Abstract
A platform for in situ and real-time measurement of protein-induced conformational changes in dsDNA is presented. We combine electrical orientation of surface-bound dsDNA probes with an optical technique to measure the kinetics of DNA conformational changes. The sequence-specific Escherichia coli integration host factor is utilized to demonstrate protein-induced bending upon binding of integration host factor to dsDNA probes. The effects of probe surface density on binding/bending kinetics are investigated. The platform can accommodate individual spots of microarrayed dsDNA on individually controlled, lithographically designed electrodes, making it amenable for use as a high throughput assay.
Keywords: DNA conformation, high throughput, optical biosensor, protein–DNA
Understanding conformational dynamics and orientation of biomolecules is critical in determining their function. For dsDNA, conformational changes result because of interactions with proteins such as histones, transcription factors, and DNA-modifying enzymes. Regulation of gene expression, for example, in both prokaryotes and eukaryotes involves formation of specialized nucleoprotein structures, or snups (1). Within these complexes, distant segments of DNA are brought into close proximity to each other and, frequently, protein-induced DNA bends or kinks are formed. Understanding the DNA conformational changes and forces responsible for bending DNA to form specific complexes are thus of considerable biological significance.
Several platforms were recently developed to study protein–DNA interactions. The most popular assays, ChIP chips and DNA adenine methyltransferase identification (DamID) have proven useful for measuring protein–DNA complexes in vivo but do not have high throughput capability. For in vitro measurements, protein-binding microarrays (PBMs) provide a high throughput approach but require cloning and expression of the proteins with an epitope tag. None of the aforementioned approaches provide information indicating the conformational changes of DNA that can result upon protein binding. Methods such as FRET and atomic force microscopy are able to measure DNA conformational changes in protein–DNA complexes but do not allow for high throughput measurements. For platforms wherein probes are surface-bound, such as spectral self-interference fluorescent microscopy, the random orientation of probes limits conformational analyses (2).
Here we introduce a platform for in situ real-time measurement of protein-induced conformational changes on immobilized DNA. The platform can accommodate individual spots of microarrayed dsDNA on individually controlled, lithographically designed pixels, making it amenable for use as a high throughput assay. We utilized the sequence-specific Escherichia coli integration host factor (IHF) to measure protein-induced dsDNA bending. IHF functions in many processes that depend on the formation of snups, e.g., DNA replication, transcriptional regulation, and site-specific recombination (3, 4). The primary function of IHF appears to be architectural; i.e., IHF introduces a sharp bend in DNA that facilitates the interaction of other components in a nucleoprotein array. DNA bending is thought to be important for transcriptional regulation of approximately 120 E. coli genes (4). High-resolution structural data of these DNA–protein complexes are limited to very few specific sequences and are still not available for a range of specific and nonspecific sequences.
In this study, all probe sequences are 80 base pairs in length and modified with alkane thiols at the 5′ surface-proximal end and Cy3 fluorophores at the 3′ distal end. Our DNA probes were designed with the H′ binding sequence, derived from the bacteriophage lambda attP recombination site and positioned at least 10 base pairs from the surface-proximal (thiol-functionalized) end so that IHF binding should be minimally affected by the presence of the induced electric field, which is localized to a few nanometers from the detector surface, as described below. To investigate the effect of binding location on the change in fluorophore height, our probes were designed with the H′ binding sequence near the proximal end (H′-Low), the probe center (H′-Mid), and the distal end (H′-High) and without the H′ cognate binding sequence (control). (See Materials and Methods for the complete probe sequences.)
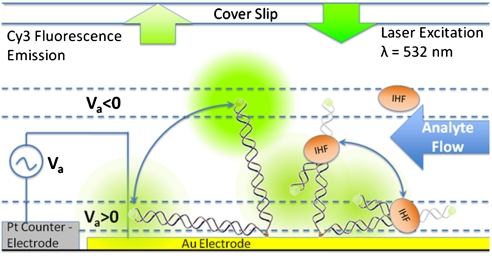
The principle for measurement of orientation and conformation of surface-bound dsDNA is depicted in Fig. 1. The efficiency of nonradiative energy transfer (ET) between the fluorophores and surface plasmons in gold follows a distance dependency ET ∝ d -3, where d is the distance between the fluorophore and the gold surface (5). This measurement allows for real-time observation of the emitter-gold distance through the fluorescence intensity. As the contour length of the tethered 80-bp dsDNA is significantly less than the persistence length (ℓ c ∼ 27 nm, ℓ p ∼ 50 nm) (6), the conformation is that of a rigid rod and the orientation of the dsDNA probes, relative to the gold surface, can be inferred (7). This orientation is controlled through a voltage applied to the gold electrode with respect to a platinum counterelectrode. The applied potential results in the formation of an ionic double layer at the electrode surface, which screens the electric field so that it is confined to a few nanometers from the electrode surface. The potential distribution as a function of distance from the surface is described by the Gouy–Chapman equation, which, for low applied potentials Ψ0, reduces to the Debye–Hückel equation Ψ(d) ∼ Ψ0 e -x/ld. The Debye length ℓ d appears as the characteristic decay length of the potential (8) and is ∼1 nm for the conditions used in our experiments (60 mM monovalent salt). Within a few Debye lengths from the surface, the double layer generates a large electric field, which exerts a force on the tethered DNA that overcomes Brownian motions and thus aligns the molecules (9). Because of the rapid decay of the electric field, the electrostatic force on the dsDNA and other charged molecules (such as charged proteins) becomes negligible at a few Debye lengths from the gold surface (9). For randomly oriented dsDNA, the determination of conformational changes through an ensemble measurement of the fluorophore-gold distance is difficult. By applying a repulsive e-field at the gold surface to induce a standing orientation in the probes, changes in their conformation due to IHF binding are more easily distinguished and can be measured in real time.
Fig. 1.
Application of a negative potential repels the DNA probe from the surface and results in high fluorescence intensity because of inefficient ET between Cy3 and the gold electrode, whereas positive potentials pull the DNA probes to the surface, resulting in reduced fluorescent intensity.
Results and Discussion
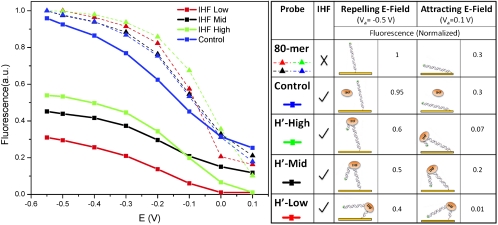
Fig. 2 shows fluorescence measurements of four dsDNA layers immobilized on separate electrodes as the applied potential is swept from -0.55 V (DNA standing) to +0.1 V (DNA lying). Red traces show measurements from electrodes having probes with the H′ binding sequence near the surface-proximal end (H′-Low), black traces are from electrodes having probes with the H′ binding sequence near the center (H′-Mid), green traces are from electrodes having probes with the H′ binding sequence near the distal end (H′-High), and blue traces are control probes without the IHF cognate binding sequence. Prior to IHF binding (dashed traces), all probes show a similar dependence of fluorophore height vs. applied field. For negative potentials (-0.55 V) we observe high fluorescence intensities, whereas for positive potentials the fluorescence decreases and saturates at low levels. We attribute this change in fluorescent intensity to the intrinsic negative charge on the DNA, which is repelled from the negatively charged and attracted to the positively charged electrode surface. A model of the dsDNA orientation for an applied potential is presented by Rant et al. (9).
Fig. 2.
Normalized fluorescence of Cy3-labeled oligonucleotide probes measured prior to and after IHF binding while the applied voltage potential is swept from negative (oligonucleotides oriented in the standing position) to positive (oligonucleotides oriented in the lying position). Probes (H′-High, H′-Mid, H′-Low, and Control) were immobilized on separate gold electrodes. IHF-induced conformation change is readily identified when oligonucleotides are oriented in the standing position with a negative applied potential. The IHF concentration is 50 nM.
Upon IHF binding to the probes, a change in the fluorophore height is observed. The application of an electric potential creates a uniform ensemble of immobilized DNA probes, which facilitates detection of conformational changes: For probes oriented in an upright position, a large drop in fluorescence is expected to occur because of IHF-induced bending upon binding. The sequence specificity of binding is then easily discernible, because bending near the proximal end of the dsDNA (H′-Low probe) pushes the fluorophore substantially closer to the gold surface than bending near the distal end (H′-High probe). In contrast, with zero or positive applied potential (dsDNA in the lying position), bending becomes less apparent and sequence-dependent bending is no longer detectable. As shown in Fig. 2, the experimentally obtained results are in excellent agreement with the expected behavior of our probes with and without bound IHF. We note an increase in the quenching of the H′-High probe in comparison to the H′-Mid probe, which is due to the variation in background levels between the electrodes on which the probe layers are immobilized. We also note that it is difficult to determine the orientation and conformation of H′-Low following IHF binding because of the fact that the bend occurs below the midpoint of the DNA sequence. Therefore, one would expect the fluorophore on the distal 3′ end to be pushed into the Au surface. The orientation in this case would also be difficult to predict, because the conformation of dsDNA within the Debye region can no longer be estimated as a stiff rod.
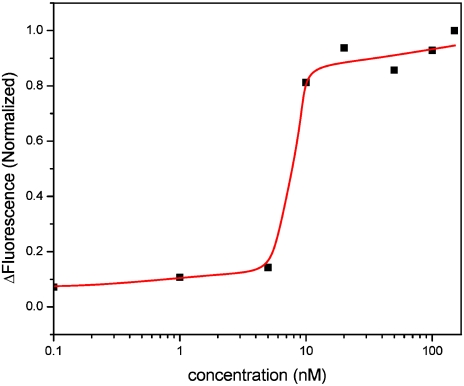
We performed titration experiments to confirm the binding affinity of IHF to the probes and found significant onset of IHF-induced bending at 5 nM IHF (Fig. 3). We assumed a direct proportionality between the fluorescence change and the extent of IHF binding. The saturation of the probe layer with IHF occurs within a surprisingly narrow concentration regime. A 1∶1 Langmuir surface adsorption model fitted to the data significantly underestimates the steepness of the transition. The abrupt onset of binding suggests that the Langmuir adsorption model is overly simplistic for this system and a more complex model should be applied to account for steric effects, coverage-dependent binding, and neighbor interactions. However, the onset of binding is in agreement with Kd values for sequence-specific IHF-oligonucleotide binding, which were found to be in the range of 0.3–2 nM (10, 11).
Fig. 3.
Plot of ΔFluorescence (fluorescence prior to IHF binding—fluorescence after IHF binding) vs. IHF concentration measured for a layer of probe 2. At least two independent experiments were conducted in the concentration range where the onset of binding is seen. The line is a guide to the eye. The onset of binding is significantly steeper than what one would expect from a Langmuir fit, which suggests that the Langmuir adsorption model is overly simplistic because it does not account for steric effects, coverage-dependent binding, and neighbor interactions.
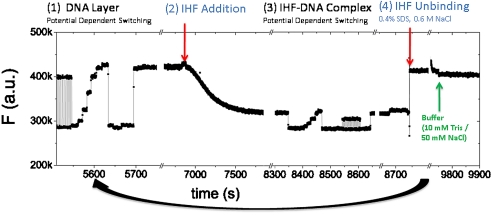
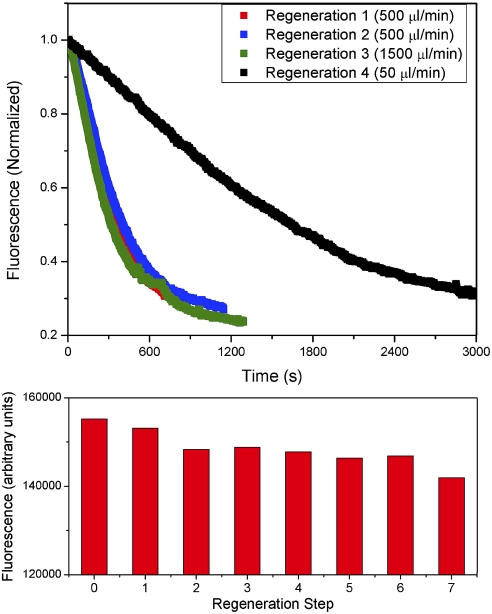
To achieve a high repeatability of the dsDNA layer surface density, we regenerate the layer to its pre-IHF-bound state by denaturing and washing away bound protein. We flow 0.4% SDS in 0.6 M NaCl to denature and dissociate IHF from our DNA probes. As a result, the fluorescence intensity recovers and a complete regeneration of the oligonucleotide layer is achieved. A sample measurement cycle is shown in Fig. 4. We measured the fluorescence intensity during successive binding and removal of IHF and found binding kinetics to be highly repeatable, as seen in Fig. 5 A. It is important to determine a suitably high flow rate so the DNA-IHF reaction rate is not limited by the diffusion of IHF to the sensor surface; therefore, measurements were made for IHF addition at flow rates of 50, 500, and 1,500 μL/ min. By assuming a parabolic flow profile in the sensor, we calculate maximum flow velocities of 0.3 × 10-3, 3 × 10-3, and 9 × 10-3 m/s, respectively. A significant change in binding kinetics is noted when the flow rate is increased from 50 to 500 μL/ min during IHF addition. For flow rates of 500 and 1500 μL/ min, the binding kinetics are nearly identical, indicating a reaction-limited regime. Consequently, all subsequent measurements were made by using a 500 μL/ min flow rate. The cycle of IHF-dsDNA binding followed by protein removal and dsDNA layer regeneration was repeated multiple times, with a small signal loss following each regeneration, as shown in Fig. 5 B.
Fig. 4.
A measurement cycle to determine the magnitude and kinetics of IHF-induced DNA binding/bending: (1) Characterization of DNA layer by measurement of fluorescence signal over a range of voltage potentials; (2) IHF is added and binding kinetics are observed while a negative potential is applied to orient DNA in the standing position; (3) characterization of IHF-bound DNA layer; (4) high salt solution with SDS denatures IHF, resulting in dissociation from and regeneration of the DNA layer.
Fig. 5.
The regeneration of the dsDNA layer through dissociation of IHF allows multiple binding experiments with high repeatability. (A) Overlaid real-time measurements of fluorescence during addition of 50 nM wild-type IHF. Oligonucleotide layers of H′-Low probes are regenerated by removal of IHF protein with 0.4% SDS in 0.6 M NaCl. Addition of protein at a flow rate of 50 μL/ min results in a decreased binding rate; however, addition of protein at 500 and 1,500 μL/ min results in nearly identical binding kinetics, indicating a reaction-limited regime. The flow rates correspond to average flow velocities of 1 × 10-3, 1 × 10-2, and 3 × 10-2 m/s, respectively. (B) The fluorescence prior to IHF binding for subsequent regeneration steps shows a 9% drop in signal after 7 layer regenerations. Flow rates are 500 μL/ min.
The observed binding time constants of a few minutes are several orders of magnitude higher than those reported for specific IHF-dsDNA binding in free solution, which are on the order of hundreds of milliseconds (12). One possible explanation is that steric hindrance of densely immobilized dsDNA alters the kinetics of IHF-DNA binding/bending. To test the effect of dsDNA probe surface density on protein binding/bending, we prepared dsDNA layers (H′-Mid probes) of varying surface densities by immobilizing a densely packed layer. Peterson et al. report a maximum monolayer coverage for thiol-modified dsDNA on Au to be 3 × 1012 cm-2 for 25-mer dsDNA (13). Steel et al. report that longer dsDNA length above 24-mers decreases surface coverage; however, they do not report the surface coverage for oligonucleotides longer than 48-mers. For an increase in the dsDNA length from 8-mers to 48-mers dsDNA, they measure a reduction of the dsDNA monolayer surface density from 2 × 1013 to 4 × 1011 cm-2, respectively (14). On the basis of this, we estimate a maximum surface coverage of roughly 1011 cm-2 for 80-mer dsDNA.
After IHF binding to a densely packed layer, we reduce the surface coverage through controlled electrical desorption, as described earlier (15). We reuse the same electrode to measure IHF-induced bending by removing the bound IHF to regenerate the dsDNA layer, as described above. The ability to regenerate the DNA layer provides a highly controlled probe layer to test the effect of a single parameter, in this case the probe surface density. The surface density is determined by monitoring the fluorescence signal and the switching efficiency. The switching efficiency corresponds to the difference in the fluorescence intensity between DNA in lying and standing orientations and is highly correlated to surface layer density because steric hindrance between adjacent probes reduces the level to which probes can be oriented in the lying position (7, 16). Therefore, as the DNA layer is desorbed gradually, the switching efficiency initially increases because of reduced steric hindrance between adjacent DNA probes, after which it plateaus (see ref. 8 for a detailed description). The point at which the fluorescence plateaus corresponds to the “free switching” regime, where the dsDNA is no longer hindered by adjacent probes. Thus, fluorescence intensity prior to IHF binding gives an indication of the probe surface density: A surface density of 5 × 1010 cm-2 can be estimated from the free switching geometry; i.e., the distance between two probes on the surface is approximately 2 × ℓ c (15). For low densities the fluorescence follows a linear relationship to surface density (17); therefore, it is possible to estimate surface densities by using fluorescent intensity values relative to the fluorescent intensity in the free switching regime.
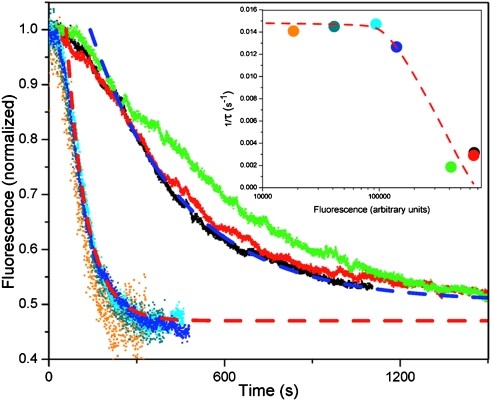
Fitting the data to single exponential decays yields time constants, τ = 320 s for high probe surface densities and τ = 70 s for low densities, a nearly fivefold increase in binding/bending rate. However, the steady state level of bending is largely unaffected by the probe density, as seen in Fig. 6. The results suggest that the dense packing of DNA sterically hinders IHF access to the DNA binding, therefore reducing the rate of protein binding. However, it appears that the conformational changes that result upon binding remain the same for the surface coverage investigated.
Fig. 6.
A plot of fluorescence vs. time for various probe surface densities shows that probe surface density significantly affects the binding/bending kinetics but does not affect the steady state level of IHF-induced bending. Fitting the data from high surface density (Blue Dashed Line) and low surface density (Red Dashed Line) to a single exponential decay yields time constants of 320 and 70 s. The Inset shows probe surface density vs. the binding rate, where τ is the time constant extracted from the exponential decay fits due to IHF-induced bending at fluorescence values prior to IHF binding. The line is a guide to the eye.
We estimate the mean separation between adjacent DNA strands in the free switching regime to be about 2xℓ c ∼ 50 nm (15). For low densities the fluorescence follows a linear relationship to surface density, and we can roughly estimate surface densities on the basis of the fluorescence values (17). The size of IHF (molecular weight 22 kDa) can be estimated to be about 3–4 nm diameter. For the regime in which slower reaction kinetics are observed, the surface density is roughly fivefold larger than the free switching regime, resulting in greater than twofold reduction in the mean separation between adjacent DNA strands, or about 20–30 nm. At these surface densities, the mean separation remains significantly larger than the protein size. It appears that the rate of penetration increases as the spacing between adjacent DNA strands increases until the spacing exceeds the protein size by about one order of magnitude. As the surface density increases and the mean separation between adjacent DNA approaches the protein size, it is likely that the layer would become impenetrable to IHF.
Whereas it is difficult to obtain exact values for the dsDNA surface density, it is clear that this variable significantly impacts the rate of protein–DNA binding. Therefore, it is important to control the surface coverage in experiments that evaluate the protein-binding rate to surface-bound probes.
Control over surface density also has physiological relevance, for example, in chromatin-bound DNA, where certain sequences are inaccessible to proteins because of steric exclusion caused by the proximity of nucleosomes and adjacent gyres of DNA (18). The ability to control the probe surface density makes this platform attractive for in vitro studies on the effects of steric access in DNA–protein binding. Such steric effects were also shown to significantly affect transcription, for example, in interactions with corepressors (19). The ability to easily control the surface probe density through electrical desorption is an added advantage of this platform.
Conclusions
We introduced a platform to measure real-time in situ protein-induced conformational changes in immobilized dsDNA probes. We demonstrated the ability to discern differences in conformational changes resulting from sequence-specific protein binding, and the measured concentration dependence of protein binding agrees with previously published values. Additionally, the binding kinetics elucidate a high dependence of the binding rate on the probe surface density. The platform allows electrical control of the probe surface density, and we show a high dependence of the binding kinetics on the probe surface density. To address this surface density dependence, we describe a method to conduct multiple binding experiments to a surface with high repeatability in the binding kinetics. The platform therefore offers a comprehensive approach to investigate protein-induced conformational changes in immobilized dsDNA. The ability to modify the technology for microarrayed measurements will allow high throughput measurements that are required to investigate the sequence dependence of DNA–protein interactions. In addition to analyzing protein–DNA interactions, we envisage that our platform can be adapted to identify and characterize compounds, which, upon DNA binding/cross-linking, induce significant DNA conformational changes, such as the chemotherapeutic agent cisplatin (20).
Materials and Methods
Au electrodes, 120 μm in diameter, were prepared by depositing Au (300 nm) on Ti (10 nm) on a single crystalline sapphire wafer. Annular Pt counterelectrodes, > 1 mm in diameter, surround the Au electrodes and are prepared by depositing Pt (300 nm) on Ti (10 nm). The Au-Pt separation is 100 μm. A silicon O-ring surrounds the Au-Pt electrodes, and the flow chamber is formed by clamping the silicon O-ring between a coverslip and the wafer. Au metal traces connect the electrodes to electrical contacts on the perimeter of the wafer. A potentiostat (Autolab PGSTAT30; Eco Chemie) was utilized to monitor and control the voltage between the Au and Pt electrodes. The substrates were cleaned with H2SO4∶H2O2 = 7∶3 piranha solution for 5 min followed by HNO3 (60%) for 15 min and rinsed with deionized water and N2 drying. DNA was obtained from IBA GmbH. The 80-base-pair-long oligonucleotides were functionalized with a thiol on the 5′ proximal end and with a Cy3 fluorophore on the 3′ distal end. Hybridization was done for 1 hr at 70 °C in buffer (20 μM DNA, 200 mM NaCl, 10 mM Tris, pH 7.3). Following hybridization, the dsDNA probes were end-grafted on the gold electrode by hand-spotting the immobilization buffer (10 mM Tris, pH 7.3, 50 mM NaCl, 10 μM dsDNA) followed by passivation of the Au electrode through coadsorption of 1 mM mercaptohexanol for 1 hr. The flow chamber is 21 mm in length, 4.2 mm in width, and 0.9 mm in height (volume ∼80 μL). All protein binding was done in 50 mM NaCl, 10 mM Tris buffer, pH 7.3. Four different probe sequences were designed and immobilized on separate electrodes. The IHF H′ binding site, derived from the phage lambda attP sequence, is highlighted in bold:
H′-Low: Proximal IHF binding site
5′ SH C6 GAT GAT AGA GAA AAA AGC ATT GCT TAT CAA TTT GTT GGC GCG GAT GTC AGT AGT AGA TAG AGT GTG TAG TGA CCT GGT GT CY3 3′
H′-Mid: IHF binding site near probe center
5′ SH C6 GAT GAT AGA AGA TAG AGT GTG TAG TGA CCT GGT GAA AAA AGC ATT GCT TAT CAA TTT GTT GGC GCG GAT GTC AGT AGT GT CY3 3′
H′-High: Distal IHF binding site
5′ SH C6 GAT GAT AGA AGA TAG AGT GTG TAG TGA CCT GGT GGC GCG GTC TGA GTT AGT AAA AAA GCA TTG CTT ATC AAT TTG TTG AG CY3 3′
Control: No H′ binding site
5′ SH C6 GAT GAT AGA AGA TAG AGT GTG TAG TGA GGT GGT GGA TGA TAG AAG ATA GAG TGT GTA GAG TGC GCG GAT GTC AGT AGT GT CY3 3′
A microscope in reflected dark field mode (Olympus BX-RLA2) was used for probe illumination (CW DPSS laser, 532 nm, Newport Spectra Physics, Pro Millennia) and fluorescence light detection. Reflected light from the laser illumination is filtered through a long wave pass filter (550 nm) and a band pass filter (Semrock, FF01-582/75-25). Probe fluorescent emission intensity (Cy3, peak emission ∼570 nm) is detected by a cooled photomultiplier (Hamamatsu, Photosensor Modules H7422) operating in the single-photon-counting mode.
Acknowledgments.
This work was supported by National Science Foundation Grants CBET-0933670 and OISE-0601631
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Echols H. Multiple DNA-protein interactions governing high-precision DNA transactions. Science. 1986;233:1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- 2.Moiseev L, Ünlü MS, Swan A, Goldberg B, Cantor C. DNA conformation on surfaces measured by fluorescence self-interference. Proc Natl Acad Sci USA. 2006;103:2623–2628. doi: 10.1073/pnas.0511214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 4.Swinger KK, Rice PA. IHF and HU: Flexible architects of bent DNA. Curr Opin Struc Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Chance RR, Prock A, Silbey R. Molecular fluorescence and energy transfer near interfaces. Adv Chem Phys. 1978;37:1–65. [Google Scholar]
- 6.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 7.Rant U, et al. Switchable DNA interfaces for the highly sensitive detection of label-free DNA targets. Proc Natl Acad Sci USA. 2007;104:17364–17369. doi: 10.1073/pnas.0703974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Israelachvili J. Intermolecular & Surface Forces. Amsterdam: Elsevier; 1991. pp. 213–254. [Google Scholar]
- 9.Rant U, et al. Electrical manipulation of oligonucleotides grafted to charged surfaces. Org Biomol Chem. 2006;4:3448–3455. doi: 10.1039/b605712h. [DOI] [PubMed] [Google Scholar]
- 10.Bao Q, Christ N, Dröge P. Single-chain integration host factors as probes for high-precision nucleoprotein complex formation. Gene. 2004;343:99–106. doi: 10.1016/j.gene.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Murtin C, Engelhorn M, Geiselmann J, Boccard F. A quantitative UV laser footprinting analysis of the interaction of IHF with specific binding sites: Re-evaluation of the effective concentration of IHF in the cell. J Mol Biol. 1998;284:949–961. doi: 10.1006/jmbi.1998.2256. [DOI] [PubMed] [Google Scholar]
- 12.Sugimura S, Crothers D. Stepwise binding and bending of DNA by Escherichia coli integration host factor. Proc Natl Acad Sci USA. 2006;103:18510–18514. doi: 10.1073/pnas.0608337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe desnsity on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steel AB, Levicky RL, Herne TM, Tarlov MJ. Immobilization of nucleic acids at solid surfaces: Effect of oligonucleotide length on layer assembly. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rant U, et al. Structural properties of oligonucleotide monolayers on gold surfaces probed by fluorescence investigations. Langmuir. 2004;20:10086–10092. doi: 10.1021/la0492963. [DOI] [PubMed] [Google Scholar]
- 16.Rant U, et al. Dynamic electrical switching of DNA layers on a metal surface. Nano Lett. 2004;4:2441–2445. [Google Scholar]
- 17.Arinaga K, et al. Controlling the surface density of DNA on gold by electrically induced desorption. Biosens Bioelectron. 2007;23:326–331. doi: 10.1016/j.bios.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 19.Zamir I, Zhang J, Lazar MA. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Gene Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]