworthy JK, Griffith GM, Scott RC, Smith KL, Oliver LR. Confirmation and control of glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in Arkansas. Weed Technol. 2008;22:108–113. [Google Scholar]
- 10.Steinrücken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980;94:1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 12.Pline-Srnic W. Physiological mechanisms of glyphosate resistance. Weed Technol. 2006;20:290–300. [Google Scholar]
- 13.Sammons RD, Heering DC, Dinicola N, Glick H, Elmore GA. Sustainability and stewardship of glyphosate and glyphosate-resistant crops. Weed Technol. 2007;21:347–354. [Google Scholar]
- 14.Heap I. The International Survey of Herbicide Resistant Weeds. 2009. Available at: www.weedscience.org. Accessed March 16, 2009. [Google Scholar]
- 15.Powles SB, Preston C. Evolved glyphosate resistance in plants: Biochemical and genetic basis of resistance. Weed Technol. 2006;20:282–289. [Google Scholar]
- 16.Preston C, Wakelin AM. Resistance to glyphosate from altered herbicide translocation patterns. Pest Manag Sci. 2008;64:372–376. doi: 10.1002/ps.1489. [DOI] [PubMed] [Google Scholar]
- 17.Baerson SR, et al. Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol. 2002;129:1265–1275. doi: 10.1104/pp.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakelin AM, Preston C. A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res. 2006;46:432–440. [Google Scholar]
- 19.Jasieniuk M, et al. Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: Distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci. 2008;56:496–502. [Google Scholar]
- 20.Rowland MW, Murray DS, Verhalen LM. Full-season Palmer amaranth (Amaranthus palmeri) interference with cotton (Gossypium hirsutum) Weed Sci. 1999;47:305–309. [Google Scholar]
- 21.Vencill WK, Grey TL, Culpepper AS, Gaines TA, Westra P. Herbicide-resistance in the Amaranthaceae. J Plant Dis Prot. 2008;XXI(Special Issue):41–44. [Google Scholar]
- 22.Rayburn AL, et al. Genome size analysis of weedy Amaranthus species. Crop Sci. 2005;45:2557–2562. [Google Scholar]
- 23.Trucco F, Jeschke MR, Rayburn AL, Tranel PJ. Promiscuity in weedy amaranths: High frequency of female tall waterhemp (Amaranthus tuberculatus) x smooth pigweed (A. hybridus) hybridization under field conditions. Weed Sci. 2005;53:46–54. [Google Scholar]
- 24.Grant WF. Cytogenetic studies in Amaranthus. III. Chromosome numbers and phylogenetic aspects. Can J Genet Cytol. 1959;1:313–328. [Google Scholar]
- 25.Jones JD, Goldsbrough PB, Weller SC. Stability and expression of amplified EPSPS genes in glyphosate resistant tobacco cells and plantlets. Plant Cell Rep. 1996;15:431–436. doi: 10.1007/BF00232070. [DOI] [PubMed] [Google Scholar]
- 26.Shah DM, et al. Engineering herbicide tolerance in transgenic plants. Science. 1986;233:478–481. doi: 10.1126/science.233.4762.478. [DOI] [PubMed] [Google Scholar]
- 27.Widholm JM, et al. Glyphosate selection of gene amplification in suspension cultures of 3 plant species. Physiol Plant. 2001;112:540–545. doi: 10.1034/j.1399-3054.2001.1120411.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu JW, et al. Isolation of rice EPSP synthase cDNA and its sequence analysis and copy number determination. Acta Bot Sin. 2002;44:188–192. [Google Scholar]
- 29.The Arabidopsis Information Resource. AraCyc 5.0. 2009. Available at: www.plantcyc.org:1555/ARA/server.html. Accessed March 19, 2009.
- 30.Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:46–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- 31.Baerson SR, et al. Investigating the mechanism of glyphosate resistance in rigid ryegrass (Lolium ridigum) Weed Sci. 2002;50:721–730. [Google Scholar]
- 32.Dinelli G, et al. Physiological and molecular bases of glyphosate resistance in Conyza bonariensis biotypes from Spain. Weed Res. 2008;48:257–265. [Google Scholar]
- 33.Dinelli G, et al. Physiological and molecular insight on the mechanisms of resistance to glyphosate in Conyza canadensis (L.) Cronq. biotypes. Pestic Biochem Physiol. 2006;86:30–41. [Google Scholar]
- 34.Trucco F, et al. Nonhybrid progeny from crosses of dioecious amaranths: Implications for gene-flow research. Weed Sci. 2007;55:119–122. [Google Scholar]
- 35.Haider JB, Vencill WK, Culpepper AS, Grey TL. Physiological response of glyphosate-resistant Palmer amaranth. Proc South Weed Sci Soc. 2007;60:180. [Google Scholar]
- 36.Field LM, Devonshire AL. Structure and organization of amplicons containing the E4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer) Biochem J. 1997;322:867–871. doi: 10.1042/bj3220867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paton MG, Karunaratne SH, Giakoumaki E, Roberts N, Hemingway J. Quantitative analysis of gene amplification in insecticide-resistant Culex mosquitoes. Biochem J. 2000;346:17–24. [PMC free article] [PubMed] [Google Scholar]
- 38.Schimke RT. Methotrexate resistance and gene amplification. Mechanisms and implications. Cancer. 1986;57:1912–1917. doi: 10.1002/1097-0142(19860515)57:10<1912::aid-cncr2820571004>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Shaner DL, Nadler-Hassar T, Henry WB, Koger CH. A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 2005;53:769–774. [Google Scholar]
- 40.Patzoldt WL, Hager AG, McCormick JS, Tranel PJ. A codon deletion confers resistance to herbicides inhibiting protoporphyrinogen oxidase. Proc Natl Acad Sci USA. 2006;103:12329–12334. doi: 10.1073/pnas.0603137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tranel PJ, Jiang WL, Patzoldt WL, Wright TR. Intraspecific variability of the acetolactate synthase gene. Weed Sci. 2004;52:236–241. [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knezevic SZ, Streibig JC, Ritz C. Utilizing R software package for dose-response studies: The concept and data analysis. Weed Technol. 2007;21:840–848. [Google Scholar]
- 45.Jiang JM, Gill BS, Wang GL, Ronald PC, Ward DC. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci USA. 1995;92:4487–4491. doi: 10.1073/pnas.92.10.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.