Abstract
Mutation rate is an important determinant of evolvability. The optimal mutation rate for different organisms during evolution has been modeled in silico and tested in vivo, predominantly through pairwise comparisons. To characterize the fitness landscape across a broad range of mutation rates, we generated a panel of 66 DNA polymerase I mutants in Escherichia coli with comparable growth properties, yet with differing DNA replication fidelities, spanning 103-fold higher and lower than that of wild type. These strains were competed for 350 generations in six replicate cultures in two different environments. A narrow range of mutation rates, 10- to 47-fold greater than that of wild type, predominated after serial passage. Mutants exhibiting higher mutation rates were not detected, nor were wild-type or antimutator strains. Winning clones exhibited shorter doubling times, greater maximum culture densities, and a growth advantages in pairwise competition relative to their precompetition ancestors, indicating the acquisition of adaptive phenotypes. To investigate the basis for mutator selection, we undertook a large series of pairwise competitions between mutator and wild-type strains under conditions where, in most cases, one strain completely overtook the culture within 18 days. Mutators were the most frequent winners but wild-type strains were also observed winning, suggesting that the competitive advantage of mutators is due to a greater probability of developing selectably advantageous mutations rather than from an initial growth advantage conferred by the polymerase variant itself. Our results indicate that under conditions where organism fitness is not yet maximized for a particular environment, competitive adaptation may be facilitated by enhanced mutagenesis.
Keywords: adaptation, competition, fidelity, mutator
Mutation rates in organisms reflect the need for accuracy to maintain critical genetic information and the requirement for flexibility to adapt to environmental changes. In eukaryotic and prokaryotic cells, spontaneous mutations occur infrequently—less than once per billion bases copied (1). A departure from this norm can be detrimental to an individual and to a population as a whole. Increased mutation rates in viruses (2, 3) and bacteria (4, 5) can lead to decreased fitness and ultimately to extinction (6). Elevated mutation frequencies are associated with human pathologies such as cancer and premature aging (7, 8). Large increases in accuracy, on the other hand, can be energetically costly (9) and also lead to decreased fitness.
Although low mutation rates benefit populations in stable environments, higher mutation rates favor adaptation. Evolution has derived mechanisms for transient changes in mutation rates to circumvent the narrow restriction imposed by the high fidelity required for long-term survival. Mutagenesis is induced by bacteria during times of stress (10), and the mammalian immune system relies on targeted hypermutation to respond to new pathogens (11). In experiments where populations of bacteria are introduced into a new environment in which they compete for resources, mutator strains usually out-compete the wild-type strain if both are initially present in comparable numbers (12–14). During long-term adaptation or passage through selective bottlenecks, mutators can arise from a population and outgrow the wild-type strain (15–17). The advantage conferred by mutator genes is indirect because they increase fitness by introducing mutations at other loci that offer selectable growth benefits (14, 15).
Experiments involving the competition of bacteria with differing mutation rates have generally used only one or a few mutators, have not included antimutators, and have typically entailed pairwise competition. Although mutators often out-compete wild-type strains in serial transfer experiments, theoretical models predict that, as mutation rates increase, a threshold is crossed where hypermutability becomes more deleterious than beneficial (18, 19). Parameters such as the periodicity or randomness of change have also been modeled (20, 21), but few in vivo data are available to validate model predictions. A more complete assessment of the optimal mutation rates in competing populations would be facilitated by a large panel of mutators and antimutators of varying fidelities.
Escherichia coli DNA polymerase I (PolI) is a high-fidelity polymerase that participates in lagging-strand replication of chromosomal DNA and in DNA repair (22). We have created a large collection of PolI mutants that exhibit either an elevated or a reduced in vivo mutation rate, with a representation of fidelities spanning six orders of magnitude (23–26). Here we report on the use of these mutants in competition experiments to investigate the relationship between replication fidelity and evolutionary survival. In our experimental strain, the endogenous PolI is temperature sensitive such that at elevated temperatures DNA synthesis by PolI becomes dependent on expression of the plasmid-encoded mutants. After 350 generations of competition, a small subset of moderate mutators predominated. Variability in the final distribution of mutants in replicate cultures and in subsequent pairwise competitions supports the “hitchhiking” model of how mutator alleles confer selective advantage by facilitating adaptation through an increase in selectable genetic diversity (18, 27). The relationship between mutation rate and adaptability is of primary importance in understanding complex processes that are contingent upon mutation such as cancer progression, emergence of drug resistance, and the evolution of species (18, 28–31).
Results
Construction of Altered Fidelity PolI Mutant Library.
The PolI mutants used in this competition experiment differ from natural isolates in two fundamental ways. First, mutants were constructed on a 3′–5′ exonuclease-deficient background. To focus on base selection, we introduced a D424A substitution in the 3′–5′ exonuclease domain to ablate DNA proofreading activity of the enzyme (32). Because the mutation was present in all clones, for the sake of simplicity, the D424A mutant will be referred to from here on as “wild-type.” This wild type actually has a fourfold increase in mutation rate in vivo compared to the native large fragment of E. coli PolI. Second, the PolI mutants used in our competition harbor amino acid substitutions in the polymerase domain that alter their accuracy in base selection. These alterations confer mutator or antimutator properties. The mutants were obtained from three libraries created by mutating specific regions—motif A, motif B, or the entire polymerase domain—and were preselected for activity on the basis of genetic complementation (33). Mutants displaying altered replication fidelities were initially identified in an in vivo screen on the basis of the reversion of a plasmid-borne β-lactamase gene. The effect of PolI mutants on elevating chromosomal mutagenesis was confirmed by using reversion of a chromosomally encoded trpE65 (ochre) allele (24) and/or a forward mutation assay for rifampicin resistance (25). The per-base-pair rate of chromosomal mutagenesis by individual PolI mutator polymerases is approximately 10-fold lower than that obtained using reporter plasmid DNA (24, 25). This is presumably due to the fact that only a portion of the E. coli genome is replicated by PolI.
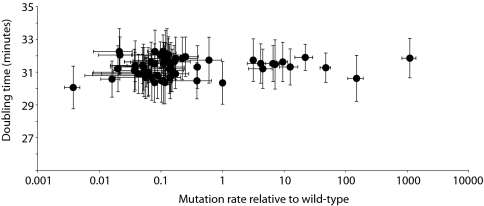
To facilitate genetic manipulation, we cloned PolI mutant genes into a plasmid (pHSG) bearing a PolI-independent origin of replication and a chloramphenicol resistance gene and transformed the reconstructed plasmid into the JS200 strain of E. coli, which harbors a temperature-sensitive endogenous PolI. Replication of the transfectant at 37 °C is dependent on the expression of the plasmid-encoded PolI (33, 34). All experiments and measurements were carried out at 37 °C to eliminate the contribution of endogenous PolI. Cell growth in liquid culture was measured by absorbance at 600 nm wavelength (OD600) and verified by plating on LB agar. Independent cultures of each of the mutants and wild-type PolI grew at similar rates (Fig. 1) for at least 19 generations.
Fig. 1.
Mutation rate and doubling time of individual PolI mutants before competition. Doubling times (mean ± SD) reflect log-phase growth in liquid culture at 37 °C. Mutation rates relative to wild type (mean ± SD) were determined by β-lactamase reversion frequency. The absolute mutation rate of cells expressing wild-type PolI was 2.0 ± 1.1 × 10−7. Mutation rates greater than one represent mutators, whereas those less than one represent antimutators.
Library Competition.
We initiated competitions with 11 mutators, 51 antimutators, and 4 wild-type (differing only by synonymous mutations) PolI clones. Overall, the mutation rate of mutants relative to wild type ranged from a 1000-fold increase to a 1000-fold decrease in accuracy (Fig. 1). Cells (1.5 × 108) of each E. coli transformant were combined to produce the starting input population. This mixture was divided equally into six tubes and cultured in 5 mL of rich (LB) media prewarmed to 37 °C with 30 μg/mL chloramphenicol. After 24 h of incubation, ≈3 × 106 cells from each tube were transferred to a new tube containing 5 mL of fresh media. The number of cells transferred and the transfer frequency is similar to protocols established by others (35). Transfer was repeated daily for 31 days to encompass a total of 350 cell generations, after which aliquots from each of the six parallel cultures were plated, and the PolI genes in the plasmids in 16 individual colonies from each culture were sequenced. An identical set of six competitions from independently harvested PolI isolates was also carried out in minimal media.
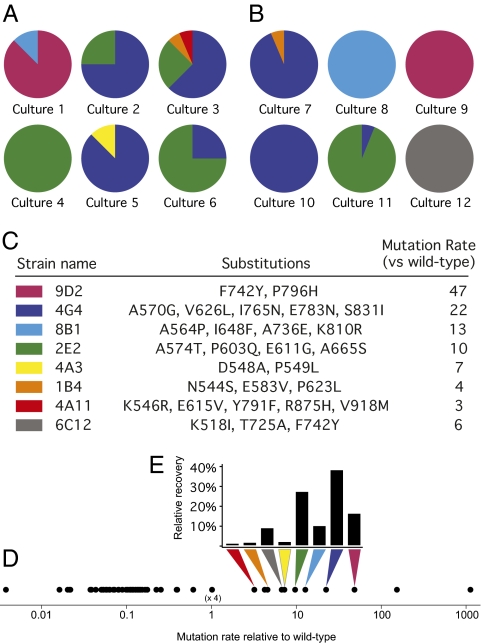
Of the 66 PolI species that were inoculated at the initiation of competition (input), only 8 genotypes were detected at the conclusion (output) (Fig. 2). The recovered mutants were all moderate mutators, with mutation rates ranging from 3- to 47-fold greater than that of the wild type. Of these, 88% had at least a 10-fold elevated mutation rate. No antimutators were detected in the output population despite constituting 77% of the input population. A total of 21 independently emerging clones (total number of pie wedges in Fig. 2 A and B) were identified at the end of the competition. The improbability that antimutators were not represented among winning clones by chance alone [P = (1–0.77)21 = 1.3 × 10−18] indicates their strong competitive disadvantage. No wild types were recovered even though they were initially present at a frequency fourfold greater than any other mutant. The null hypothesis that wild-type clones share an equal probability of surviving through competition because mutators can also be rejected (P = [1–4/(4 + 11)]21 = 1.5 × 10−3). The extreme mutators, I709N and I709N, A759R, with mutation rates of 150-and 1100-fold above that of wild type, respectively, were similarly not recovered.
Fig. 2.
Library competition outcome. (A) Mutants recovered after 31 days of competition in rich media. Sixteen isolates from each culture were sequenced. Each mutant identified is depicted as a different color. Pie charts reflect the relative abundance of mutants in each culture. (B) Mutants recovered after competition in minimal media. (C) Substitutions and relative mutation rate of recovered strains. (D) Distribution of fidelities represented among the input population. (E) Fidelity of mutants recovered at the end of competition and their relative frequency among the 196 isolates sequenced. Among 12 independent cultures, only moderate mutators survived to the end of competition.
The profiles of mutants recovered after competition in rich media (Fig. 2A) and minimal media (Fig. 2B) were similar. The limited number and unique set of mutants recovered in each culture is consistent with a process of ongoing evolution whereby mutations are stochastically acquired and undergo selection for advantageous growth phenotypes. The predominant survivor in each culture after competition was reassayed for mutation rate, which was not significantly different from the rate before competition, suggesting that the fidelity of the plasmid-encoded PolI gene was not altered during serial transfer.
Survivors Exhibit Different Growth Properties from Ancestors.
To investigate the degree to which survival of winning mutants correlated with novel phenotypes acquired during competition, we compared the growth properties of the most abundant winner in each rich media competition (from day 31) with the ancestor from which it originated (from day 0). Doubling time was calculated from OD600 measurements taken during exponential growth, and maximum culture density was assessed after 24 h of incubation (Table 1). In five of the six winners, significant differences existed between the winner and its ancestor. Four of these manifested as higher maximum culture densities whereas one winner exhibited a significantly shorter doubling time. No statistically significant difference in either parameter existed among ancestral strains.
Table 1.
Growth properties of competition survivors and their ancestors
| Mutation rate (relative to wild type) | Doubling time (min) | Maximum culture density (OD600) | ||||||
| Tube | Survivor | Precompetition | Postcompetition | P value | Precompetition | Postcompetition | P value | |
| 1 | 9D2 | 47 | 31.2 ± 0.7 | 32.1 ± 0.4 | — | 3.10 ± 0.10 | 4.45 ± 0.14 | 0.002 |
| 2 | 4G4 | 22 | 31.9 ± 0.5 | 31.3 ± 0.8 | — | 2.98 ± 0.07 | 3.71 ± 0.10 | 0.005 |
| 3 | 4G4 | 22 | 31.9 ± 0.5 | 31.6 ± 0.4 | — | 2.98 ± 0.07 | 3.23 ± 0.07 | — |
| 4 | 2E2 | 10 | 31.6 ± 0.5 | 29.2 ± 0.3 | 0.03 | 3.07 ± 0.08 | 3.10 ± 0.06 | — |
| 5 | 4G4 | 22 | 31.9 ± 0.5 | 31.4 ± 0.5 | — | 2.98 ± 0.07 | 3.56 ± 0.10 | 0.01 |
| 6 | 2E2 | 10 | 31.6 ± 0.5 | 31.8 ± 0.6 | — | 3.07 ± 0.08 | 3.62 ± 0.11 | 0.02 |
Cultures and measurements were made in triplicate with reported values given as the mean ± SE. P values were calculated by the Student’s t test.
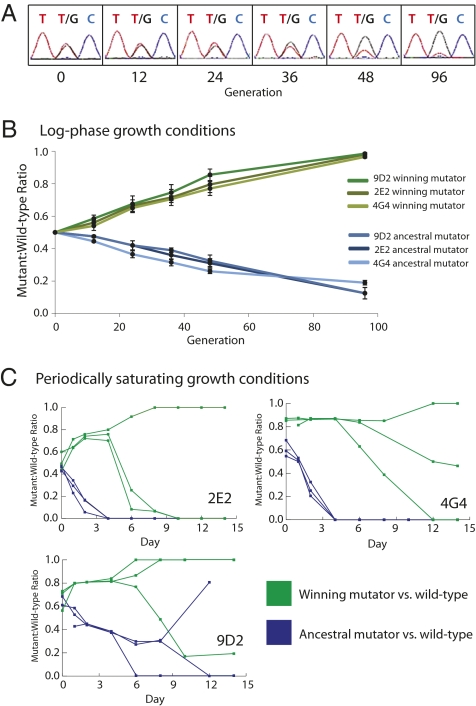
To empirically assess differences in relative growth between winning mutator clones and their corresponding ancestral strains, a series of pairwise experiments were carried out in which members of three winner/ancestor sets were separately competed against the same wild-type strain under continuous exponential growth. Each ancestor and each winner [strains 2E2 (culture 4), 4G4 (culture 5), and 9D2 (culture 1)] were mixed with an equivalent number of wild-type cells and sequentially passaged in rich media. The relative frequency of the bacterial strains at sequential time points was ascertained from nucleotide peak-height ratios obtained from capillary DNA sequencing chromatograms (Fig. 3A). All three ancestral mutants exhibited a clear and persistent growth disadvantage relative to wild type, whereas the mutants that were selected after 350 generations in the original competition exhibited a relative growth advantage (Fig. 3B).
Fig. 3.
Competitive efficiency of library competition winners relative to their precompetition ancestors. Winners and ancestors were individually competed against a single wild-type strain. After a 50:50 mixing, each culture was repeatedly passaged and the relative abundance of each strain determined at periodic time points by DNA sequencing and assessment of peak height ratios at sites differing between the mutant and wild-type strains. (A) Representative chromatograms at different time points in a pairwise competition demonstrating a temporal shift in culture composition. (B) Mutant:wild-type ratio as a function of generations of log-phase growth of winning mutators (green) and corresponding ancestors (blue) competed against wild type. Data points reflect triplicate cultures ± SE. (C) Mutant:wild-type ratio as a function of days of passage under library competition conditions. Data series within each panel represent individual cultures. Under both conditions, winners demonstrated a consistent growth advantage relative to wild type in contrast to the consistent disadvantage of their ancestors.
We then repeated the pairwise competitions under the same conditions used in the original library competition (daily passage, with stationary growth; Fig. 3C). Growth for the first 3 days was similar to that under continuous growth conditions with winners outgrowing wild type and wild type outgrowing ancestors at similar rates in replicate cultures. Beginning around the fourth day, however, the composition of individual cultures began to stochastically change with some dominant winner populations collapsing and other minority ancestor populations rebounding. Under these conditions, the later variability likely reflects ongoing evolution in response to the periodically saturated growth environment. It is interesting to note that, under both growth conditions, ancestral mutators appear to be at a growth disadvantage relative to wild type yet, in the original competition, were able to survive long enough to adapt and out-compete the wild-type strains by 31 days. The rebound of one ancestral 9D2 clone on day 7 (Fig. 3C) illustrates the occurrence of such an event. The absence of a preexisting growth advantage in ancestral mutators, along with the empirical fitness advantage and quantifiable new growth properties present in winning mutators, argue that second-site mutations arising during competition, rather than growth benefits conferred by the PolI alleles themselves, form the basis for mutator selection.
Moderate Mutators Evolve More Efficiently than Wild Type in Pairwise Competition.
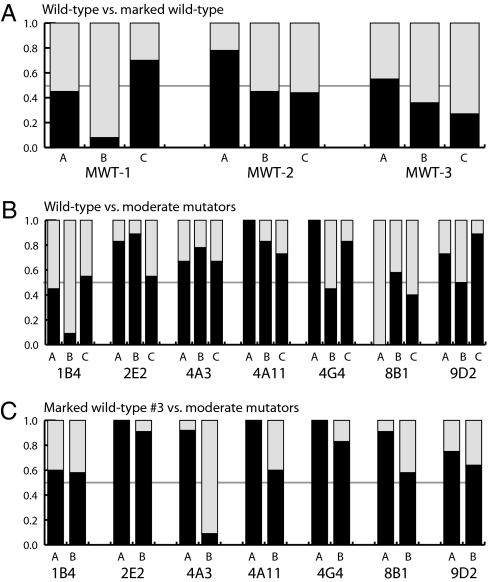
To confirm that the repeated selection for moderate mutators was the result of a reproducible adaptive advantage provided by the mutator PolI genes rather than by a few chance jackpot mutations that arose during the growth period prior to library competition, we carried out a large series of pairwise competitions using retransformed clones. Original stocks of plasmids containing the seven PolI moderate mutator variants recovered from the rich media competitions and four wild-type strains (differing only by synonymous SNPs) were retransformed into a fresh JS200 background. Five individual colonies (denoted A–E) from each transformation were picked and grown to midlog phase, quantified by OD600, and mixed 50:50 with one of five independent clonal isolates of a competitor strain. Approximately 107 cells of each mixture was used to seed 12 identical 1-mL rich media cultures. Daily passage of ≈4 × 106 cells into fresh media was carried out for 18 days (∼166 generations) after which time the PolI composition of each culture was determined by DNA sequencing. In 98% of competitions, one strain was found to have completely overtaken the other to “win” by 18 days. A detectable mixture of both competing strains remained in only 11 cultures. These appeared to be randomly distributed among different types of competitions and were excluded from analysis.
We first competed our experimental wild-type strain against each of three other wild-type strains marked by synonymous base substitutions to see how the distribution of winners varied under a neutral competition scenario (Fig. 4A). Each triplet cluster of bars in Fig. 4 represents competitions between wild type and one of the marked variants. Each bar within a cluster represents a group of approximately 12 identical competitions between a particular pair of clonal isolates of the wild type and the wild-type variant strains. The gray portion of each bar represents the fraction of the identical competitions in which the wild-type strain won, and the black portion represents the fraction won by the marked variant. The overall frequency of wins by the wild type and the marked wild-type strains was not significantly different (P = 0.47, two-tailed Fisher’s exact test), consistent with a neutral scenario whereby the probability of an advantageous mutation arising and leading to a clonal sweep is equal among competing strains.
Fig. 4.
Pairwise competitions. Each bar represents a series of approximately 12 identical cultures. Gray indicates the fraction won by the wild-type strain by day 18, and black indicates the fraction won by the strain noted beneath it. Clusters of bars indicate identical types of competitions carried out with independently transformed isolates of each competitor strain. (A) Competitions between wild-type and three neutrally marked wild-type (MWT) strains. As expected, the probability of winning was not statistically different among the different wild-type strains. (B) Competitions between wild-type and moderate mutators. (C) Competitions between a neutrally marked wild-type strain and moderate mutators. Mutator strains 2E2, 4A11, 4G4, and 9D2 were statistically more likely to win pairwise competitions than wild type.
We next carried out a similar series of competitions between wild type or a neutrally marked wild-type variant and each of the seven types of moderate mutators that survived through the original rich-media library competitions. As a group, the frequency of wins by moderate mutators was significantly greater than 50% (a neutral scenario) in both the wild-type competitions (Fig. 4B; P = 3.8 × 10−3) and the marked wild-type competitions (Fig. 4C, P = 6.0 × 10−6) by a one-tailed Fisher’s exact test. Merging data from Fig. 4 B and C to consider all of the ∼60 competitions with each type of moderate mutator, 4 of 7 mutators (2E2, 4A11, 4G4, and 9D2) won statistically more than half the time (P = 3.1 × 10−4, = 3.9 × 10−4, = 1.3 × 10−4, and = 4.1 × 10−2, respectively, by one-tailed Fisher’s exact test). Included among these were the three most prominent winners of the original rich-media library competition, entailing 94% of the population recovered there (Fig. 2 A and B). Significance was also met for strain 4A3 when excluding outlying data from isolate E. Both wild-type and moderate mutator strains won a subset of every type of competition, consistent with a model of winners emerging through natural selection of stochastically occurring mutations rather than from an initial growth advantage of one strain. For mutators with a significant pairwise adaptive advantage over wild type, the advantage was generally consistent across the five independent clonal isolates, indicating that it was dependent on the specific PolI variant rather than on stochastic differences in the background strain.
Mutator Phenotype Is Intrinsic to PolI Mutator Enzyme.
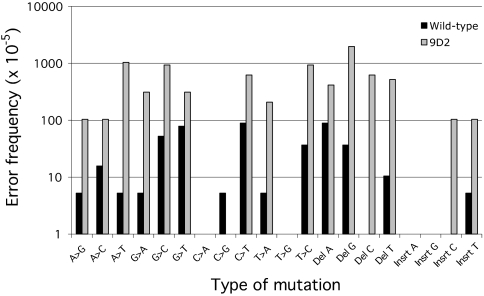
The strongest mutator that was recovered from the initial competitions was mutant 9D2, which produced a 47-fold elevated in vivo mutation rate. This PolI mutant harbors two mutations, F742Y and P796H, which are located in the fingers domain at the C terminus of the ends of the N-helix and P-helix, respectively, flanking the O-helix (motif B). Among family A polymerases, F742 has 63% identity and 67% similarity, whereas P796 has 70% identity and 70% similarity. To confirm that the in vivo mutator activity of this mutant was intrinsic to the PolI enzyme, we characterized its DNA replication fidelity in vitro. We first subcloned 9D2 into a high-expression pLEX vector that encoded an additional six N-terminal histidines. The mutator and a wild-type control were expressed and purified in tandem to apparent homogeneity using a histidine-binding nickel column as previously described (36). No 3′–5′ exonuclease activity was detected in either the wild-type or mutant protein. The in vitro replication fidelity of the mutator was measured using the M13 gap-filling assay (37). The 9D2 mutator exhibited a 20-fold increase in overall mutation frequency, with an elevation of all types of mutations except for C → A, C → G, and T → G substitutions (Fig. 5). There was an ≈1000-fold increase in C deletions and 100-fold increases in both T deletions and C insertions. The error rates that we determined for the wild-type control are consistent with previously published data (38).
Fig. 5.
In vitro mutation frequencies of specific types of errors determined by M13 gapped plasmid assay. Del, deletion; Insrt, insertion.
Discussion
If the results that we obtained with E. coli PolI can be generalized, they indicate that selective growth advantages can be conferred by enhanced mutagenesis in certain environments and that this advantage is conferred only within a narrow range of mutation rates. In our library competition experiments, the survivors were invariably mutators, the most prevalent having frequencies 10- to 47-fold greater than that of wild-type. Although we cannot completely rule out the possibility that certain PolI mutants may have a direct beneficial effect on bacterial fitness, several lines of evidence suggest that selection for mutators is the result of an increased probability of developing selectably advantageous mutations rather than of a preexisting growth advantage. First, the ancestors of winning mutator clones demonstrated a decreased, rather than an increased, fitness relative to wild type under pairwise competition (Fig. 3). Second, the distribution of survivors differed in both sets of independently passaged cultures originating from the same founding mixtures (Fig. 2 A and B). If one or a few PolI variants had a preexisting advantage, one would expect the composition of winners to be similar in all 12 cultures. Third, the three most prolific winners of the library competitions (entailing more than 80% of the populations recovered) won pairwise competitions statistically more frequently than wild type, but did not win in every culture (Fig. 4 B and C). In the absence of an adaptive advantage, a mutator would be expected to win no more frequently than wild type (Fig. 4A) whereas a preexisting growth advantage would manifest as a win in every culture. The consistency of adaptive advantage across independent isolates of each mutator strain argues against random differences in genomic background being responsible for the advantage. Fourth, measurements of growth rate and maximum culture density after the competition revealed that dominant survivors (in five of six rich-media cultures) differed significantly from their ancestors and from each other (Table 1). This observation suggests that the growth advantage of the survivors was gained during serial passages in culture. The diversity in phenotype further supports a “hitchhiking” model of how mutator alleles confer selective advantage (18, 27). Fifth, pairwise competition of winning mutators and their ancestors against wild type demonstrated a growth advantage of strains recovered after 350 generations that was not present initially (Fig. 3). This finding similarly indicates an increase in fitness acquired during competition.
No wild-type PolI was detected following competition. Although this is not a new result (13, 16), it is noteworthy that it has been observed in a system where a large diversity of fidelity mutants was competed. No antimutators were detected, even though 51 were included in our starting population. The absence of antimutators in the final population argues that they harbor decreased fitness relative to the rest of the population, despite their apparently normal growth rates when measured individually.
Extreme mutators did not fare any better in our competition. Two mutators with rates of ≈150- and 1100-fold greater than that of wild type failed to survive to the end of competition. This result was unanticipated. We had expected their high mutation rate to provide a selective advantage, as had previously been demonstrated in antibiotic-facilitated bottlenecks with this degree of mutator (25). These results demonstrate a limit to the selective advantage that can be conferred by enhanced mutagenesis.
Previous studies in the literature have established the importance of mutability for adaptation. Pairwise competition of bacteria demonstrated that mutators often out-compete their wild-type counterparts if both were originally present in comparable numbers (12–14). Long-term cultures of bacteria that were originally isogenic produced mutators that eventually out-competed the wild-type species (16, 17). Populations that were subjected to genetic bottlenecks also became dominated by mutators (39). Additionally, bacteria lacking endogenous error-prone DNA polymerases exhibited loss of fitness when grown in competition with a wild-type strain (40). We have now shown that a specific range of mutational rates is selected during competition of a large panel of polymerase mutants with fidelities spanning six orders of magnitude. Mutants within this range of elevated mutation rates are able to efficiently acquire beneficial mutations that allow them to outgrow the rest of the population without exceeding the threshold of error catastrophe (6, 18).
The range of favorable mutation rates is likely to depend on the environmental context of competition. Our experiments were carried out under periodically fluctuating conditions of rapid growth followed by nutrient depletion and confluence. Such a dynamic environment would be expected to produce many new selective pressures and favor strains with the ability to adapt. Competition in rich (LB) and minimal (M9) media selected transformants exhibiting a similar range of mutation rates. A different panel of mutators had been expected due to the assumption that a more stringent nutrient environment (minimal media) would necessitate a lower mutation rate. It is possible that selection for stability by minimal media may have been masked by a stronger selection for adaptation due to saturating conditions.
Our analysis of the mutant 9D2 confirmed that the mutator phenotype that it conferred in vivo is, indeed, determined by the PolI enzyme itself (Fig. 5). This mutator exhibited a 20-fold increase in mutation rate when replicating the M13 gapped plasmid. This elevation of mutation rate is in accord with the 47-fold increase observed in vivo, considering differences in sequence context and the likely effects of interacting proteins (24). The 20-fold overall increase in mutation rate of 9D2 is the largest of any of the library mutants with amino acid substitutions outside of the active site. Interestingly, this variant exhibited a 26-fold increase in the rate of insertions and deletions (Fig. 5), which are likely to cause frameshifts throughout the genome. Frameshifts may be well-tolerated in the short term as a species adapts to a new environment, given that loss of some nonessential genes may be energetically advantageous (41).
In these studies, we have empirically determined the optimal window of mutation rates for an evolving bacterial population. Our results indicate that under conditions where a clone’s fitness is not yet maximized for its environment, competitive adaptation may be facilitated by moderately enhanced mutagenesis. Although an increase in accuracy can be observed in vitro, this confers a handicap to competing cells. Within a population of mutants of varying fidelity, moderate mutators possess an advantage over antimutators, which are slow to acquire beneficial mutations, and over extreme mutators, which suffer a relative loss of fitness due to the accumulation of deleterious mutations (42). The enrichment of mutators in bacterial populations competing under stress may mimic selection in analogous situations, such as in human pathogens being confronted with an immune response or drug treatment or in tumors under growth-limiting conditions. Identifying ways to alter growth conditions in this E. coli model to select against mutators may offer insight into approaches that can be used to slow or arrest progression of microbial and neoplastic disease.
Materials and Methods
E. coli strain JS200 [recA718 polA12(ts) uvrA155 trpE65 lon-11 sulA1] was first described as SC18-12 (43). PolI mutants were constructed on pHSG-based plasmid pECpolI-3′exo− (24, 25), which contains a chloramphenicol-resistance selectable marker and a PolI-independent pSC100 origin of replication. All cultures were grown aerobically at 37 °C in prewarmed LB media supplemented with 30 μg/mL chloramphenicol unless otherwise noted. Details of competition culture conditions and mutation rate measurements are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Bradley Preston for suggesting competition experiments, to Manel Camps and Mike Schmitt for insightful comments and reagents, and to Lucy Kwong and Ayu Rahardjo for technical assistance. Research was supported by National Institutes of Health grants CA102029 and CA115802 (to L.A.L.) and AG033485 (to J.J.S). E.C.L. was supported by a scholarship from the Cora May Poncin Foundation and J.J.S. by a fellowship from the Achievement Rewards for College Scientists (ARCS) foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912451107/DCSupplemental.
References
- 1.Drake JW. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann NY Acad Sci. 1999;870:100–107. doi: 10.1111/j.1749-6632.1999.tb08870.x. [DOI] [PubMed] [Google Scholar]
- 2.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: Direct molecular test by using ribavirin. Proc Natl Acad Sci USA. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeb LA, et al. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Negishi K, Loakes D, Schaaper RM. Saturation of DNA mismatch repair and error catastrophe by a base analogue in Escherichia coli. Genetics. 2002;161:1363–1371. doi: 10.1093/genetics/161.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eigen M. Error catastrophe and antiviral strategy. Proc Natl Acad Sci USA. 2002;99:13374–13376. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldsby RE, et al. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 8.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 9.Furió V, Moya A, Sanjuán R. The cost of replication fidelity in human immunodeficiency virus type 1. Proc Biol Sci. 2007;274:225–230. doi: 10.1098/rspb.2006.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg SM. Evolving responsively: Adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- 11.Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao L, Cox E. Competition between high and low mutating strains of Escherichia. Evolution. 1983;37:125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibson TC, Scheppe ML, Cox EC. Fitness of an Escherichia coli mutator gene. Science. 1970;169:686–688. doi: 10.1126/science.169.3946.686. [DOI] [PubMed] [Google Scholar]
- 14.Giraud A, et al. Costs and benefits of high mutation rates: Adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 15.Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in A cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 17.Barrick JE, et al. Genome evolution and adaption in a long-term experiment with Escherichia coli. Nature. 2009;461:1219–1221. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 18.Radman M, Matic I, Taddei F. Evolution of evolvability. Ann NY Acad Sci. 1999;870:146–155. doi: 10.1111/j.1749-6632.1999.tb08874.x. [DOI] [PubMed] [Google Scholar]
- 19.Taddei F, et al. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 20.Ishii K, Matsuda H, Iwasa Y, Sasaki A. Evolutionarily stable mutation rate in a periodically changing environment. Genetics. 1989;121:163–174. doi: 10.1093/genetics/121.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornberg A, Baker T. DNA Replication. New York: W.H. Freeman; 1992. [Google Scholar]
- 23.Patel PH, Loeb LA. DNA polymerase active site is highly mutable: Evolutionary consequences. Proc Natl Acad Sci USA. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinkai A, Loeb LA. In vivo mutagenesis by Escherichia coli DNA polymerase I. Ile(709) in motif A functions in base selection. J Biol Chem. 2001;276:46759–46764. doi: 10.1074/jbc.M104780200. [DOI] [PubMed] [Google Scholar]
- 25.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh E, Choe J, Loeb LA. Highly tolerated amino acid substitutions increase the fidelity of Escherichia coli DNA polymerase I. J Biol Chem. 2007;282:12201–12209. doi: 10.1074/jbc.M611294200. [DOI] [PubMed] [Google Scholar]
- 27.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 28.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: Origin and consequences. Annu Rev Pathol. 2009;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orgel LE. Evolution of the genetic apparatus: A review. Cold Spring Harb Symp Quant Biol. 1987;52:9–16. doi: 10.1101/sqb.1987.052.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Johns GC, Joyce GF. The promise and peril of continuous in vitro evolution. J Mol Evol. 2005;61:253–263. doi: 10.1007/s00239-004-0307-1. [DOI] [PubMed] [Google Scholar]
- 32.Derbyshire V, et al. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 33.Sweasy JB, Loeb LA. Mammalian DNA polymerase beta can substitute for DNA polymerase I during DNA replication in Escherichia coli. J Biol Chem. 1992;267:1407–1410. [PubMed] [Google Scholar]
- 34.Camps M, Loeb LA. Use of PolI-deficient E. coli for functional complementation of DNA polymerase. Methods Mol Biol. 2003;230:11–18. doi: 10.1385/1-59259-396-8:11. [DOI] [PubMed] [Google Scholar]
- 35.Lenski R, Rose M, Simpson S, Tadler S. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 36.Shinkai A, Patel PH, Loeb LA. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J Biol Chem. 2001;276:18836–18842. doi: 10.1074/jbc.M011472200. [DOI] [PubMed] [Google Scholar]
- 37.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 38.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J Biol Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 39.Miller JH, et al. Direct selection for mutators in Escherichia coli. J Bacteriol. 1999;181:1576–1584. doi: 10.1128/jb.181.5.1576-1584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci USA. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong S, Khodursky A, Dykhuizen DE, Dean AM. Evolutionary genomics of ecological specialization. Proc Natl Acad Sci USA. 2004;101:11719–11724. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funchain P, et al. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witkin EM, Roegner-Maniscalco V. Overproduction of DnaE protein (alpha subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J Bacteriol. 1992;174:4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.