Abstract
Purpose
The retinal homeobox (Rx) gene plays an essential role in retinal development. An Rx-like (Rx-L) gene from Xenopus laevis has been identified. The purpose of this study was to analyze the function of Rx-L in the developing retina.
Methods
DNA-binding properties of Rx-L were analyzed by electrophoretic mobility shift assay (EMSA), with in vitro–translated proteins and radiolabeled oligonucleotide probe. The Rx-L expression pattern was analyzed by in situ hybridization using whole or sectioned embryos and digoxigenin-labeled antisense riboprobes. Rx-L loss of function was studied by using antisense morpholino oligonucleotides targeted to the Rx-L translation initiation site. Embryos injected with control or Rx-L morpholinos were analyzed at stage 41 or 45.
Results
Rx-L shares homology with Rx at the homeo-, OAR, and Rx domains, but lacks an octapeptide motif. Rx-L is expressed in the developing retina beginning in the early tailbud stage. In the maturing retina, Rx-L expression is restricted primarily to the developing photoreceptor layer and the ciliary marginal zone. Rx-L can bind a photoreceptor conserved element-1 (PCE-1) oligonucleotide, an element conserved among all known photoreceptor gene promoters. In a promoter activity assay, Rx-L functions as a stronger transcriptional activator than Rx. Antisense morpholino-mediated knockdown of Rx-L expression resulted in a decrease in rhodopsin and red cone opsin expression levels in Xenopus retinas. Injection of the Rx-L antisense morpholino oligonucleotide also resulted in a decrease in the length of both rod and cone outer segments.
Conclusions
The results suggest that Rx-L functions to regulate rod and cone development by activating photoreceptor-specific gene expression.
The retina is a multilayered neural tissue and is derived from the inner surface of the optic vesicle through cell migration and cell differentiation.1–3 Retinal development begins when the diencephalon evaginates during the neural tube stages of embryogenesis and contacts the presumptive lens ectoderm. A complex set of reciprocal cell and tissue interactions ensues, resulting in the thickening and invagination of the retinal neuroepithelium to form the eyecup. The cells of the retinal neuroepithelium undergo expansion, cell cycle exit, stratification, and differentiation, and ultimately form the mature neural retina. The processes of retinal cell specification and differentiation are regulated by many transcription factors (reviewed by Marquardt4). The earliest developmental processes of specification involve the actions of a set of transcription factors known as the eye field transcription factors (EFTFs).5 These include Pax6, ET, Six3, Optx2, Tlx, and Lhx2. These genes form a genetic network that is largely conserved from flies to humans.6 Many of the EFTFs are mutually inter-dependent for initiation or maintenance of expression. Proper expression and function of these gene products is essential for normal eye development. Another member of this group of EFTFs is the retinal homeobox gene, Rx.7 Rx is expressed in the anterior neural plate and the presumptive eye fields, coinciding with the initiation of expression of other EFTFs but before the onset of morphologic signs of retinal development, and continues throughout retinal maturation and differentiation.8–11 In zebrafish and Xenopus, misexpression of Rx results in the formation of ectopic retinal tissue at the expense of the forebrain.8,12,13 Inhibition of Rx expression or function results in severe deficiencies in eye development in several vertebrate species.8,12–17 This body of work indicates that Rx is essential for normal eye development.
Rx gene products share the following conserved domains with other aristaless-related homeobox gene family members: an N-terminal octapeptide domain, a highly conserved paired-like homeodomain, and a C-terminal orthopedia-aristaless-rx (OAR) domain.8,18 Rx has been shown to bind to PCE-1 (photoreceptor conserved element-1) elements found in functionally important regulatory regions of all known photoreceptor cell-specific genes,19 including the Xenopus laevis rhodopsin and red cone opsin genes.20–22 Rx has been reported to activate reporter constructs containing multimerized PCE-1 elements, albeit weakly.19,23,24 In addition to PCE-1 elements (also known as Ret1 elements), vertebrate rhodopsin promoters contain additional conserved elements, such as BAT/Ret2, NRE/Ret3, and Ret4.20,25 BAT elements interact primarily with members of the otx family, such as otx2, crx, and otx5b26,27 and can also weakly interact with Rx.19,24 The NRE is the site of interaction for the leucine zipper protein Nrl.28 Crx and Nrl (and frog analogues such as otx5b and XLmaf) can synergize to activate rhodopsin promoters.20,25–27,29,30 The Ret4 site is presumed to interact with homeobox transcription factors, since it contains a canonical TAAT core homeobox recognition sequence and has been reported to interact with crx and members of the Rx family.24
Some vertebrates encode a Rx-related gene product that functions as a stronger transcriptional activator than Rx, such as chicken RaxL23,31 and human QRX.24 Mice do not appear to encode a RaxL/QRx-like gene.24 The RaxL and QRX gene products share similar homeodomains and OAR domains with Rx but do not encode an octapeptide domain.23,24,31 Regions outside of these two domains share little identity. As is the case with Rx, QRX and RaxL have been shown to bind PCE-1 elements.23,24,31 It has been suggested that QRX also binds the BAT and Ret4 sites.24 QRX can activate a rhodopsin promoter reporter construct by co-operating with CRX and NRL. However, investigations of RaxL/QRx-like gene product function in retinal development have been hampered by the absence of loss-of-function data. We report the identification of a new member of the Rx family, Rx-L (Rx-like), from Xenopus laevis. We show that it is orthologous or analogous to QRX and RaxL in structure, expression, and molecular function and that it plays an important role in modulating the expression of opsin genes and photoreceptor development.
Methods
We confirm that this research adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Embryo Injection
Embryos were obtained by in vitro fertilization.32 Morpholinos (MOs; 0.1 or 0.25 mM) and green fluorescent protein (GFP) RNA (0.03 μg/μL) were co-injected into one prospective dorsal blastomere at the four-cell stage in a 10-nL volume. Embryos with GFP expression in only one eye were selected for further studies. The RxL antisense MO (5′-CTAGAAACATCCCTTGTGCTGACAG-3′), was designed by the manufacturer (Gene-Tools, Philomath, OR; http://www.gene-tools.com) to target a region including the translation start codon (in italic), 16 nucleotides upstream, and 5 nucleotides downstream. As a control, an MO with random sequence was obtained from Gene-Tools. Capped RNAs for microinjection were prepared (mMessage mMachine kits; Ambion, Austin, TX). RNAs were purified by gel filtration chromatography (RNA Mini Columns; Roche Diagnostics, Indianapolis, IN) to remove unincorporated nucleotides and cap analogue.33 Rx and Rx-L RNAs were prepared from plasmids pSP64T/Rx1A and pCS2/Rx-L, respectively.
DNA Constructs
The RxL expressed sequence tag (EST; XL073a16) was obtained from The National Institute of Basic Biology, Japan (http://xenopus.nibb.ac.jp) and will be referred to as pBS/Rx-L. The coding region (CDS) was amplified from the EST by using specific primers, RxL CDS F and R containing EcoRI and XhoI sites, respectively (F: 5′- GATCGAATTCGCAGCTAAGTGCAGTTCAGG, R: 5′-GATCTCAGATCAGATTGGCTGCCATG; restriction enzyme sites used for subcloning are in italic). The amplified product was subcloned into pCS234 using corresponding restriction sites to prepare pCS2/Rx-L. To prepare RxL-GFP, cDNA encoding the final 39 nucleotides of the 5′-untranslated region (UTR) and the first 38 amino acids of RxL after the start codon, which includes the target site for RxL MO, were amplified (primers: RxL CDS F, above, and RxL SGP R: 5′-GATCACTAGTCCTGCGGTGTTTCTTTTTAG), digested using EcoRI and SpeI (recognition sites were incorporated into PCR primers, in italic) and cloned in-frame with the GFP coding sequence in the super GFP (SGP) plasmid using EcoRI and XbaI sites.35 Amplification errors were minimized by using a high-fidelity polymerase (Platinum Pfx; Invitrogen, Carlsbad, CA). Integrity of the amplified products was ensured by sequencing (DNA Sequencing Core, Columbus Children’s Research Institute [CCRI]). For preparation of in situ hybridization probes, the Rx1A coding region was liberated from pSP64T/Rx1A using BglII and EcoRI, and ligated with a cloning vector (pBlueScriptII KS; Stratagene, La Jolla, CA) digested with BamHI and EcoRI, resulting in the plasmid pBS/Rx1A. XOP-Luc was prepared by inserting the BamHI-BglII fragment of pXOP-EGFP (the gift of Barry Knox)20 into the BglII site of pGL3 (Promega, Madison, WI). The Pax6 plasmid (pCS2/Pax6) was the gift of Yi Rao,36 and the Rx plasmid (pSP64T/Rx1A) was the kind gift of Milan Jamrich.8
Luciferase Assay
RNA encoding Rx or RxL (100 pg) and XOP-Luc reporter plasmid (25 pg) was injected into Xenopus laevis embryos (one blastomere at the two-cell stage). As an internal control, pRLtk, a plasmid containing a Renilla luciferase expression cassette under the control of the thymi-dine kinase promoter, (50 pg) was co-injected. For each set of effectors and reporters, 20 embryos were injected and cultured to approximately st 10.5. Embryos were divided into three groups of at least three embryos each, and lysed in PLB (20 μL/embryo; Stop ‘n Glo Dual Luciferase Assay Kit; Promega). Lysates were clarified by centrifugation37 and firefly, and Renilla luciferase activities were measured using 20 μL of lysate, according to the manufacturer’s instructions. Firefly luciferase levels were normalized against Renilla luciferase levels. The statistical significance of the results was determined with the Student’s group t-test.
Electrophoretic Mobility Shift Assays
Proteins were synthesized in vitro (Quick TNT Linked In Vitro Transcription/Translation Kit; Promega) and verified by SDS-PAGE. Electro-phoretic mobility shift assays using 3 μL of in vitro translated Rx or Rx-L were performed as described previously15 using a radiolabeled PCE-1 probe19 and unlabeled PCE-1, BAT-1,19 and Ret-420 oligonucleotide competitors. Rx and Rx-L were synthesized from plasmids pSP64T/Rx1A and pCS2/Rx-L, respectively.
Immunohistochemistry
Stage (st)-41 or -45 tadpoles having GFP expression in only one eye were fixed in 4% paraformaldehyde (MEMFA32) at room temperature for an hour, dehydrated with 100% methanol for at least 1 hour, embedded in paraffin, and serially sectioned at 8 μm. Staining of paraffin-embedded sections with antibodies was performed as previously described.38 Primary antibodies were used at the following dilutions: mouse anti-rhodopsin (RetP1; Biomeda, Foster City, CA) 1:50; mouse anti-islet 1, 1:50 (39.4D5; Developmental Studies Hybridoma Bank [DSHB], University of Iowa). Biotinylated-peanut agglutinin (PNA; Vector Laboratories, Burlingame, CA) was used at 5 μg/mL.
Quantitation of Rod Length
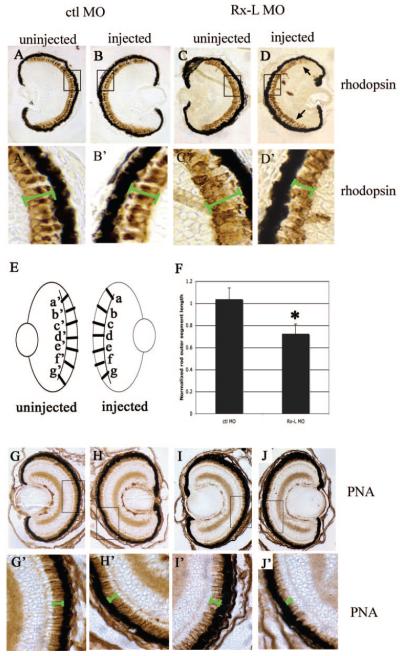
To determine the effect of Rx-L knockdown on rod length, we measured the width of the photoreceptor layer in digital images of injected or uninjected retinas immunostained with RetP1 antibody. Using image management software (Photoshop; Adobe Systems, Mountain View, CA), bars were drawn across the photoreceptor layer of the injected and uninjected retina of the same embryo. Each bar was drawn so that it spanned the region of Ret-P1 staining, along the schlerovitreal axis (parallel to the long axis of the rods, perpendicular to the retinal pigmented epithelium). Six or seven bars were drawn per section, at approximately equal intervals around the retina (Fig. 5E). The bars were drawn at similar positions of each uninjected and injected retina pair. The length of each bar was determined by the software. Bar lengths from injected retinas were normalized against the lengths of corresponding bars from uninjected retinas and averaged. In the example illustrated in Figure 5E, the lengths of bars a– g and a’–g’ would be determined and the following ratios would be calculated and averaged: a/a’, b/b’,… , g/g’. The average normalized bar lengths from Rx-L and control MO oligonucleotide-injected retinas were compared by a Student group t-test.
Figure 5.
The Rx-L antisense MO oligonucleotide resulted in perturbations in photoreceptor development. (A–D) Analysis of rod photoreceptor morphology visualized by immunohistochemistry with an antibody to rhodopsin (Ret-P1). (E) Diagram of measurements used to analyze changes in photoreceptor layer thickness. (F) Quantification of the effect of micro-injection of control or Rx-L antisense MO oligonucleotide on rod photoreceptor layer thickness. (G–J) Analysis of cone morphology visualized by binding to PNA. (A’–D’) and (G’–J’): high-magnification (100×) views of boxed area indicated in (A–D) and (G–J), respectively. Green bars indicate the width of the photoreceptor layer. All images were captured at 40× magnification. *P < 0.00013.
In Situ Hybridization
Wholemount and section in situ hybridizations were performed with digoxygenin-labeled antisense riboprobes, as described previously.32 Section in situs were performed on 8-μm sections of paraffin-embedded, paraformaldehyde-fixed embryos.39 Rx antisense probe was prepared from pBS/Rx1A linearized with EcoRI and transcribed with T7. Rx sense probe was prepared from pBS/Rx1A linearized with BamHI and transcribed with T3. To prepare Rx-L antisense probe, we linearized pBS/RxL with EcoRI and transcribed it with T7 RNA polymerase. To prepare Rx-L sense probe, we linearized pBS/RxL with XhoI and transcribed it with T3 RNA polymerase. To prepare the Pax6 antisense riboprobe, we linearized pCS2/Pax6 with HindIII and transcribed it with T3 RNA polymerase. Rhodopsin cDNA was amplified from st 37 tadpole RNA by using specific primers (F: 5′-GCTGCTACCATGAACGGAAC-3′; R: 5′-GAAGCTCTTATCCAGGAGACAC-3′) and was sub-cloned by using the TOPO TA system (Invitrogen). Riboprobe was synthesized from BamHI-linearized DNA using T7 RNA polymerase. Probes for red cone opsin (RCO) were prepared as described previously.8,40
Real-Time PCR
Total RNA was extracted (TriZOL; Invitrogen) from isolated heads of st-41 embryos bilaterally injected with Rx-L or control MO. Three embryos were used in each group. RNA from each head was reversed transcribed to cDNA (Superscript III First-Strand Synthesis System for RT-PCR; Invitrogen) using random hexamer oligonucleotides as primers, according to the manufacturer’s protocol. Levels of rhodopsin and red cone opsin cDNAs were quantified by real-time PCR. Real-time PCR was performed in a 25-μL amplification mixture containing 1 μLof template cDNA, 12.5 μL of 2× PCR master mix (SYBR Green; Applied Biosystems, Foster City, CA), and 100 nM forward and reverse primers, respectively (5′-TCCTGATCTGTTGGGTGCC-3′, 5′-TGAAGACTGGGCCAAAGTCG-3′ for rhodopsin; 5′-TCTTGCGTTTGGTCTGCAGG-3′, 5′-TGATGTCTTCAAGCCGTGAGG-3′ for red cone opsin; and 5′-CAGATTGGTGCTGGATATGC-3′, 5′-ACTGCCTTGATGACTCCTAG-3′ for EF-1α as an internal control). The PCR conditions included a polymerase activation step at 95°C for 10 minutes followed by 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds and run on a sequence detector (model 7500; Applied Biosystems). The relative change of rhodopsin or red cone opsin in the injected embryos was calculated by 2−ΔΔCT, using EF-1α as an internal control and uninjected embryos as a reference. The statistical significance of relative differences in expression levels was determined by Student’s group t-test.
Results
Sequence Analysis of Rx-L
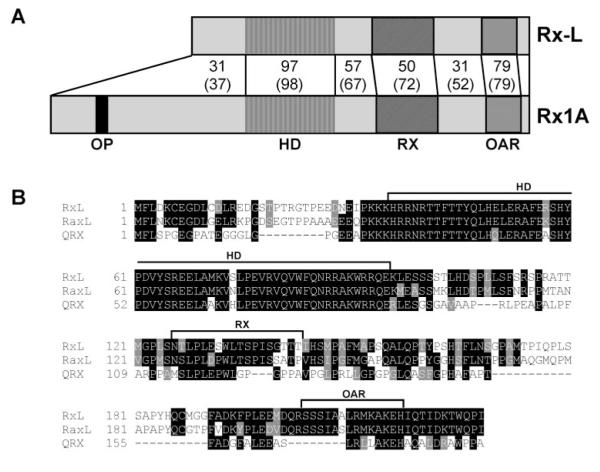
We identified an EST encoding an Rx-like gene product by BLAST search of Xenopus ESTs. Figure 1 shows the alignment between Xenopus laevis Rx1A with Rx-L. The overall identity is 62% and similarity is 75%. The homeodomains of Rx-L and Rx1A are nearly identical, with only one mismatch (97% amino acid identity and 98% similarity). The C-terminal OAR domains of the two protein sequences also share high homology. The region between two domains is relatively dissimilar, except for the Rx-domain, which shows moderate identity and similarity. Rx-L does not encode an octapeptide motif, found near the N terminus of Rx and many related proteins.
Figure 1.
Rx-L is related to Rx and similar to other Rx-like proteins. (A) Comparison of the predicted protein sequence of Xenopus laevis Rx-L and Rx1A. Rx1A and Rx-L sequences were aligned by using ClustalW (http://www.ebi.ac.uk/clustalw; European Bioinformatics Institute, European Molecular Biology Laboratory, Heidelberg, Germany). The percentage of amino acid identity and similarity (with respect to Rx-L) are summarized in the figure. Canonical domains are indicated. (B) Alignment of Xenopus laevis (Rx-L), chicken (RaxL), and human (QRX) Rx-like gene product sequences by ClustalW. Canonical domains are denoted with brackets above the alignment. OP, octapeptide; HD, homeodomain; RX, Rx domain; OAR, orthopedia-aristaless-Rx domain.
Similar Rx-like gene products, RaxL and QRX, have been identified in chick and humans, respectively.23,24,31 Rx-L is also similar to QRX and RaxL (Fig. 1B). Of note, the Rx domains of the Rx-like proteins are more similar to each other than they are to that of Rx.
Expression of Rx-L in Xenopus Retina
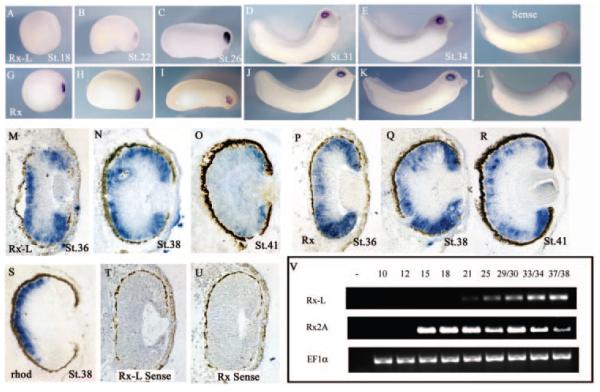
The expression of Rx-L during early Xenopus eye development was analyzed by in situ hybridization and RT-PCR (Fig. 2). Rx-L is not expressed at neurula stages, and extremely faint Rx-L expression can be detected in the early tailbud embryo stage (Figs. 2A, 2B), whereas Rx is expressed robustly in the developing eye in these stages (Figs. 2G, 2H). In late tailbud stages, Rx-L expression is observed in the developing eye, and becomes excluded from the developing lens (Figs. 2C–E), as is the case for Rx (Figs 2I–K). No staining was observed when embryos are probed using sense control riboprobes encoding Rx-L (Fig. 2F) or Rx (Fig. 2L). These results suggest that Rx-L is expressed in the developing retina but that Rx-L expression begins later than that of Rx.
Figure 2.
Rx-L was expressed in the developing retinas of tailbud embryos and tadpoles. (A, L) Whole-mount in situ hybridization (ISH) with an Rx-L (A–E) or an Rx(G–K) antisense riboprobe. (F, L) ISH with Rx-L (F) or Rx (L) sense control probes. (M–U) ISH on sections of paraffin-embedded tadpoles with antisense riboprobes for Rx-L (M–O), Rx (P–R), or rhodopsin (S) at st 36 (M, P), st 38 (N, Q, S), or st 41 (O, R). (T, U) ISH with sense control riboprobes for Rx-L (T) or Rx (U) at st 36. All wholemounts and sections are oriented with dorsal side to the top of the images. Embryo staging is indicated in each panel. (V) Analysis of expression of Rx and Rx-L in early development by RT-PCR with RNA purified from whole embryos (at indicated stages). EF-1α was used as a control for RT-PCR.
The spatial pattern of Rx-L expression was analyzed in further detail by in situ hybridization using sections of Xenopus tadpoles. At st 36 and 38, when photoreceptors have begun differentiating and the nuclear layers of the retina are becoming defined, Rx-L expression was found primarily in the peripheral and outer portion of the central regions of the neural retina (Figs. 2M, 2N), including the ciliary marginal zone (CMZ), where retinal progenitor cells are found at this stage.41 The expression of Rx-L in the central retina is comparable to that of rhodopsin, which is restricted to cells in the differentiating photoreceptor layer (Fig. 2S). At these stages, Rx is also expressed in the CMZ and the outer neural retina (Figs. 2P, 2Q). Rx is expressed in a thicker layer than Rx-L and rhodopsin in the central neural retina, comprising the developing outer nuclear layer and at least a portion of the inner nuclear layer. At st 41 when retinal cell types have become morphologically defined, Rx-L expression can no longer be detected in the central retina (Fig. 2O). Residual expression can be detected in the CMZ. By contrast, Rx is expressed in the photoreceptor layer, intermittently in the inner nuclear layer, and robustly in the CMZ (Fig. 2R). Staining was not observed when Rx or Rx-L sense control riboprobes were used (Figs. 2S, 2T). These results suggest that Rx-L expression is restricted to the differentiating photoreceptor layer and the CMZ in the developing retina at st 36 to 38, when photoreceptor differentiation is initiated.
To pinpoint the initiation of Rx-L expression, we performed RT-PCR using RNA isolated from whole embryos at different stages (Fig. 2V). Low levels of Rx-L expression can be detected as early as st 21 (neural tube/early tailbud stage) and increases by st 25 and 30. Rx expression is detectable at st 13 to 14, the late gastrula/early neural plate stages8 when Rx-L expression is not detectable. Rx-L expression continues through st 38, when photoreceptors are differentiating. Taken together, these results indicate that Rx-L is expressed during stages of retinal development and differentiation, after the initial specification of the eye, until retinal maturation.
Binding of the PCE-1/Ret1 Site
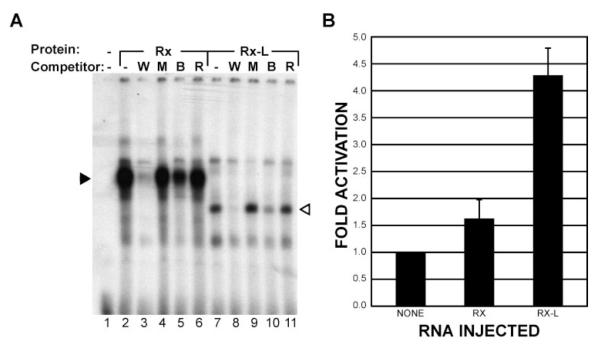
The DNA-binding properties of the Rx-L gene product were studied by EMSA using a radiolabeled PCE-1 oligonucleotide probe and synthetic protein. As has been reported for Rx,19 Rx-L bound the PCE-1 oligonucleotide. A shifted band was observed when the PCE-1 probe was incubated with Rx-L protein, indicating interaction between Rx-L protein and the PCE-1 oligonucleotide (Fig. 3A). The radiolabeled complex was not observed in the presence of an excess of unlabeled wild-type but not mutated PCE-1 oligonucleotide, indicating that binding of Rx-L to the PCE-1 probe was specific. It has been suggested that QRX binds other conserved elements found in the rhodopsin promoter containing core homeodomain binding motifs, such as BAT and Ret4.24 As reported previously, Rx and Rx-L had less affinity for the BAT element, as the BAT competitor decreased the binding between Rx-L and the PCE-1 probe but failed to completely compete for binding (Fig. 3A, lanes 5, 10). The Ret-4 oligonucleotide did not compete significantly for binding of either Rx or Rx-L to the PCE-1 probe (Fig. 3A, lanes 6, 11), indicating that neither Rx-L nor Rx bind appreciably to the RET-4 element. Taken together, our data suggest that the primary binding site for Rx-L and Rx is the PCE-1 site.
Figure 3.
Function of Rx-L as a transcription factor. (A) Rx-L binds the same oligonucleotide as RLamx. Synthetic Rx1A (lanes 2, 6) and Rx-L (lanes 7, 11) were used in an EMSA with a radiolabeled PCE-1 probe and various oligonucleotide competitors. W, wt PCE-1; M, mutated PCE-1; B, BAT-1; R, Ret4. The major specific Rx and Rx-L complexes are indicated (solid and open arrowheads, respectively). (B) Rx-L is a stronger transcriptional activator than Rx. Lucif-erase assay performed with lysates from embryos coinjected with XOP-Luc reporter plasmid and Rx or Rx-L RNAs, as shown.
Rx-L as a Transcriptional Activator
To investigate the function of Rx-L, we generated a DNA reporter construct, XOP-Luc, containing a minimal XOP promoter and a luciferase expression cassette. The XOP promoter contains four highly conserved transcriptional binding sites: PCE-1, BAT1, NRE, and Ret4.20 The ability of Rx and Rx-L to activate the XOP-Luc reporter was assayed in Xenopus embryos (Fig. 3B). When Rx RNA was co-injected with the XOP reporter construct, there was approximately a 1.5-fold increase in XOP-Luc reporter activity; however, injection of a similar amount of Rx-L RNA elicited a 4.2-fold increase in reporter activity. Rx-L activated expression of the reporter construct to a significantly greater degree than did Rx (P < 0.0097), indicating that it is a stronger activator than Rx.
Effect of the Knockdown of Rx-L
To study the function of Rx-L in Xenopus retinal development, an antisense MO oligonucleotide targeted to the Rx-L translation start (Rx-L MO) was used to knock down Rx-L expression in Xenopus embryos. The specificity of the Rx-L MO was assayed in two ways. First, the Rx-L MO inhibited in vitro translation of Rx-L in a dose-dependent manner, whereas a control MO oligonucleotide (ctl MO) did not affect Rx-L translation (Fig. 4A). To further confirm the efficacy of the Rx-L MO in Xenopus embryos, we made a DNA construct (Rx-L-GFP) containing the 5′-end of the Rx-L cDNA, including the MO target, fused in-frame with a GFP expression cassette. Every embryo injected with Rx-L-GFP exhibited GFP expression (Fig. 4C). Co-injection of Rx-L-GFP with control (ctl) MO at different concentrations (Figs. 4D, 4E) did not affect GFP expression. The Rx-L MO knocked down the GFP expression to nearly undetectable levels at both concentrations (Figs. 4F, 4G). From these results, we determined that the Rx-L MO could specifically knock down the translation of Rx-L.
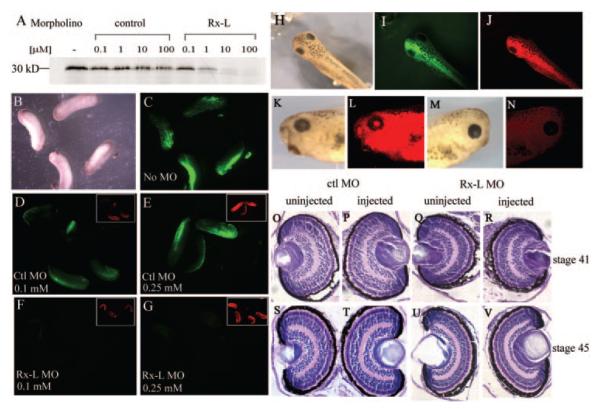
Figure 4.
Injection of the Rx-L anti-sense MO oligonucleotide specifically inhibited translation of Rx-L but did not result in gross changes in eye morphology. (A) The Rx-L antisense MO oligonucleotide (Rx-L MO), but not the control MO oligonucleotide (ctl MO), can inhibit Rx-L translation in vitro. (B–G) The Rx-L MO, but not a control MO, can inhibit expression of a Rx-L-GFP fusion protein containing the Rx-L MO target in Xenopus embryos. (B, C) bright-field (B) and fluorescent (C) views of embryos injected with Rx-L-GFP plasmid. (D, E) GFP fluorescence in embryos injected with Rx-L-GFP plasmid and 0.1 (D) or 0.25 (E) mM ctl MO. (F, G) GFP fluorescence in embryos injected with Rx-L-GFP plasmid and 0.1 (F) or 0.25 (G) mM Rx-L MO. Insets: visualization of lissamine tag of MO (red fluorescence). (H–N) Microinjection of 0.25 mM Rx-L MO did not affect external morphology of the eye. Embryos were unilaterally co-injected with lissamine-conjugated Rx-L MO and GFP RNA at the four-cell stage. (H–J) Dorsal views of injected embryos with bright-field illumination (H), green fluorescence to detect GFP tracer (I) or red fluorescence to detect lissamine-conjugated MO (J). (K, L) lateral view of the injected side of the same embryo viewed with bright-field illumination (K) or red fluorescence (L). (M, N) lateral view of the uninjected side of the same embryo viewed with bright-field illumination (M) or red fluorescence (N). (O–V) Retinas from embryos injected with Rx-L MO appeared histologically similar to those from embryos injected with ctl MO at st 41 and 45 (as indicated).
Next, we knocked down expression of Rx-L, by injecting Rx-L MO unilaterally into Xenopus embryos along with a GFP lineage tracer. No outward changes were observed in these embryos (Figs. 4H–N). Injected tadpoles with normal ocular appearance and unilateral GFP expression were fixed and analyzed. No obvious histologic changes were observed in these retinas (Figs. 4O–V).
The effects of injection of the Rx-L MO on rod photoreceptors were analyzed by immunostaining with RetP1 (anti-rhodopsin) antibody (Figs. 5A–D). No changes in Ret-P1 immuno-staining were observed in retinas of ctl MO–injected embryos (compare Figs. 5A, 5B). Retinas of Rx-L MO–injected embryos appeared to have shorter outer segments (compare Figs. 5C and 5D). In two independent experiments, the rods of Rx-L MO–injected eyes were significantly shorter than those of ctl MO–injected eyes. In one experiment (Fig. 5F), the rods of MO-injected eyes were 72.13% ± 9.01% the length of those of the contralateral uninjected side, whereas rods of ctl MO–injected eyes were nearly the same length (103.38% ± 10.55%). In a second, independent experiment, we found that rods of MO-injected eyes were 78.06% ± 10.66% the length of those of the contralateral uninjected side, whereas rods of ctl MO–injected eyes were nearly the same length (100.34% ± 3.66%). In both cases, Rx-L MO injection resulted in a significant decrease in rod length compared with the ctl MO (experiment 1: P < 0.00013; experiment 2: P < 0.00014). The rhodopsin RNA level was also studied directly by in situ hybridization using a rhodopsin antisense riboprobe (Figs. 5A–5D). We found that rhodopsin expression was reduced in retinas injected with the Rx-L MO (compare Figs. 6C, 6D). The control MO had no apparent effect on rhodopsin expression (compare Figs. 6A, 6B). We verified these results by quantitative RT-PCR (Fig. 6I). We found that injection of Rx-L MO reduced rhodopsin expression by nearly 90% compared with when ctl MO was injected, a statistically significant reduction (P < 0.002). Our results suggest that the rhodopsin level of the retina on the MO-injected side of the embryo was much lower than on the uninjected side, suggesting that Rx-L expression is necessary for rhodopsin gene expression and proper rod photoreceptor development.
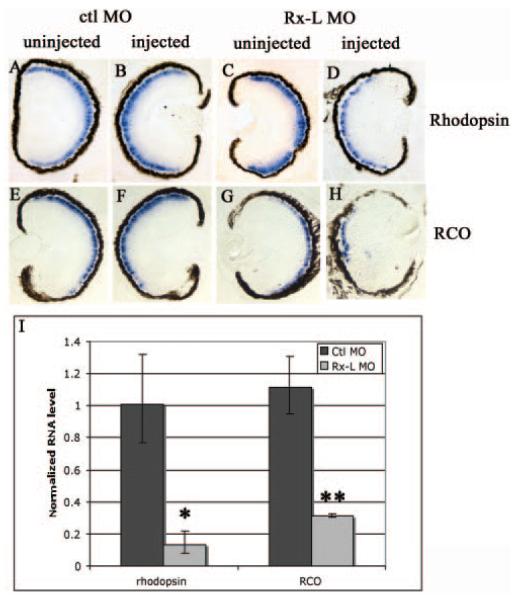
Figure 6.
Rx-L knockdown decreased rod and cone opsin expression. (A–D) Analysis of rhodopsin gene expression by in situ hybridization performed on sectioned retinal tissue. (E–H) Analysis of red cone opsin expression by in situ hybridization. (I) Quantatative analysis of red cone opsin expression levels in control and Rx-L antisense MO oligonucleotide–injected embryos by real-time RT-PCR. *P < 0.002; **P < 0.001.
The red cone opsin (RCO) promoter also contains a conserved PCE-1 element,22 suggesting that it may be a target of Rx-like transcription factors. We investigated the expression of RCO in Rx-L knockdown embryos by in situ hybridization (Figs. 6E–6H). We found that Rx-L knockdown resulted in a decrease in RCO expression (compare Figs. 6G, 6H). Using quantitative RT-PCR, we found that injection of the Rx-L MO reduced expression of RCO by more than 70% compared with injection of ctl MO, a statistically significant reduction (P < 0.001). To determine the effect of Rx-L knockdown on cone development, we used PNA, which binds to the cone outer sheath and labels essentially the entire cell (Figs. 5G– J).42,43 Injection of the Rx-L MO diminished the width of the cone photoreceptor layer as visualized by PNA labeling (Figs. 5I, 5J), whereas injection of ctl MO did not affect PNA labeling (Figs. 5G, 5H). These data suggest that the knockdown of Rx-L reduces expression of RCO and has a deleterious effect on cone development.
The effects of Rx-L knockdown seemed to be specific to photoreceptors. We analyzed the expression of Pax6 in Rx-L MO–injected embryos by in situ hybridization (Fig. 7A–D). In st-41 tadpoles, Pax6 is expressed broadly in the ganglion cell layer and the inner nuclear layer and is unaffected by injection of the Rx-L MO (compare Figs. 7C and 7D). We also analyzed, by immunohistochemistry, the expression of Isl-1, a transcription factor expressed primarily in retinal ganglion cells and to a lesser degree in amacrine and horizontal cells, (Figs. 7E–H). Injection of the Rx-L MO did not noticeably affect the expression of Isl-1 (compare Figs. 7G, 7H). In addition, injection of the Rx-L MO did not affect expression of other homeodomain transcription factors expressed in the developing retina, such as Rx and otx5b (data not shown). These results verify that the effect of Rx-L knockdown on photoreceptors was specific and not the result of a general deficiency in retinal development.
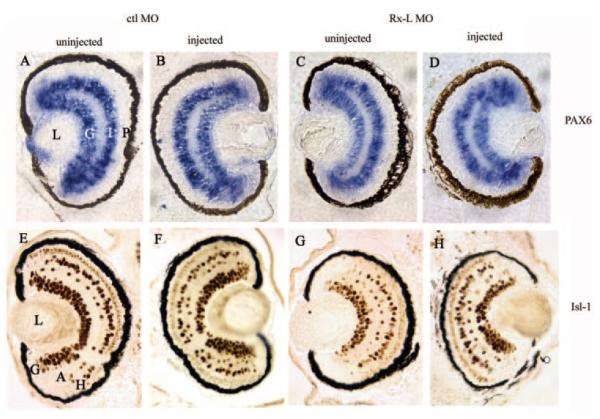
Figure 7.
Microinjection of the Rx-L MO oligonucleotide was not broadly deleterious to retinal development. (A–D) The expression of Pax6 in the ganglion cell and inner nuclear layers of the neural retina are unaffected by injection of the Rx-L MO. (E–H) Injection of the Rx-L MO did not affect the development of nonphotoreceptor cell types such as ganglion, amacrine, and horizontal cells, visualized by immunohistochemistry with an antibody to Isl-1. (A), (E) and (C), (G): retinas from uninjected sides of embryos injected with control or Rx-L MO, respectively. (B), (F) and (D), (H): retinas from injected sides of embryos injected with control or Rx-L MO, respectively. A, amacrine cells; G, ganglion cell layer; H, horizontal cells; I, inner nuclear layer; L, lens; P, photoreceptor layer.
Discussion
Rx-L is a newly discovered member of the Rx family. Rx-L and Rx belong to the aristaless-related paired-like homeobox gene family.18 Members of this subfamily of homeobox proteins contain a characteristic homeodomain and a conserved domain of unknown function located near the C terminus, called the aristaless or OAR domain. Most members of this family also have an octapeptide motif related to the eh-l sequence at N-terminal region. The octapeptide motif mediates transcriptional repression through interaction with corepressors of the groucho-TLE family.44,45 Our results suggest that Rx-L and Rx have different transcriptional activities. Xenopus Rx is a weak activator, whereas Rx-L is a relatively potent activator. This is in agreement with comparisons of transcriptional activities of Rax/RaxL23 and RX/QRX.24 The ability of Rx-L to function as a stronger activator than Rx is largely due to the lack of an octapeptide motif in Rx-L (El-Hodiri et al., unpublished data, 2006). Rx-L is one of very few members in aristaless-related gene family that does not have an octapeptide motif, along with Prx and Otp, which are known to function as transcriptional activators.46
Why are Rx-like gene products needed in addition to Rx/Rax gene products? Perhaps these gene products function to activate expression of PCE-1-containing promoters during development. Because both Rx and Rx-L bind the PCE-1 site and Rx-L is a stronger activator than Rx, the function of the Rx-like family members may be to boost, rather than initiate, promoter activity. There are examples of control of target gene activity during different phases of development by differential expression of transcriptional regulators. For example, in anterior pituitary development, the transcriptional repressor Rpx/Hesx1 is replaced by the activator Prop-1 when pituitary development shifts from specification and patterning to differentiation.47
Our results indicate that one primary function of Rx-L is to regulate photoreceptor-specific gene expression. Our EMSA results suggest that Rx-L binds to the Ret-1/PCE-1 site, a site usually upstream of photoreceptor related genes. QRX was found to bind to the Ret4 site as well as the PCE-1 site.24 Our data indicate that Rx-L primarily interacts with the PCE-1 site and weakly, if at all, with the Ret4 site in EMSA. This apparent difference in binding specificity is difficult to understand in light of the high degree of identity between the homeodomains of Xenopus Rx-L and human QRX (93.4%). However, it is possible that the few differences between the two proteins result in subtle difference in DNA-binding characteristics.
Our in situ hybridization results show that Rx-L is primarily expressed in the maturing photoreceptor layer in Xenopus tailbud embryos. In contrast, QRX is expressed in inner retinal layers in addition to the outer retina.24 This may reflect a real difference between the expression patterns of Rx-like genes in frogs and mammals or may reflect differences in photoreceptor development between these organisms. Chicken RaxL is expressed in both inner and outer retinal layers during retinal maturation but becomes restricted to the photoreceptor layer by E14.23 Overall, then, Rx-like genes are expressed in developing photoreceptors.
Our results suggest that Rx-L is involved in regulating photoreceptor development by controlling transcription of photo-receptor-specific genes. Studies in chicken demonstrated that expression of a putative dominant negative allele of a chicken Rx-like gene, cRaxL, causes a decrease in expression of early photoreceptor markers.23 However, expression of a dominant negative or interfering form of a transcription factor does not necessarily result in loss of function. Mutations in QRX have been found in some human retinal diseases probably due to a role for QRX in photoreceptor function and/or survival,24 although these studies do not conclusively demonstrate that QRX mutations result in these diseases. Mice do not appear to encode a QRX/Rx-L gene,24 making it impossible to study QRX/Rx-L function by gene knockout in mammals. Xenopus is thus an ideal animal model to study Rx-L loss of function. Our data suggest that an Rx-L MO can knock down the expression of Rx-L both in vitro and in Xenopus embryos, and that Rx-L is required for the normal spatial expression of photoreceptor-specific genes at appropriate levels. Our results also indicate that loss of Rx-L expression leads to defects in photoreceptor development.
Several vertebrate species have the Rx-L gene in addition to the Rx gene, as discussed above. A notable exception seems to be the mouse, whose genome does not encode an Rx-like gene. This suggests that, in mice, Rx can function as both Rx and Rx-like gene products do in other species. Specifically, this suggests that mouse Rx can act as a strong transcriptional activator, at least under some circumstances, despite the presence of an octapeptide motif and high levels of expression of groucho family members in the developing Xenopus eye.48 One possibility is that Rx-groucho interactions are regulated at the physical or functional level, to allow mouse Rx to function as a strong transcriptional activator during some periods of development, such as the initial phases of photoreceptor development, to promote increased levels of transcription of Rx-L target genes such as rhodopsin and red cone opsin. Additional experiments are necessary to explore this possibility and determine conditions under which Rx may function as a strong activator.
Acknowledgments
The authors thank Xiu-Lan Qi for technical assistance in the early phases of this project; William Harris, Milan Jamrich, Barry Knox, and Yi Rao for kindly providing the plasmid; Tiffany Cook, David Cunningham, Andy Fischer, Milan Jamrich, Peter Mathers, Orson Moritz, Donald Zack, and members of the El-Hodiri laboratory for critical discussions and technical suggestions; and Andy Fischer, Holly Moose, and Amy Sater for critical reading of the manuscript.
Supported by National Eye Institute Grant EY015480 (HME). The 39.4D5 (anti-Isl-1) antibody, developed by T. Jessell, was obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, Iowa.
Footnotes
Disclosure: Y. Pan, None; S. Nekkalapudi, None; L.E. Kelly, None; H.M. El-Hodiri, None
References
- 1.Zaghloul NA, Yan B, Moody SA. Step-wise specification of retinal stem cells during normal embryogenesis. Biol Cell. 2005;97:321–337. doi: 10.1042/BC20040521. [DOI] [PubMed] [Google Scholar]
- 2.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 3.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 4.Marquardt T. Transcriptional control of neuronal diversification in the retina. Prog Retin Eye Res. 2003;22:567–577. doi: 10.1016/s1350-9462(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 5.Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- 6.Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- 7.Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- 8.Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 9.Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- 10.Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev. 1997;61:187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang JC, Raymond PA. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev Biol. 2001;231:13–30. doi: 10.1006/dbio.2000.0125. [DOI] [PubMed] [Google Scholar]
- 13.Andreazzoli M, Gestri G, Angeloni D, Menna E, Barsacchi G. Role of Xrx1 in Xenopus eye and anterior brain development. Development. 1999;126:2451–2460. doi: 10.1242/dev.126.11.2451. [DOI] [PubMed] [Google Scholar]
- 14.Loosli F, Winkler S, Burgtorf C, et al. Medaka eyeless is the key factor linking retinal determination and eye growth. Development. 2001;128:4035–4044. doi: 10.1242/dev.128.20.4035. [DOI] [PubMed] [Google Scholar]
- 15.Voronina VA, Kozhemyakina EA, O’Kernick CM, et al. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 16.Tucker P, Laemle L, Munson A, et al. The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis. 2001;31:43–53. doi: 10.1002/gene.10003. [DOI] [PubMed] [Google Scholar]
- 17.Loosli F, Staub W, Finger-Baier KC, et al. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meijlink F, Beverdam A, Brouwer A, Oosterveen TC, Berge DT. Vertebrate aristaless-related genes. Int J Dev Biol. 1999;43:651–663. [PubMed] [Google Scholar]
- 19.Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- 20.Mani SS, Batni S, Whitaker L, Chen S, Engbretson G, Knox BE. Xenopus rhodopsin promoter: identification of immediate upstream sequences necessary for high level, rod-specific transcription. J Biol Chem. 2001;276:36557–36565. doi: 10.1074/jbc.M101685200. [DOI] [PubMed] [Google Scholar]
- 21.Batni S, Scalzetti L, Moody SA, Knox BE. Characterization of the Xenopus rhodopsin gene. J Biol Chem. 1996;271:3179–3186. doi: 10.1074/jbc.271.6.3179. [DOI] [PubMed] [Google Scholar]
- 22.Moritz OL, Peck A, Tam BM. Xenopus laevis red cone opsin and Prph2 promoters allow transgene expression in amphibian cones, or both rods and cones. Gene. 2002;298:173–182. doi: 10.1016/s0378-1119(02)00923-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen CM, Cepko CL. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development. 2002;129:5363–5375. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- 24.Wang QL, Chen S, Esumi N, et al. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum Mol Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- 25.Su CY, Lim J, Tsai HJ. Structural characterization and transcriptional pattern of two types of carp rhodopsin gene. Comp Biochem Physiol B Biochem Mol Biol. 2000;125:37–45. doi: 10.1016/s0305-0491(99)00149-2. [DOI] [PubMed] [Google Scholar]
- 26.Chen S, Wang QL, Nie Z, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 27.Whitaker SL, Knox BE. Conserved transcriptional activators of the Xenopus rhodopsin gene. J Biol Chem. 2004;279:49010–49018. doi: 10.1074/jbc.M406080200. [DOI] [PubMed] [Google Scholar]
- 28.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain: a possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 30.Ma GC, Wang TM, Su CY, Wang YL, Chen S, Tsai HJ. Retina-specific cis-elements and binding nuclear proteins of carp rhodopsin gene. FEBS Lett. 2001;508:265–271. doi: 10.1016/s0014-5793(01)03058-7. [DOI] [PubMed] [Google Scholar]
- 31.Ohuchi H, Tomonari S, Itoh H, Mikawa T, Noji S. Identification of chick rax/rx genes with overlapping patterns of expression during early eye and brain development. Mech Dev. 1999;85:193–195. doi: 10.1016/s0925-4773(99)00094-5. [DOI] [PubMed] [Google Scholar]
- 32.Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- 33.Krieg PA, Melton DA. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 34.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 35.Rubenstein A, Merriam J, Klymkowsky MW. Localizing the adhesive and signaling functions of plakoglobin. Dev Genet. 1997;20:91–102. doi: 10.1002/(SICI)1520-6408(1997)20:2<91::AID-DVG2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Tierney C, Wen L, Wu JY, Rao Y. A single morphogenetic field gives rise to two retina primordia under the influence of the prechordal plate. Development. 1997;124:603–615. doi: 10.1242/dev.124.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SW, Fang X, Ji H, et al. Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J Biol Chem. 2002;277:8202–8208. doi: 10.1074/jbc.M109508200. [DOI] [PubMed] [Google Scholar]
- 38.El-Hodiri HM, Shou W, Etkin LD. xnf7 functions in dorsal-ventral patterning of the Xenopus embryo. Dev Biol. 1997;190:1–17. doi: 10.1006/dbio.1997.8692. [DOI] [PubMed] [Google Scholar]
- 39.Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- 40.Chang WS, Harris WA. Sequential genesis and determination of cone and rod photoreceptors in Xenopus. J Neurobiol. 1998;35:227–244. [PubMed] [Google Scholar]
- 41.Perron M, Harris WA. Retinal stem cells in vertebrates. Bioessays. 2000;22:685–688. doi: 10.1002/1521-1878(200008)22:8<685::AID-BIES1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 42.Johnson LV, Hageman GS, Blanks JC. Interphotoreceptor matrix domains ensheath vertebrate cone photoreceptor cells. Invest Ophthalmol Vis Sci. 1986;27:129–135. [PubMed] [Google Scholar]
- 43.Uehara F, Sameshima M, Muramatsu T, Ohba N. Localization of fluorescence-labeled lectin binding sites on photoreceptor cells of the monkey retina. Exp Eye Res. 1983;36:113–123. doi: 10.1016/0014-4835(83)90094-5. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 45.Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 46.Galliot B, de Vargas C, Miller D. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- 47.Dasen JS, Barbera JP, Herman TS, et al. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev. 2001;15:3193–3207. doi: 10.1101/gad.932601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molenaar M, Brian E, Roose J, Clevers H, Destree O. Differential expression of the Groucho-related genes 4 and 5 during early development of Xenopus laevis. Mech Dev. 2000;91:311–315. doi: 10.1016/s0925-4773(99)00259-2. [DOI] [PubMed] [Google Scholar]