Abstract
The phospholipase neutral sphingomyelinase (N-SMase) has been recognized as a major mediator of processes such as inflammation, development and growth, differentiation and death of cells, as well as in diseases such as Alzheimer’s, atherosclerosis, heart failure, ischemia/reperfusion damage, or combined pituitary hormone deficiency. Although activation of N-SMase by the proinflammatory cytokine TNF was described almost two decades ago, the underlying signaling pathway is unresolved. Here, we identify the Polycomb group protein EED (embryonic ectodermal development) as an interaction partner of nSMase2. In yeast, the N terminus of EED binds to the catalytic domain of nSMase2 as well as to RACK1, a protein that modulates the activation of nSMase2 by TNF in concert with the TNF receptor 1 (TNF-R1)-associated protein FAN. In mammalian cells, TNF causes endogenous EED to translocate from the nucleus and to colocalize and physically interact with both endogenous nSMase2 and RACK1. As a consequence, EED and nSMase2 are recruited to the TNF-R1•FAN•RACK1-complex in a timeframe concurrent with activation of nSMase2. After knockdown of EED by RNA interference, the TNF-dependent activation of nSMase2 is completely abrogated, identifying EED as a protein that both physically and functionally couples TNF-R1 to nSMase2, and which therefore represents the “missing link” that completes one of the last unresolved signaling pathways of TNF-R1.
Keywords: embryonic ectodermal development, immune response, inflammation
Neutral sphingomyelinases (N-SMases) mediate stress-induced ceramide generation and participate in inflammation, development, cellular growth, differentiation and death, heart failure, ischemia/reperfusion damage, atherosclerosis, and Alzheimer’s disease (1, 2). They are acutely activated by TNF, a major mediator of inflammatory and immunoregulatory responses (3, 4). Out of the three N-SMase genes cloned in mammals, nSMase2 corresponds to the biochemically characterized N-SMase. It is a membrane-bound protein with two putative N-terminal hydrophobic membrane-anchoring domains, a collagen-like linker region, and a C-terminal catalytic domain (Fig. 1A) (5). nSMase2 has been linked to cell cycle regulation and contact inhibition (1), late embryonal and postnatal development (5, 6), exosome secretion (7), and Alzheimer’s disease and combined pituitary hormone deficiency (1, 6). In response to TNF, nSMase2 is important for inflammatory signaling, cell adhesion and migration, endothelial regulation, cell death, and cutaneous barrier repair (5, 8–11). Even though N-SMase activation by TNF has been reported since almost 20 years ago (3), the corresponding signaling pathway is not fully resolved. In response to TNF, nSMase2 is activated exclusively by the 55-kDa receptor TNF receptor 1 (TNF-R1) (10). We have previously defined a neutral sphingomyelinase activation domain (NSD) within TNF-R1 (12) that serves as a binding site for the protein FAN (factor associated with N-SMase activation) (13). FAN recruits the WD repeat protein RACK1 to TNF-R1 and in complex modulates the activity of nSMase2 (14). However, the subsequent course of the N-SMase pathway and the involved components have remained elusive.
Fig. 1.
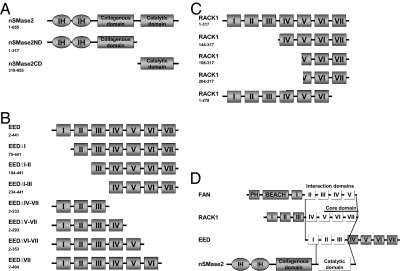
Mapping of the interaction domains of nSMase2, EED, and RACK1. Schematic representation of the used nSMase2 (A), EED (B), and RACK1 (C) constructs. The range of amino acids encoded by each construct is given below its name. (D) Overview of the interaction domains of nSMase2, EED, and RACK1. The minimal core binding domain of RACK1 is additionally indicated, together with the interaction domain of FAN (14). BEACH, Beige and Chédiak-Higashi domain; IH, intramembranous hydrophobic domain; PH, pleckstrin-homology domain. WD repeats are marked by roman numerals.
Here, we have identified embryonic ectoderm development (EED) as an interaction partner of nSMase2. EED is a nuclear WD repeat protein of the Polycomb group and participates in lymphocyte proliferation/differentiation, as well as in organogenesis and embryonic development (15, 16). EED has been independently cloned as WAIT-1, a protein that interacts with integrins at the plasma membrane (17), and thus couples to molecules that carry out vital functions in the immune system (18). Following integrin activation or by the HIV-1 protein Nef, EED is rapidly recruited from the nucleus to the plasma membrane (15). Other nonnuclear functions of EED encompass actin polymerization-dependent processes, such as antigen receptor signaling in T cells (19). Our data strongly suggest that EED is the last “missing link” in the signaling pathway from TNF-R1 to neutral sphingomyelinase.
Results
EED Interacts with nSMase2 in Yeast.
An initial yeast interaction trap screen using a cDNA expression library from HeLa cells suggested EED as a potential interaction partner of nSMase2. To map the binding domains of nSMase2 and EED in more detail, bait and prey constructs for full-length nSMase2, its N-terminal domain (nSMase2ND), or its C-terminal region (nSMase2CD) (Fig. 1A) were cotransformed in combination with full-length EED or EED deletion mutants (Fig. 1B) and tested for interaction. Full-length nSMase2 and nSMase2CD bound to full-length EED and to select deletion mutants of EED, whereas nSMase2ND did not interact with any EED construct (Tables S1 and S2), demonstrating that nSMase2 interacts with EED through its C-terminal catalytic domain (Fig. 1D). EED bound to full-length nSMase2 and nSMase2CD as long as WD repeats I to III were present (Tables S1 and S2), defining this region as its nSMase2-interaction domain. Of note, deletion of WD repeat I still allowed for residual interaction of EED with nSMase2CD (but not full-length nSMase2), indicating that the nSMase2-interaction domain of EED does not comprise the entire WD repeat I (Fig. 1D).
Mapping of the EED/RACK1 Interaction Sites.
Next, we investigated the capacity of EED to interact with FAN and RACK1, the other known proteins of the N-SMase pathway. Full-length FAN did not bind to full-length EED nor to full-length nSMase2. Full-length RACK1, however, interacted with all EED constructs (but not with full-length nSMase2) that contained WD repeats I to III, and with full-length EED (Tables S3 and S4), identifying WD repeats I to III of EED as its RACK1-binding domain (Fig. 1D).
To delineate the EED-binding region of RACK1, we analyzed a panel of RACK1 deletion mutants (Fig. 1C). Deletion of WD repeats I to III (RACK1144–317) did not yet alter the binding of RACK1 to EED (Tables S3 and S4). If the deletion was extended into WD repeat V (RACK1198–317), interaction with full-length EED was lost but still detectable for all other EED mutants containing the RACK1 binding domain (Tables S3 and S4). This interaction was further decreased by deletion of six additional amino acids (RACK1204–317) (Tables S3 and S4). Finally, deletion of the last C-terminal WD repeat of RACK1 (RACK11–278) completely abolished interaction with all tested EED constructs (Tables S3 and S4), suggesting that the full EED-interaction domain of RACK1 spans WD repeats IV to VII of RACK1, with a core binding domain encompassing parts of WD repeat V to WD repeat VII (Fig. 1D).
In summary, our data show that in yeast, EED interacts with both RACK1 and nSMase2 and therefore may be able to physically link nSMase2 to TNF-R1, FAN, and RACK1 in the N-SMase pathway.
TNF Induces Colocalization of EED and nSMase2.
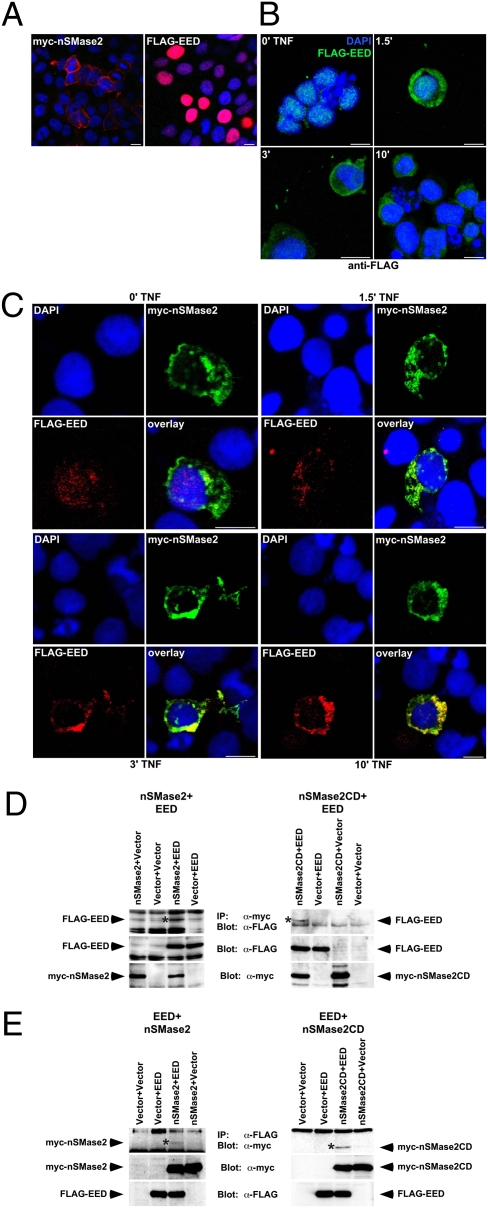
We further analyzed the interaction of EED and nSMase2 in higher eukaryotic cells by intracellular colocalization studies. In confluent 293 cells expressing myc-tagged full-length nSMase2 (pMYC.nSMase21–655), the enzyme localized in the cytoplasm, as well as at the plasma membrane (Fig. 2A) (5). In contrast, FLAG-tagged full-length EED (pFLAG.EED2–441) showed a predominantly nuclear localization (Fig. 2A) (15). An interaction of EED and nSMase2 would therefore require the translocation of nuclear EED to the cytoplasm or to the plasma membrane. In line, EED is identical to WAIT-1, a protein that interacts with integrins at the plasma membrane (17). Moreover, in cell types such as Jurkat, the HIV-1 protein Nef or integrin activation actively recruit EED from the nucleus to the plasma membrane (15).
Fig. 2.
Overexpressed EED and nSMase2 colocalize and coimmunoprecipitate in mammalian cells. (A) Transfected pMYC.nSMase21–655 (Left, red) or pFLAG.EED2–441 (Right, red) were detected in 293 cells. (B) Jurkat cells were treated with TNF for the indicated times before transfected pFLAG.EED2–441 was detected (green). (C) Jurkat cells were transfected with pFLAG.EED2–441 (red) and pMYC.nSMase21–655 (green) and stimulated with TNF for the indicated times. Colocalization of EED and nSMase2 is indicated by yellow staining (overlay). Cell nuclei were stained with DAPI (blue). (Scale bars, 10 μm.) (D) Immunoprecipitation of myc-tagged full-length nSMase2 (Left) or nSMase2CD (Right) and vector controls from transfected 293 cells. (E) Parallel experiment showing the reverse association of full-length nSMase2 (Left) and nSMase2CD (Right) with EED. For both (D) and (E), coimmunoprecipitating FLAG-EED, myc-nSMase2, or myc-nSMase2CD are shown (Top, asterisk); (Middle and Bottom) Expression of the fusion proteins in total lysates.
Therefore, we used Jurkat cells to investigate whether TNF elicited a similar translocation of EED in the timeframe required for activation of nSMase2. After confirming the predominantly nuclear localization of transfected pFLAG.EED2–441 in untreated cells (Fig. 2B) (0’ TNF), we observed a rapid and pronounced redistribution of EED from the nucleus into the cytoplasm and to the plasma membrane within 1.5 min of TNF-treatment, being largely complete at 3 min and still persisting after 10 min (Fig. 2B). These results were confirmed with an independent antibody that recognizes the EED-portion instead of the FLAG-tag (Fig. S1). In Jurkat cells expressing both pFLAG.EED2–441 and pMYC.nSMase21–655, we found that the translocation of EED was not unspecifically directed to the entire cytoplasm or plasma membrane, but confined to the sites where nSMase2 was expressed, resulting in clear colocalization of both proteins (Fig. 2C). Of note, the colocalization of EED fully coincided with the activation pattern of nSMase2 (see below), implicating a role of EED in the activation of nSMase2 by TNF. However, deactivation of nSMase2 may not depend on presence or absence of EED, as EED remained still colocalized at 10 min (Fig. 2C), when the activity of nSMase2 had already returned to basal levels (see below).
EED and nSMase2 Physically Interact in Mammalian Cells.
For coimmunoprecipitation experiments, 293 cells were transiently transfected with myc-tagged full-length nSMase2 or nSMase2CD (pMYC.nSMase2318–655) in combination with FLAG-tagged full-length EED or the parental empty vectors. After verifying the expression of the constructs (Fig. 2 D and E, Middle and Bottom), we could detect EED in immunoprecipitates of full-length nSMase2 and nSMase2CD but not in control immunoprecipitates lacking one or both proteins (Fig. 2D, Top). In reverse experiments, bands corresponding to the exact size of full-length nSMase2 and nSMase2CD were detected specifically in EED-immunoprecipitates, but not in control immunoprecipitations (Fig. 2E, Top), confirming that EED and nSMase2 physically interact in mammalian cells. Of note, this interaction was already observed in untreated cells, in the absence of TNF (but clearly increased by treatment with TNF; Fig. 3E). As an explanation, EED is inherently present at low levels in the cytosol of 293 cells and has been similarly coimmunoprecipitated from unstimulated 293 cells together with the protein Ezh2 (19). Low levels of intrinsic cytosolic EED have also been observed in other cells (15) and most likely result from the reported inherent nucleo-cytoplasmic shuttling activity of EED (15).
Fig. 3.
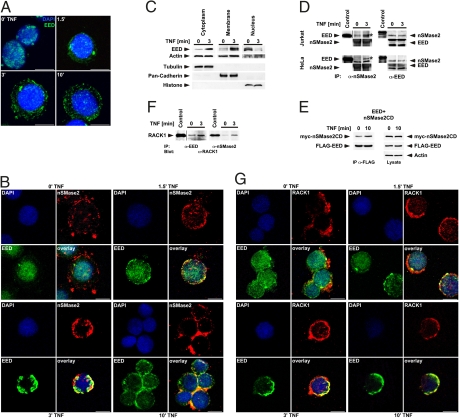
Interaction of the endogenous EED, nSMase2 and RACK1 proteins. (A) Jurkat cells were treated with TNF for the indicated times followed by detection of the endogenous EED protein (green). (B) Jurkat cells were analyzed for TNF-induced colocalization of endogenous EED (green) and nSMase2 (red). (C) Endogenous EED was detected by Western blot in cytoplasmic, membrane and nuclear fractions from Jurkat cells after stimulation with TNF for 3 min or not. The multiple bands represent different isoforms of endogenous EED that have been previously described (25). Equal loading and successful fractionation were verified by detection of actin, tubulin, pan-cadherin, and histone. (D) Coimmunoprecipitation of endogenous EED with endogenous nSMase2 (Left) and vice versa (Right) from Jurkat (Upper) or HeLa cells (Lower). nSMase2 or EED were immunoprecipitated from cells treated with TNF for 3 min or not and coimmunoprecipitating endogenous EED (asterisk) or nSMase2 were detected by Western blot. Lysates from 293 cells overexpressing pFLAG.EED2–441 or pMYC.nSMase21–655 (migrating slightly above endogenous nSMase2) were used for control. Blots depicting immunoprecipitated nSMase2 or EED are shown below the corresponding coimmunoprecipitations. (E) pFLAG.EED2–441 and pMYC.nSMase2318–655 (nSMase2CD) were overexpressed in 293 cells followed by treatment with TNF for 0 and 10 min. Subsequently, FLAG-EED was immunoprecipitated and coimmunoprecipitating myc-nSMase2CD was detected by Western blot (Upper Left). The immunoprecipitated FLAG-EED protein is shown for control in the lower left panel. Total lysates from untreated and TNF-treated cells were probed for equal expression of myc-nSMase2CD, FLAG-EED, together with actin as a loading control (Right). (F) The immunoprecipitates from Jurkat cells shown in (D) were reanalyzed for the presence of coimmunoprecipitating endogenous RACK1 (arrow). As a control, the abundant endogenous RACK1 from 293 cell lysates is shown. (G), Jurkat cells were analyzed for TNF-induced colocalization of endogenous EED (green) and RACK1 (red). Cell nuclei were stained with DAPI (blue). (Scale bars, 10 μm.)
TNF Induces Interaction of Endogenous EED and nSMase2.
Because results obtained with overexpressed proteins have to be interpreted with caution, we confirmed both the predominantly nuclear localization of endogenous EED in Jurkat, 293, COS-7, and NIH 3T3 cells (Fig. 3A, 0’ TNF, and Fig. S2) and its cytoplasmic/membrane translocation in response to TNF (Fig. 3A and Fig. S3). Endogenous EED translocated identically in activated primary human T cells, underscoring its relevance in an in vivo physiologic cell model (Fig. S4). Furthermore, endogenous EED colocalized with endogenous nSMase2 (whose cytoplasmic/membrane localization was not changed by TNF-treatment) (Fig. 3B and Fig. S5), in an identical timeframe as seen for the overexpressed proteins (Fig. 3B).
In purified cytoplasmic and membrane fractions from Jurkat cells, TNF caused a clearly detectable increase of endogenous EED, whereas a corresponding decrease was evident in the nuclear fraction (Fig. 3C), thus confirming its translocation by an independent approach.
In coimmunoprecipitations of the endogenous proteins from Jurkat cells, bands identical to the predicted size of EED were detected in immunoprecipitates of nSMase2 that clearly increased in response to TNF (Fig. 3D, Upper Left). In the reverse approach, endogenous nSMase2 was likewise detected in EED immunoprecipitates (Fig. 3D, Upper Right). Most likely because of limitations in antibody affinity and abundance of endogenous nSMase2, a TNF-induced increase was, however, not unambiguously detectable. Yet, this was clearly the case when nSMase2CD was overexpressed and coimmunoprecipitated together with EED in 293 cells (Fig. 3E). As another cell system for which shuttling of endogenous EED has been reported (15), we additionally analyzed HeLa cells and obtained identical results (Fig. 3D, Lower).
EED Couples nSMase2 to RACK1.
As outlined in Fig. 1D, in yeast, the same binding region of EED mediates interaction with nSMase2 as well as with RACK1. To clarify whether EED can actually bind to both proteins simultaneously, and thus indeed link nSMase2 to RACK1, we reanalyzed the EED-immunoprecipitations containing coimmunoprecipitated nSMase2 from Fig. 3D (Upper Right) and additionally found coimmunoprecipitating RACK1 (Fig. 3F, Left). To exclude the possibility of mutually exclusive RACK•EED- and nSMase2•EED-complexes in the same immunoprecipitation, we analyzed the nSMase2-immunoprecipitates from Fig. 3D (Upper Left), again finding bands of the size predicted for RACK1 (Fig. 3F, Right). Because RACK1 does not directly bind to nSMase2, its presence in the nSMase2-immunoprecipitates must result from indirect binding of RACK1 to EED which then coimmunoprecipitates together with nSMase2. In conclusion, these results strongly suggest that EED has the ability to simultaneously interact with both RACK1 and nSMase2. In both experiments, the amount of coimmunoprecipitated RACK1 increased in TNF-treated cells, once more consistent with a TNF-dependent translocation of EED from the nucleus to the cytoplasm or the plasma membrane where it then may couple nSMase2 to RACK1, and thus to the TNF-R1•FAN•RACK1-complex. In line with this concept, immunofluorescence analyses demonstrated the translocation of endogenous EED and its colocalization with endogenous RACK1 in response to TNF in Jurkat (Fig. 3G) as well as in HeLa cells (Fig. S6).
TNF Recruits EED and nSMase2 to the TNF-R1 Complex.
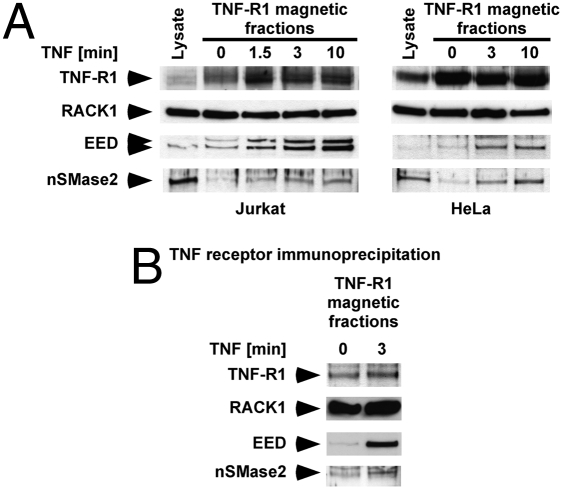
To determine whether the TNF-induced interaction of EED with both nSMase2 and RACK1 indeed resulted in their recruitment to the TNF-R1 complex, we employed immunomagnetic sorting. In this unique experimental approach, TNF receptors are labeled with biologically active biotin-TNF coupled to streptavidin-coated magnetic microbeads. Subsequently, intact TNF/receptor complexes as well as receptor-associated proteins are isolated within their native membrane environment using a custom-made high-gradient magnetic chamber. The obtained magnetic fractions are then analyzed for enriched proteins (e.g., by conventional Western blot) (20). As shown in Fig. 4A, TNF rapidly recruited endogenous EED and nSMase2 into magnetic fractions from Jurkat and HeLa cells as early as 1.5 min. TNF-R1 and RACK1 were uniformly present in all magnetic fractions, coherent with previous studies showing a constitutive and TNF-independent association of FAN and RACK1 with TNF-R1 (13, 14).
Fig. 4.
TNF-dependent recruitment of EED and nSMase2 to the TNF-R1 complex. (A) Magnetic fractions harboring labeled TNF•TNF-R1-complexes were derived from Jurkat or HeLa cells after stimulation with TNF for the indicated times and immunoblotted for presence/recruitment of endogenous TNF-R1, RACK1, EED, and nSMase2. Protein cell extracts were used as control (lysate). (B) Coimmunoprecipitation of RACK1, EED, and nSMase2 with magnetic TNF-R1. TNF receptor magnetic fractions isolated from untreated or TNF-treated Jurkat cells were solubilized, TNF-R1 was precipitated using biotin-TNF/streptavidin-coated magnetic microbeads and analyzed for coimmunoprecipitating RACK1, EED, and nSMase2.
Next, we investigated whether RACK1, EED, and nSMase2 detected in the isolated magnetic fractions were directly associated with TNF-R1. Magnetic fractions from nonstimulated or Jurkat cells stimulated with TNF for 3 min were solubilized, and TNF-R1 was precipitated together with associated proteins using biotin-TNF and streptavidin-coated magnetic microbeads. As shown in Fig. 4B, RACK1, EED, and nSMase2 were detected along with TNF-R1 in the precipitates, demonstrating the direct association of these proteins with the TNF-R1-complex and excluding the possibility of simple cocompartmentalization with TNF-R1. Again, RACK1 was constitutively present in the complex, whereas both EED and nSMase2 were recruited to the TNF-R1 complex in a TNF-dependent manner. To determine the domains of TNF-R1 that mediate this recruitment, we analyzed 70Z/3TR55∆212–308/346 pre-B cells expressing a deletion mutant of TNF-R1 that still carries the NSD but is unable to signal through its death domain (12) and found that translocation of EED was readily detectable. In contrast, TNF was unable to mobilize EED in 70Z/3TR55∆308–340 cells expressing a TNF-R1 deletion mutant that lacks the NSD (12), thus ascribing EED-signaling to the NSD and thus to the N-SMase pathway rather than to the death domain pathway of TNF-R1 (Fig. S7).
In summary, our data demonstrate that in mammalian cells, TNF causes EED to translocate from the nucleus to the cytoplasm and to the plasma membrane, leading to colocalization and interaction with both nSMase2 and RACK1 and thus to recruitment of EED and nSMase2 to the TNF-R1•FAN•RACK1-complex. Moreover, the translocation of EED occurs in a timeframe that coincides with the activation of nSMase2 by TNF, suggesting that EED may functionally participate in this process.
EED Is Required for Activation of nSMase2 by TNF.
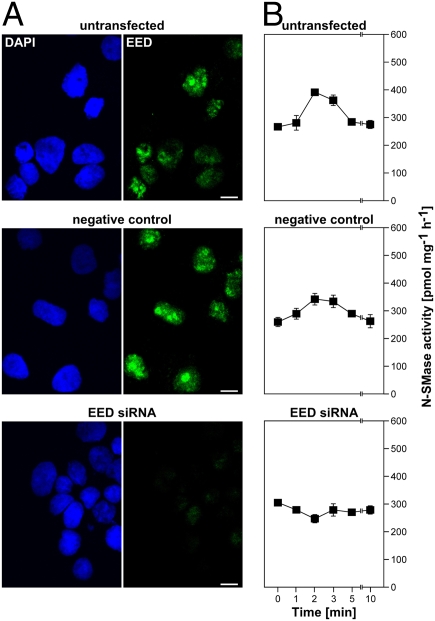
To test this hypothesis, we selectively targeted EED by RNA interference. In a pilot experiment, even high levels of pFLAG.EED2–441 were effectively down-regulated in 293 cells by three different small interfering (si)RNAs specific for EED (EED_1, -_2, and -_3), with siRNAs EED_2 and EED_3 being most effective (Fig. S8). For the subsequent experiments (and also to avoid unspecific off-target effects of a single siRNA), we therefore employed a pool of the siRNAs EED_2 and EED_3. As shown in Fig. 5A, the pooled siRNAs EED_2 and EED_3 efficiently and specifically down-regulated the endogenous EED protein of Jurkat cells, whereas a negative control siRNA did not affect endogenous EED levels. When we measured nSMase2 activity in these cells after 0, 1, 2, 3, 5, and 10 min of TNF-stimulation, nSMase2 showed a maximal activation at 2 to 3 min in untransfected cells, and returned to basal activity within 10 min (Fig. 5B), in agreement with previous data (11). In Jurkat cells transfected with the negative control siRNA, nSMase2 displayed an essentially identical activation pattern. However, in Jurkat cells transfected with the pooled siRNAs EED_2 and EED_3, the TNF-dependent activation of nSMase2 was completely abrogated (Fig. 5B), providing strong evidence that EED not only interacts with nSMase2 but is actually required for nSMase2 activation by TNF-R1, thus defining EED as a unique and essential component of the signaling pathway from TNF-R1 to nSMase2.
Fig. 5.
EED is required for activation of nSMase2 by TNF. (A) Untransfected Jurkat cells or Jurkat cells nucleofected with negative control siRNA or with siRNAs specific for EED were analyzed for expression of endogenous EED. Cell nuclei were stained with DAPI (blue). (Scale bars, 10 μm.) (B) Alternatively, cells were treated with TNF for the indicated times before N-SMase activity was measured. The values shown represent the means from triplicate determinations performed in parallel; error bars indicate the respective standard deviations. One out of two independent experiments with similar results is shown.
Of note, an EED construct constitutively targeted to the plasma membrane (H-2Kk-EED) did not increase the basal or the TNF-dependent activity of nSMase2 (Fig. S9 A and B, Top), suggesting that recruitment of EED to the plasma membrane has to occur in the context of the TNF-R1•FAN•RACK1-complex for efficient activation of nSMase2. Identical results were observed with the isolated nSMase2-interaction domain of EED (FLAG-EED∆IV–VII) or the C-terminal region (FLAG-EED∆I–III), which had lost their nuclear localization and constitutively localized to the cytoplasm (Fig. S9 A and B, Middle and Bottom), implicating that all domains of EED are required for nucleo-cytoplasmic shuttling as well as for the activation of nSMase2 by TNF-R1. All three constructs even suppressed the TNF-dependent activation of nSMase2 in a dominant negative manner (Fig. S9), probably by interference with the assembly of the endogenous TNF-R1•FAN•RACK1•EED•nSMase2 complex, and in line with similar effects of overexpressed RACK1 that we have previously observed (14).
Discussion
In this study, we have established EED as a binding partner of nSMase2, representing the missing link that physically and functionally couples nSMase2 to TNF-R1, thereby completing the N-SMase pathway. In this pathway (Fig. 6), TNF triggers activation of TNF-R1, leading to the NSD-dependent translocation of EED from the nucleus. EED then simultaneously interacts with both RACK1 and nSMase2. This interaction couples EED and nSMase2 to the TNF-R1•FAN•RACK1-complex, and results in the activation of nSMase2.
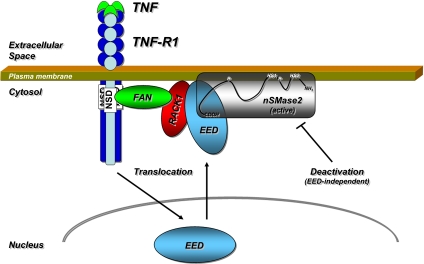
Fig. 6.
EED is the missing link that couples TNF-R1 to nSMase2. The scheme depicts the proposed role of EED in the N-SMase pathway. Activation of TNF-R1 in response to TNF induces translocation of EED from the nucleus to the plasma membrane, where it binds to RACK1 and becomes part of the TNF-R1•FAN•RACK1 complex. Concurrently, EED binds to the catalytic C-terminal domain of nSMase2, which is more freely accessible than the tightly membrane-integrated N-terminal region. This interaction physically links nSMase2 to the TNF-R1•FAN•RACK1•EED complex, resulting in activation of nSMase2. Subsequently, nSMase2 is deactivated by EED-independent mechanisms. The membrane topology of nSMase2, including the palmitoylation clusters (P) and the two N-terminal hydrophobic segments (HS1, HS2) is adapted from ref. 21.
We have found that nSMase2 binds to EED through its C-terminal catalytic domain, but not through its N terminus. Tani and Hannun (21) have put forward a model in which the N terminus of nSMase2 closely integrates into the membrane by its two hydrophobic segments, whereas its C terminus is more loosely associated with the plasma membrane through a palmitoylation site. Because of this tight integration, the N-terminal region of nSMase2 may be inaccessible for interacting proteins such as EED, whereas the more freely available catalytic domain could serve as an interface by which nSMase2 interacts with cytoplasmic proteins. EED—once bound to the catalytic domain—may therefore directly contribute to the TNF-R1-triggered activation of nSMase2 as a stimulatory cofactor (e.g., by affecting the conformation of the catalytic domain or by facilitating its accessibility for substrate). The deactivation of nSMase2 most likely occurs by EED-independent mechanisms, as we have not observed a concurrent dissociation of EED from nSMase2 (Fig. 6).
Several lines of evidence suggest that the TNF-induced N-SMase pathway may couple to integrin signaling. First, TNF can activate integrins through inside-out signaling (22). Second, as an integrin-interacting protein, RACK1 has been functionally linked to cell adhesion and cell migration (23). Third, EED was independently isolated and named WAIT-1 because of its interaction with integrins (17) and is rapidly recruited to the plasma membrane by integrin activation (15). Therefore, a crosstalk between the N-SMase pathway and integrin signaling might include a role of integrins in the TNF-R1-mediated activation of nSMase2 (e.g., by guiding EED into the vicinity of nSMase2 or by stabilizing a TNF-R1•FAN•RACK1•EED•nSMase2 complex at the plasma membrane). Alternatively, activation of nSMase2 by TNF-R1 might have impact on integrin signal transduction, for example by modulating membrane fluidity (24), thus facilitating the clustering of integrin molecules. Future experiments will help to further clarify the role of EED and nSMase2 in these processes, as well as the contribution of EED to the downstream effects of TNF-R1-FAN-RACK1-EED-nSMase2 signaling.
In conclusion, the identification of EED as the potentially last missing link in the pathway from TNF-R1 to nSMase2 not only completes one of the last remaining major signaling pathways of TNF-R1 after almost 20 years, but will hopefully also contribute to a better understanding of the context of nSMase2 activation in both physiology and disease: for example, in the regulation of major inflammatory responses to TNF.
Materials and Methods
Yeast Interaction Trap System, Interaction Site Mapping.
Bait and prey constructs of nSMase2, EED and RACK1 (Fig. 1) were generated by standard procedures and tested for interaction as previously outlined (14). See SI Materials and Methods for details.
Cell Culture, Intracellular Colocalization Studies, Coimmunoprecipitations.
Preparation of Membrane, Cytoplasmic, and Nuclear Fractions.
Membrane, cytoplasmic, and nuclear fractions were generated using ProteoJET extraction kits (Fermentas) and analyzed as outlined in SI Materials and Methods.
Magnetic Labeling of TNF Receptors, Isolation of TNF Receptor Magnetic Fractions, and Immunoprecipitation of TNF Receptor-Associated Proteins.
Magnetic fractions were prepared as described in detail in SI Materials and Methods and analyzed using antibodies against TNF-R1, RACK1, EED, and nSMase2.
RNA Interference, Measurement of nSMase2 Activity.
Jurkat cells were left untransfected or nucleofected with a negative control siRNA or pooled EED-specific siRNAs EED_2 and EED_3 and analyzed for expression of endogenous EED by immunofluorescence. In parallel, cells were stimulated in triplicates with 100 ng/mL hTNF for the indicated times and nSMase2 activity in the lysates was measured as described in ref. 11. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kyoung-Ae Yoo-Ott, Sandra Ussat, and Parvin Davarnia for excellent technical assistance. This work was supported in part by National Institutes of Health Grant GM 43825 (to Y.A.H.), by a grant from the Forschungskommission der Medizinischen Fakultät (to D.A.), and by Grants SCHU 733/9-1 and SFB415 project A11 (to S.S.) and AD 133/4-1 (to D.A.) from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908486107/DCSupplemental.
References
- 1.Clarke CJ, Hannun YA. Neutral sphingomyelinases and nSMase2: bridging the gaps. Biochim Biophys Acta. 2006;1758:1893–1901. doi: 10.1016/j.bbamem.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Pavoine C, Pecker F. Sphingomyelinases: their regulation and roles in cardiovascular pathophysiology. Cardiovasc Res. 2009;82:175–183. doi: 10.1093/cvr/cvp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MY, Linardic C, Obeid L, Hannun YA. Identification of sphingomyelin turnover as an effector mechanism for the action of tumor necrosis factor alpha and gamma-interferon. Specific role in cell differentiation. J Biol Chem. 1991;266:484–489. [PubMed] [Google Scholar]
- 4.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 5.Clarke CJ, et al. The extended family of neutral sphingomyelinases. Biochemistry. 2006;45:11247–11256. doi: 10.1021/bi061307z. [DOI] [PubMed] [Google Scholar]
- 6.Stoffel W, Jenke B, Blöck B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci USA. 2005;102:4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 8.Clarke CJ, Truong TG, Hannun YA. Role for neutral sphingomyelinase-2 in tumor necrosis factor alpha-stimulated expression of vascular cell adhesion molecule-1 (VCAM) and intercellular adhesion molecule-1 (ICAM) in lung epithelial cells: p38 MAPK is an upstream regulator of nSMase2. J Biol Chem. 2007;282:1384–1396. doi: 10.1074/jbc.M609216200. [DOI] [PubMed] [Google Scholar]
- 9.Kreder D, et al. Impaired neutral sphingomyelinase activation and cutaneous barrier repair in FAN-deficient mice. EMBO J. 1999;18:2472–2479. doi: 10.1093/emboj/18.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolesnick RN, Krönke M. Regulation of ceramide production and apoptosis. Annu Rev Physiol. 1998;60:643–665. doi: 10.1146/annurev.physiol.60.1.643. [DOI] [PubMed] [Google Scholar]
- 11.Neumeyer J, et al. TNF-receptor I defective in internalization allows for cell death through activation of neutral sphingomyelinase. Exp Cell Res. 2006;312:2142–2153. doi: 10.1016/j.yexcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Adam D, Wiegmann K, Adam-Klages S, Ruff A, Krönke M. A novel cytoplasmic domain of the p55 tumor necrosis factor receptor initiates the neutral sphingomyelinase pathway. J Biol Chem. 1996;271:14617–14622. doi: 10.1074/jbc.271.24.14617. [DOI] [PubMed] [Google Scholar]
- 13.Adam-Klages S, et al. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86:937–947. doi: 10.1016/s0092-8674(00)80169-5. [DOI] [PubMed] [Google Scholar]
- 14.Tcherkasowa AE, et al. Interaction with factor associated with neutral sphingomyelinase activation, a WD motif-containing protein, identifies receptor for activated C-kinase 1 as a novel component of the signaling pathways of the p55 TNF receptor. J Immunol. 2002;169:5161–5170. doi: 10.4049/jimmunol.169.9.5161. [DOI] [PubMed] [Google Scholar]
- 15.Witte V, et al. HIV-1 Nef mimics an integrin receptor signal that recruits the polycomb group protein EED to the plasma membrane. Mol Cell. 2004;13:179–190. doi: 10.1016/s1097-2765(04)00004-8. [DOI] [PubMed] [Google Scholar]
- 16.Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- 17.Rietzler M, Bittner M, Kolanus W, Schuster A, Holzmann B. The human WD repeat protein WAIT-1 specifically interacts with the cytoplasmic tails of beta7-integrins. J Biol Chem. 1998;273:27459–27466. doi: 10.1074/jbc.273.42.27459. [DOI] [PubMed] [Google Scholar]
- 18.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su IH, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Tchikov V, Schütze S. Immunomagnetic isolation of tumor necrosis factor receptosomes. Methods Enzymol. 2008;442:101–123. doi: 10.1016/S0076-6879(08)01405-5. [DOI] [PubMed] [Google Scholar]
- 21.Tani M, Hannun YA. Analysis of membrane topology of neutral sphingomyelinase 2. FEBS Lett. 2007;581:1323–1328. doi: 10.1016/j.febslet.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouaouina M, Blouin E, Halbwachs-Mecarelli L, Lesavre P, Rieu P. TNF-induced beta2 integrin activation involves Src kinases and a redox-regulated activation of p38 MAPK. J Immunol. 2004;173:1313–1320. doi: 10.4049/jimmunol.173.2.1313. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zha XH, et al. Sphingomyelinase treatment induces ATP-independent endocytosis. J Cell Biol. 1998;140:39–47. doi: 10.1083/jcb.140.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.