Abstract
The Lost City Hydrothermal Field, an ultramafic-hosted system located 15 km west of the Mid-Atlantic Ridge, has experienced at least 30,000 years of hydrothermal activity. Previous studies have shown that its carbonate chimneys form by mixing of ∼90 °C, pH 9–11 hydrothermal fluids and cold seawater. Flow of methane and hydrogen-rich hydrothermal fluids in the porous interior chimney walls supports archaeal biofilm communities dominated by a single phylotype of Methanosarcinales. In this study, we have extensively sampled the carbonate-hosted archaeal and bacterial communities by obtaining sequences of >200,000 amplicons of the 16S rRNA V6 region and correlated the results with isotopic (230Th) ages of the chimneys over a 1,200-year period. Rare sequences in young chimneys were commonly more abundant in older chimneys, indicating that members of the rare biosphere can become dominant members of the ecosystem when environmental conditions change. These results suggest that a long history of selection over many cycles of chimney growth has resulted in numerous closely related species at Lost City, each of which is preadapted to a particular set of reoccurring environmental conditions. Because of the unique characteristics of the Lost City Hydrothermal Field, these data offer an unprecedented opportunity to study the dynamics of a microbial ecosystem’s rare biosphere over a thousand-year time scale.
Keywords: biofilm, preadaptation, rare biosphere, geochronology
Mucilaginous biofilms coat the porous carbonate mineral matrix of actively venting chimneys of the Lost City Hydrothermal Field (LCHF), an ultramafic-hosted system located 15 km west of the spreading axis of the Mid-Atlantic Ridge at a depth of ∼750 m (1). Previous studies have shown that the biofilms contain 106–109 cells per gram of carbonate mineral and that >80% of the cells belong to a single phylotype of archaea known as Lost City Methanosarcinales (LCMS) (2). It is presumed that the abundant LCMS cells have adapted to the ∼90 °C, pH 9–11 fluids and that they use high concentrations of dissolved hydrogen (H2) and methane (CH4) in the vent fluids (3, 4), but their physiology is unknown. As chimneys become less active, changes in mineralogy and fluid chemistry cause other organisms to become abundant (5). For example, chimneys with very little visible hydrothermal venting do not contain the LCMS biofilm. Instead, a single phylotype belonging to the ANME-1 group of anaerobic methanotrophic archaea is the dominant archaeon (5). The outer walls of carbonate chimneys, where mineralogy and fluid chemistry can be substantially different compared with the chimney interiors (2, 6), harbor bacteria with high similarity to sulfur- and methane-oxidizers, but archaea are much more abundant in chimneys venting up to 90 °C, high pH fluid (2).
Unlike typical black smoker systems that are commonly fueled by heat from underlying magmatic activity, hydrothermal flow at the LCHF is driven by cooling of ultramafic rocks and lesser gabbroic material that underlie the field (1). Fluid chemistry is governed by exothermic geochemical reactions in the subsurface known as serpentinization (1). These reactions commonly occur in ultramafic environments on Earth where water reacts with the mineral olivine, and they are expected to occur on other planetary bodies hosting aqueous fluids (7). Serpentinization of subsurface ultramafic minerals is one of the most likely sources of CH4 on Mars (8). At the LCHF, serpentinization reactions produce alkaline (pH 9–11) fluids that are rich in calcium (up to 30 mmol/kg), low in metals, and have near-zero concentrations of CO2 (1, 4, 6). Serpentinization also provides an abiogenic source of H2 (14 mmol/kg), CH4 (2 mmol/kg), and low-molecular-weight hydrocarbons (3, 4). The ability of Lost City–like environments to generate reduced organic compounds exothermically and abiotically makes them plausible settings for the origin and early evolution of life (9).
Lost City also differs from most magma-driven systems in that it is very long-lived (10). Carbon isotopic measurements indicate that venting has been ongoing for at least 30,000 years, with individual chimneys active for at least 300 years, and modeling results indicate the system could remain active for up to 1 million years (10). Therefore, conditions within Lost City chimneys could have been conducive to the growth of LCMS for tens or hundreds of thousands of years, and it is possible that it has remained the dominant member of the ecosystem throughout that time. To our knowledge, biology has no precedent for such a low diversity, long-lived ecosystem. Characterizations of microbial community dynamics with respect to time are typically performed within the context of habitat monitoring. Numerous studies have described seasonal changes of particular communities (11, 12), documented initial colonizations of substrates (13), and observed short-term responses to environmental changes (14, 15). The time scales of these studies are on the order of months or at most years, practical time periods for observational studies. No direct evidence exists for changes in microbial communities occurring over longer time scales.
Technological advances in nucleic acid sequencing (16) are increasing the sensitivity at which changes in microbial communities can be detected. Recent high-sensitivity surveys of diverse environmental samples including those from shallow and deep ocean waters (17), hydrothermal fluids (17, 18), arctic tundra (19), and various soils (19, 20) have revealed that the vast majority of a microbial habitat’s diversity is comprised by taxa present at very low abundances—so low that they were previously undetectable. The most abundant organisms represent only a fraction of the total diversity. The high diversity of rare sequences indicates that they have been evolving over long periods of time and that at least some of them belong to organisms that are preadapted to environmental conditions that have occurred in the past and may reoccur in the future. A fundamental prediction of the “Rare Biosphere” model (17) is that when environmental conditions change, some of these rare, preadapted taxa can rapidly exploit the new conditions, increase in abundance, and out-compete the once abundant organisms that were adapted to the past conditions. No studies have tested this prediction by examining a shift in species composition involving extremely rare taxa occurring during a known time interval.
The longevity and extremely low diversity of the LCHF carbonate chimneys provide a unique test of the Rare Biosphere model. In this study, we use pyrosequencing technology to generate an extensive sequence dataset of the 16S rRNA V6 region from four carbonate chimney samples collected from structures varying from a massive edifice venting 88 °C, high pH fluid to a small, inactive chimney bathed in seawater. We investigate whether these low diversity communities harbor highly diverse but rare species and attempt to correlate our biological data with chimney sample ages derived from coregistered 230Th isotopic measurements. We show that rare sequences in young chimneys samples are commonly much more abundant in older samples, suggesting that organisms can remain rare for long time periods until environmental changes allow them to proliferate.
Results
230Th Dating of Carbonate Chimney Samples.
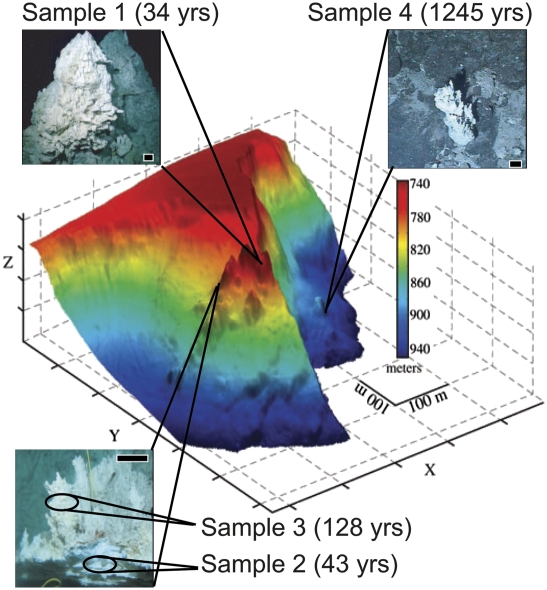
Uranium–thorium (238U-234U-230Th-232Th or U-Th or 230Th) disequilibrium dating is a powerful geochronological tool used to date inorganic and biogenic materials ranging in age from modern to 600 kyr (21). In this study, four discrete chimney samples (Fig. 1) were dated using 230Th age-dating techniques. Sample 1 (230Th age = 34.3 ± 8.1 yr) was collected from a site known as marker 3 or Poseidon where emitting fluids reach 88 °C (4). Samples 2 and 3 are from a flange named marker C that is bathed by fluids exhibiting a temperature gradient of 9–70 °C because of mixing of hydrothermal fluids with ambient seawater. Samples 2 and 3 were located 20 cm apart (Fig. 1) but yielded very different ages (230Th ages = 43 ± 23 yr and 128 ± 57 yr, respectively). Sample 4 (230Th age = 1245 ± 257 yr) was collected from a chimney with no visible venting and contained coral polyps on its exterior, typical of inactive chimneys at the LCHF (6). Complete descriptions of the samples and the 230Th age-dating technique are available in SI Methods.
Fig. 1.
Location within the Lost City Hydrothermal Field of carbonate chimney samples from which sequences were collected. Sample 1 was obtained from the pinnacle of the main edifice known as Poseidon, 50 m above the flange from which samples 2 and 3 were collected. Sample 4 is from a small, isolated chimney with no apparent venting activity found near the bottom of a cliff face. Scale bars in all photographs are 10 cm.
Archaeal Community Structure.
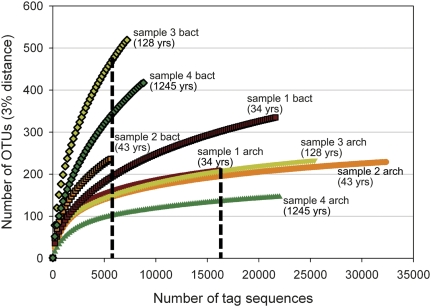
Of 167,031 total archaeal V6 amplicons (“tags”) sequenced from the four carbonate chimney samples (Fig. 1 and Table S1), we identified 2,635 unique sequence types that clustered into 817 operational taxonomic units (OTUs) at the 3% distance threshold. Sample 4 (230Th age = 1245 ± 257 yr) contained significantly fewer OTUs than samples 1–3 (230Th ages = 34–128 yr), as shown by the rarefaction curves of observed OTUs vs. the total number of tags sequenced (Fig. 2). The near-asymptotic shape of the rarefaction curves as well as the small difference between the observed numbers of OTUs and the values from two richness estimators (Table 1 and Table S1) indicate that our study has nearly completely sampled the archaeal V6 amplicon libraries from these samples.
Fig. 2.
Rarefaction analysis of archaeal and bacterial V6 tag sequences for each of the four chimney samples. OTUs, operational taxonomic units defined by clustering sequences with a 3% pairwise distance threshold. Bacterial diversity (curves outlined in black) is clearly greater than archaeal diversity in all samples. Rarefaction curves for archaeal sequences are near-asymptotic, indicating nearly complete sampling of archaeal V6 amplicon libraries. Dashed line indicates the point to which data were subsampled to compare diversity values among samples in Table 1.
Table 1.
Comparison of 230Th ages and V6 tag sequence diversity measurements for carbonate chimney samples from the Lost City Hydrothermal Field
| Archaea* | Bacteria* | ||||||||||
| Chimney sample† | Max fluid temperature, °C | Age, yr | OTUs‡ | ACE richness estimator4 | Chao1 richness estimator§ | Simpson’s Reciprocal Index§ | OTUs‡ | ACE richness estimator§ | Chao1 richness estimator§ | Simpson’s reciprocal index§ | |
| 1 | 88 | 34.3 ± 8.1 | 342 | 448–568 | 441–599 | 2.04–2.13 | 177 | 283–388 | 217–319 | 5.6–5.9 | |
| 2 | 70 | 43 ± 23 | 265 | 336–438 | 310–405 | 1.56–1.61 | 270 | 458–587 | 360–522 | 8.3–9.1 | |
| 3 | 70 | 128 ± 57 | 265 | 322–410 | 301–386 | 1.64–1.69 | 428 | 859–1,061 | 648–925 | 20.0 | |
| 4 | Nd | 1,245 ± 257 | 135 | 131–170 | 129–191 | 1.22–1.25 | 307 | 582–744 | 434–630 | 16.7 | |
*Archaeal diversity is reported for OTU relative abundances normalized down to 16,260 tags; bacterial diversity is reported for OTU relative abundances normalized down to 5,567 tags. Table S1 compares both archaeal and bacteria diversity at 5,567 tags.
† DSV Alvin numbers for samples 1–4 are 3881–1408, 3869–1404, 3869–1443, and 3876–1133, respectively.
‡OTUs (operational taxonomic units) defined at 3% sequence difference.
§ Ranges of diversity indices include 95% confidence intervals as calculated by DOTUR (39).
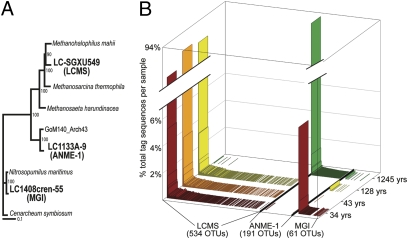
Figure 3 presents normalized profiles of archaeal OTUs for each sample. A single OTU corresponding to the previously published 16S rRNA sequence of the putative methanogen Lost City Methanosarcinales (LCMS; Fig. 3A) (2, 5) comprised >90% of all tag sequences from samples 1–3 (Table S2). An additional 533 OTUs, none of which contained >5% of any sample’s total tags, were also assigned to the order Methanosarcinales (Fig. 3B) by the GAST automated taxonomic assignment process (22). The relative abundance of variant sequences generally decreased with increasing distance from the dominant sequence, a trend consistent with that expected from artificial diversity generated by pyrosequencing error. Several sequences occur much more frequently than predicted by random error, however, and probably represent genuine diversity. Furthermore, we have recently reported that variation in the V6 region of these Methanosarcinales sequences is correlated with greater variation in the intergenic transcribed spacer (ITS) region, a marker that often reflects physiological variation (23). Therefore, the number of OTUs measured in the V6 region by this study may underestimate the total genetic diversity.
Fig. 3.
Relative abundance distribution of archaeal OTUs among samples. (A) Bootstrapped phylogenetic tree of full-length archaeal 16S rRNA clones showing the relationship of three Lost City clones (in bold) to previously published close relatives. (B) Relative normalized abundance (as percentage of total tag sequences per sample) of each archaeal OTU (clustered at 3% distance threshold) in each of the four chimney samples. The OTUs are labeled according to which of the three groups they belong according to pairwise similarity with the V6 region of the full-length 16S rRNA clones. ANME-1, anaerobic methanotrophic archaea; LCMS, Lost City Methanosarcinales; MGI, Marine Group I Crenarchaeota.
The oldest sample (1245 ± 257 yr) was also dominated by a single OTU, but it corresponds to the previously published (5) 16S rRNA clone LC1133A-9, which is phylogenetically similar to the ANME-1 group of anaerobic methanotrophic archaea (Fig. 3A). Interestingly, the same OTU that dominates sample 4 is also present at low abundance in sample 1 (Fig. 3B), indicating that ANME-1 organisms are present in chimneys as young as 34 yr but are only dominant in much older chimneys. A third major archaeal group was represented by a set of sequences primarily present only in sample 1 and matching 16S rRNA clone LC1408cren-55, which is 97% similar over its full length to the ammonia-oxidizing crenarchaeon Nitrosopumilus maritimus (24). The presence of these sequences in the youngest, hottest chimney sample and, in hot chimney fluids (5), indicates that they probably represent thermophilic organisms, although their physiology is unclear because ammonia oxidation is not expected to be thermodynamically favorable in the anoxic fluids of LCHF chimneys.
The patterns visualized in Fig. 3 are quantified in two Venn diagrams illustrating the percentage of tags and numbers of OTUs shared among samples (Fig. S1). We also calculated overall community similarities (Fig. S2) based on the relative abundance of each OTU (Bray-Curtis similarity coefficient) as well as the presence/absence of each OTU (Jaccard similarity coefficient). As expected from Fig. 3, the three samples aged 34–128 yr were all very similar to each other (>85% Bray-Curtis, >48% Jaccard) and very different from sample 4 (<0.3% Bray-Curtis, <2% Jaccard). Remarkably, the archaeal community similarity between samples 2 and 3 (95% Bray-Curtis, 56% Jaccard) was much higher than the similarity between sample 1 replicates, so the small differences between samples 2 and 3 may not reflect any natural variation (additional details on estimation of variation between replicates in SI Methods).
Bacterial Community Structure.
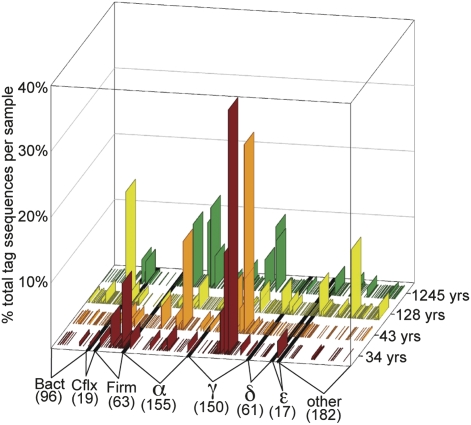
Our study identified most of the diversity present in the bacterial V6 amplicon library from the youngest chimney sample (34.3 ± 8.1 yr), as shown by the nearly asymptotic rarefaction curve of OTUs discovered in sample 1 (Fig. 2). The rarefaction curves of samples 3 and 4 (128 ± 57 and 1245 ± 257 yr), in contrast, have very steep slopes and are, therefore, likely to contain much more diversity than we were able to discover with >7000 tag sequences from each sample. A few, highly abundant sequence types dominated the bacterial communities in each sample with rare sequences accounting for most of the observed diversity (Fig. 4). This pattern is very similar to that seen in the archaeal communities (Fig. 3B), although the bacterial communities have a more even distribution (reflected in the much higher Simpson’s reciprocal index of diversity, Table 1 and Table S1) and have greater diversity, especially at large taxonomic distances (Fig. S3).
Fig. 4.
Relative abundance distribution of bacterial OTUs among samples. OTUs are labeled according to their taxonomic assignment by the GAST process (22). α, Alphaproteobacteria; Bact, Bacteroides; Cflx, Chloroflexus; δ, Deltaproteobacteria; ε, Epsilonproteobacteria; Firm, Firmicutes; γ, Gammaproteobacteria.
The taxonomic groups to which V6 tag sequences were assigned (Fig. 4) were consistent with previous work using different techniques (5). Notably, sequences assigned to Gammaproteobacteria, Alphaproteobacteria, Firmicutes, and Chloroflexi were the most abundant and diverse. The most abundant phylotypes in each group (Table S3) probably represent nonphotosynthetic organisms oxidizing reduced sulfur species (e.g., Thiotrichales, Rhodobacterales) or H2 (e.g., Clostridia, Dehalococcoidetes) from the vent fluids. The low but significant concentrations of abiotically-derived organic carbon in Lost City hydrothermal fluids (4) may be an important carbon source for some of the bacteria, particularly Clostridia with high sequence similarity to Desulfotomaculum species, which occur in the youngest, most hydrothermally influenced sample (Table S3). Many Clostridia use low-molecular-weight organic acids, often in syntrophic association with methanogens (25 –27). The possible ecological roles of these organisms in LCHF chimneys have been discussed previously (5).
Although the shifts in bacterial OTU composition are not as obvious as those in the archaeal communities, there are clear differences among samples with different ages (Fig. 4). Samples 1 and 2 (34.3 ± 8.1 and 43 ± 23 yr) have high overall community similarity (45% Bray-Curtis, 23% Jaccard), and samples 3 and 4 (128 ± 57 yr and 1245 ± 257 yr) are very similar to each other (34% Bray-Curtis, 23% Jaccard; Fig. S2). Note that samples 2 and 3, which were only ∼20 cm apart in situ (Fig. 1) and had nearly identical archaeal communities, have only moderate bacterial community similarity (18% Bray-Curtis, 17% Jaccard). Interestingly, the distribution of OTUs among samples was roughly constant regardless of whether all OTUs are included in the analysis, or if only abundant, or if only rare OTUs are considered (Fig. S4 and S5). In short, samples of similar ages contain similar bacterial communities; with few exceptions, this trend is true for dominant as well as rare organisms.
Diversity Within Thiomicrospira.
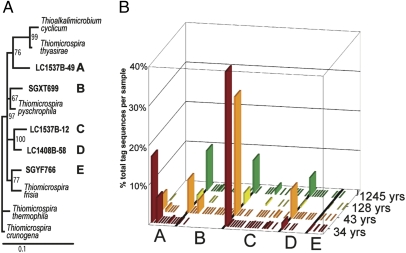
Previous work has shown that sequences most closely related to genus Thiomicrospira (order Thiotrichales; Table S3) are widely distributed throughout the LCHF (5). A single OTU assigned to genus Thiomicrospira corresponding to previously published full-length 16S rRNA clone LC1537B-12 comprised 37% of all bacterial V6 tags from the youngest sample (34.3 ± 8.1 yr). The same OTU also occurred in the other three samples but at lower relative abundance (28% at 43 yr, 3% at 128 yr, and 8% at 1,245 yr). An additional 23 OTUs (group C in Fig. 5), none of which comprised more than 2% of any sample’s total tags, were at least 93% similar to clone LC1537B-12. Similarly, group A contained two dominant and several rare OTUs, all of which were at least 96.7% similar to the V6 region of clone LC1537B-49. The high occurrence of these OTUs in the younger, higher pH samples and absence in the two older, more neutral pH samples is consistent with the physiology of their closest relative (Fig. 5A), the alkaliphile Thioalkalimicrobium cyclicum (28). Group B OTUs, in contrast, were the most dominant Thiomicrospira-like sequences in sample 4, which showed no signs of venting and was likely the same temperature as the background seawater. All of these OTUs were at least 95% similar to a clone with 95.3% similarity over the full gene length to the pyschrophile Thiomicrospira psychrophila (29).
Fig. 5.
Relative abundance distribution of Thiomicrospira-related OTUs among samples. (A) Bootstrapped phylogenetic tree of full-length 16S rRNA clones showing the relationship of Lost City clones (in bold) to previously published Thiomicrospira clones. (B) Relative normalized abundance of each OTU assigned to genus Thiomicrospira by GAST (22) in each of the four chimney samples. Pairwise similarities between sequences representative of each OTU and the V6 region of each clone were calculated, and OTUs were grouped according to which clone they were most closely related, as indicated by A–E labels. In groups A–D, similarities between sequences and clones ranged from 93 to 100%; in group E, 86–100%. OTUs within each group that have the highest similarity to the clone are sorted to the left.
Although all samples have several Thiomicrospira OTUs, samples of different ages are dominated by different OTUs (Fig. 5). The most abundant Thiomicrospira OTU in the oldest sample, for example, is extremely rare in the youngest sample, and the overall similarity of the Thiomicrospira community between these two samples is low (24% Bray-Curtis, 22% Jaccard). As seen with the whole bacterial community, the two youngest samples have the most similar Thiomicrospira OTUs (61% Bray-Curtis, 32% Jaccard), and the two oldest samples are also very similar (39% Bray-Curtis, 31% Jaccard).
Discussion
Dominance by a Few Species.
Over the past >30,000 years of venting at the LCHF (10), extreme environmental conditions created by mixing of <150 °C, pH 11 fluids enriched in dissolved H2 and CH4 with oxygen-rich seawater have selected for an unusual assemblage of microorganisms. Previous studies have shown that >80% of detectable cells in the warm, anoxic interior zones of carbonate chimneys are dominated by biofilms formed by Lost City Methanosarcinales (LCMS, 2). This study highlights the extremely low archaeal diversity of Lost City chimneys: >90% of all V6 tags in each sample clustered into a single OTU. Such extreme dominance of single phylotypes within hydrothermal chimneys is unprecedented; in other hydrothermal environments where methanogens dominate, diverse species adapted to various metabolic substrates are typically present (30, 31).
A small number of species also dominate the bacterial communities, and the identity of the dominant species often differs among chimney samples (Fig. 4). This trend is true even within narrow taxonomic groups. For example, different OTUs representing genus Thiomicrospira dominated different chimney samples (Fig. 5). These organisms probably require reduced sulfur species from hydrothermal fluid, but they also need access to oxygen (the electron acceptor for all known Thiomicrospira) from seawater. Inorganic carbon is scarce in high pH hydrothermal fluid, so organisms in chimneys with little exposure to seawater may be carbon-limited, as evidenced by the unprecedented enrichment of 13C in lipids extracted from Lost City chimneys (32). Thus, the diversity of Thiomicrospira sequences detected in this study (Fig. 5) probably resulted from many similar species adapting to slightly different niches within concentration gradients of reduced sulfur species, oxygen, and inorganic carbon, as well as the associated temperature and pH gradients.
Species Shift Between Rare and Dominant.
The most notable differences in the archaeal community involve the shift to a community completely dominated by ANME-1 sequences in sample 4 (1245 ± 257 yr) and the relative lack of the Marine Group I Crenarchaeota in samples 2–4. Although ANME-1 sequences are nearly absent from samples 1–3, the identical sequence that dominates sample 4 is present, but rare, in sample 1 (34.3 ± 8.1 yr), which was collected from a chimney 100 m distant and shallower in depth. Deeper sequencing of samples 2 and 3 would most likely also recover this sequence at low abundance. Previous work has shown that ANME-1 sequences are present in multiple chimneys throughout the LCHF (5) including one chimney that has been dated to 10,500 years (33). Therefore, ANME-1 organisms are well dispersed throughout the vent field but are able to thrive only in chimneys that are not venting high-temperature, high-pH fluids, conditions typical of old chimneys (1, 6). It is likely that these chimneys previously vented high-temperature, high-pH fluids and were therefore inhabited by microbial communities more similar to that of the younger chimneys reported here.
Many more examples of sequences that are rare in young samples but abundant in older samples are evident in the bacterial V6 tag pyrosequences (Fig. S6). Taxonomic differences among chimney samples of varying ages are most likely caused by differences in mineralogy and fluid chemistry, and both of these factors are expected to change with somewhat predictable trends during the cycle of chimney formation, growth, and senescence (6). Although fluid chemistry is less directly correlated to chimney sample age than mineralogy, it is clear, that over long time scales, fluid chemistry strongly determines the mineralogy of chimneys (6). On short time scales, fluctuations in chemistry may affect microbial communities independently of mineralogy and chimney sample age, and it can be expected that these factors will have varying influences on different organisms.
Further work is necessary to confidently distinguish the influences of mineralogy, fluid chemistry, and age on each archaeal and bacterial species; but in general our results show that chimney sample age can be a useful indicator of bacterial community composition (Fig. 4). For example, samples 2 and 3 were collected from the same chimney flange, separated by ≈20 cm in situ, but have remarkably different bacterial communities (18% Bray-Curtis similarity). Sample 2 (43 ± 23 yr) was closer to the vent flange opening and is composed of aragonite (CaCO3) and brucite (Mg(OH)2) minerals that are stable in vent fluid–dominated environments. In contrast, sample 3 is older (128 ± 57 yr), contains more calcite than aragonite (calcite is a polymorph of metastable aragonite) and minor brucite, and was more exposed to ambient seawater (6). Clearly, the distribution of bacterial species correlates more strongly with sample age and mineralogy than with physical distance, as sample 2 has high similarity (45% Bray-Curtis) with sample 1, which is from a different chimney but of similar mineralogy and age (34.3 ± 8.1 yr). Moreover, sample 3 is dominated by the same bacterial sequences as sample 4 (1245 ± 257 yr), even though those two samples were separated by at least 100 m laterally and in depth. The four samples examined in this study do not constitute strong statistical evidence, but the initial data from this study, together with previous surveys of microbial diversity and geochemistry of additional samples (5, 6), are consistent with a correlation of mineralogy and age with microbial community composition.
The archaeal communities, in contrast, appear to be more strongly influenced by fluid chemistry when comparing samples younger than ∼150 yr. Although samples 2 and 3 have different ages and bacterial communities, their archaeal populations are nearly identical (95% Bray-Curtis similarity). Both samples were exposed to the same hydrothermal fluid, which may have fueled the same archaeal population, while bacteria in the older sample adapted over time to the increased influence of cold seawater that also resulted in mineralogical and porosity changes (6). Both samples have high cell densities and high proportions of archaea (Table S4), indicating that the archaeal populations are strongly stimulated by the conditions at this chimney flange. Thus, the archaeal community may not be influenced by mineralogical changes as long as fluid chemistry (e.g., H2 and CH4 concentrations) remains constant. Indeed, minor differences in fluid chemistry (3, 4) between chimneys may explain the presence of Crenarchaeota in sample 1 but not sample 2, which is of nearly identical age (Table 1).
The Lost City Rare Biosphere.
The archaeal and bacterial communities inhabiting carbonate chimneys at the LCHF are not “frozen in time” and do not record the history of the chimneys. Nevertheless, many chimneys must have formed, grown, and become inactive during the >30,000 years that the LCHF has been active (10). Furthermore, it is clear that mineralogy and fluid chemistry are correlated over long time scales with chimney growth stages (6) and chimney sample ages (33). Therefore, we infer that old, inactive chimneys formerly had environmental characteristics typical of younger chimneys. There is no reason to expect important differences between young chimneys active today and young chimneys that were active thousands of years ago. If environmental conditions broadly determine microbial community composition, microbial communities inhabiting chimneys today should resemble those that inhabited chimneys of similar ages hundreds or thousands of years ago. Although we have been able to collect V6 tag pyrosequences from only four samples, previous studies of LCHF chimneys have shown that archaeal and bacterial communities change when chimneys become less active (5). Furthermore, 230Th ages of additional LCHF chimney samples are consistent with the trend that chimney samples of similar ages have similar mineralogy and fluid chemistry (33). In conclusion, the results from this study and others (1, 5, 6, 10, 33) support a connection between microbial community composition and the geochemical conditions characteristic of a chimney sample’s age over long time scales.
The shifts in community composition observed in this study do not reflect new speciation events. Our results show that organisms favorably selected by new conditions already existed at low levels before the environmental change occurred (Fig. S6). Therefore, the genomes of the favorably selected organisms must have already encoded the necessary adaptations before the change. They were preadapted to the new conditions. Alternatively, every community shift may have resulted from a rare organism acquiring a useful gene via lateral gene transfer. Another possibility is that mutation and/or recombination occurred much more quickly in genes required for the adaptation but not in the V6 hypervariable region measured in this study. These processes may occur, but a more parsimonious explanation of the many shifts detected in this study where a rare organism rapidly increased its relative abundance is that these organisms had been preadapted for hundreds or thousands of years to the particular niche created by the environmental change. Therefore, the rare biosphere of the Lost City microbial community represents a large repository of genetic memory created during a long history of past environmental changes that selected for new species within a small pool of organisms that originally colonized the extreme environment. The rare organisms were able to rapidly exploit new niches as they arose because they had been previously selected for the same conditions in the past.
The Lost City Hydrothermal Field has been active for at least 30,000 years and probably much longer (10). Ultramafic environments such as Lost City have surely existed throughout Earth’s history and were probably much more widespread on the early Earth when ultramafic lavas were more common (1, 34). Indeed, the high concentrations of hydrogen generated at ultramafic-hosted systems like Lost City (4) may have supported the earliest ecosystems on Earth (35, 36). Therefore, the ecological dynamics described in this study may have been occurring in similar environments for most of Earth’s history, and the large pool of rare organisms present today may reflect that long history.
Methods
Pyrosequencing and Diversity Calculations.
Carbonate chimney samples were collected from the LCHF with DSV Alvin during cruise AT07-34 aboard the R/V Atlantis in April/May 2003 (http://www.lostcity.washington.edu). Detailed descriptions of 230Th dating techniques and the DNA extraction protocol are available in SI Methods. V6 amplicon libraries were constructed and sequenced as in refs. 17 and 18 with a 454 Life Sciences GS20 pyrosequencer and a 454 Life Sciences FLX pyrosequencer for the archaeal data. Tag sequences were screened for quality as recommend by (37). Archaeal and bacterial sequence alignments were constructed by submitting to the NAST aligner (http://greengenes.lbl.gov) all unique sequences pooled from all four samples, including primers to ensure full-length alignment. Obvious alignment errors were manually corrected, and primers were trimmed, resulting in final archaeal and bacterial alignments in which most sequences contained 60–65 bp. Similar results as those reported here were achieved by aligning with MUSCLE (38). The distance matrix for each alignment was calculated with quickdist as described elsewhere (17). Sequences were clustered into OTUs, and rarefaction curves and diversity estimators were calculated with DOTUR (39). Some of the rare sequences detected in this study may be artifacts of pyrosequencing technology (40, 41), but this possibility does not affect the main conclusions of our study. Because many of the rare sequences are much more abundant in other samples, these sequences are clearly not artifacts. Furthermore, we have recently reported that diversity in V6 tag pyrosequences is correlated to diversity at another marker and may actually underestimate the total genetic diversity (23). Additional methods for pyrosequencing and error determination are available in SI Methods.
Comparison of OTU Membership Among Samples.
After sequences were clustered into OTUs with DOTUR, the program SONS (42) was used to determine the relative abundance distribution of each OTU in each sample. The SONS output informed creation of the plots in Figs. 3–5 and Figs. S1–S6. Samples 1–4 yielded 16260, 32345, 25471, and 21983 archaeal tags and 21582, 5567, 7162, and 8716 bacterial tags, respectively. To normalize relative abundances of each OTU among samples, tags were randomly resampled down to the sample with the fewest tags using Daisy-Chopper (available from www.genomics.ceh.ac.uk/GeneSwytch/) after clustering OTUs. Consequently, normalized numbers of OTUs reported in Table 1 and Figs. 3–5 are lower than the total nonnormalized values reported in Table S1 and Fig. 2. Bray-Curtis and Jaccard similarities between samples were calculated with Primer 6 (http://www.primer-e.com) without any further data transformation. Taxonomies were assigned to each tag by the GAST process (22) via the VAMPS website.
Data Availability.
All V6 sequence data are available at the VAMPS database (http://vamps.mbl.edu), under dataset names ICM_LCY_Av6 and ICM_LCY_Bv6 and in the NCBI Short Read Archive under submission number SRP000912. See Supplementary Information for sample names in the databases.
Supplementary Material
Acknowledgments
We express our appreciation to the crews of the R/V Atlantis and DSV Alvin and the scientific party of the 2003 Lost City Expedition. We thank J. Huber and S. Huse for helpful discussions. This research was supported by the W.M. Keck Foundation to M.L.S., the NASA Astrobiology Institute through the Carnegie Institution for Science to J.A.B. and through the MBL to M.L.S., and NSF Grant OCE0137206 and NOAA Ocean Exploration support to D.S.K.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Tag sequences are available in the VAMPS database (http://vamps.mbl.edu), and in the NCBI Short Read Archive under accession number SRP000912.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905369107/DCSupplemental.
References
- 1.Kelley DS, et al. A serpentinite-hosted ecosystem: The Lost City hydrothermal field. Science. 2005;307:1428–1434. doi: 10.1126/science.1102556. [DOI] [PubMed] [Google Scholar]
- 2.Schrenk MO, Kelley DS, Bolton SA, Baross JA. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ Microbiol. 2004;6:1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- 3.Proskurowski G, Lilley MD, Kelley DS, Olson EJ. Low temperature volatile production at the Lost City Hydrothermal Field, evidence from a hydrogen stable isotope geothermometer. Chem Geol. 2006;229:331–343. [Google Scholar]
- 4.Proskurowski G, et al. Abiogenic hydrocarbon production at Lost City Hydrothermal Field. Science. 2008;319:604–607. doi: 10.1126/science.1151194. [DOI] [PubMed] [Google Scholar]
- 5.Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. Methane- and sulfur-metabolizing microbial communities dominate the Lost City Hydrothermal Field ecosystem. Appl Environ Microbiol. 2006;72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig KA, Kelley DS, Butterfield DA, Nelson BK, Früh-Green G. Formation and evolution of carbonate chimneys at the Lost City Hydrothermal Field. Geochim Cosmochim Acta. 2006;70:3625–3645. [Google Scholar]
- 7.Schulte M, Blake D, Hoehler T, McCollom T. Serpentinization and its implications for life on the early Earth and Mars. Astrobiology. 2006;6:364–376. doi: 10.1089/ast.2006.6.364. [DOI] [PubMed] [Google Scholar]
- 8.Oze C, Sharma M. Have olivine, will gas: Serpentinization and the abiogenic production of methane on Mars. Geophys Res Lett. 2005;32:L10203. [Google Scholar]
- 9.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6:805–814. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 10.Früh-Green GL, et al. 30,000 Years of hydrothermal activity at the Lost City Vent Field. Science. 2003;301:495–498. doi: 10.1126/science.1085582. [DOI] [PubMed] [Google Scholar]
- 11.Dimitriu PA, Pinkart HC, Peyton BM, Mormile MR. Spatial and temporal patterns in the microbial diversity of a meromictic soda lake in Washington State. Appl Environ Microbiol. 2008;74:4877–4888. doi: 10.1128/AEM.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moisander PH, Morrison AE, Ward BB, Jenkins BD, Zehr JP. Spatial-temporal variability in diazotroph assemblages in Chesapeake Bay using an oligonucleotide nifH microarray. Environ Microbiol. 2007;9:1823–1835. doi: 10.1111/j.1462-2920.2007.01304.x. [DOI] [PubMed] [Google Scholar]
- 13.Pagé A, Tivey MK, Stakes DS, Reysenbach A-L. Temporal and spatial archaeal colonization of hydrothermal vent deposits. Environ Microbiol. 2008;10:874–884. doi: 10.1111/j.1462-2920.2007.01505.x. [DOI] [PubMed] [Google Scholar]
- 14.Goffredi SK, Wilpiszeski R, Lee R, Orphan VJ. Temporal evolution of methane cycling and phylogenetic diversity of archaea in sediments from a deep-sea whale-fall in Monterey Canyon, California. ISME J. 2008;2:204–220. doi: 10.1038/ismej.2007.103. [DOI] [PubMed] [Google Scholar]
- 15.Lear G, Anderson MJ, Smith JP, Boxen K, Lewis GD. Spatial and temporal heterogeneity of the bacterial communities in stream epilithic biofilms. FEMS Microbiol Ecol. 2008;65:463–473. doi: 10.1111/j.1574-6941.2008.00548.x. [DOI] [PubMed] [Google Scholar]
- 16.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sogin ML, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;5:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 19.Neufeld JD, Li J, Mohn WW. Scratching the surface of the rare biosphere with ribosomal sequence tag primers. FEMS Microbiol Lett. 2008;283:146–153. doi: 10.1111/j.1574-6968.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- 20.Roesch LFW, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards RL, Chen JH, Wasserburg GJ. 238U -234U-230Th-232Th systematics and the precise measurement of time over the past 500,000 years. Earth Planet Sci Lett. 1986/1987;81:175–192. [Google Scholar]
- 22.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brazelton WJ, Sogin ML, Baross JA. Multiple scales of diversification within natural populations of archaea in hydrothermal chimney biofilms. Environ Microbiol Reports. 2009 doi: 10.1111/j.1758-2229.2009.00097.x. DOI:10.1111/j.1758-2229.2009.00097.x. [DOI] [PubMed] [Google Scholar]
- 24.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 25.Cao XH, Liu XL, Dong XZ. Alkaliphilus crotonatoxidans sp. nov., a strictly anaerobic, crotonate-dismutating bacterium isolated from a methanogenic environment. Int J Syst Evol Microbiol. 2003;53:971–975. doi: 10.1099/ijs.0.02373-0. [DOI] [PubMed] [Google Scholar]
- 26.Imachi H, et al. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol. 2006;72:2080–2091. doi: 10.1128/AEM.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lueders T, Pommerenke B, Friedrich MW. Stable-isotope probing of microorganisms thriving at thermodynamic limits: Syntrophic propionate oxidation in flooded soil. Appl Environ Microbiol. 2004;70:5778–5786. doi: 10.1128/AEM.70.10.5778-5786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorokin DY, et al. Thioalkalimicrobium cyclicum sp. nov. and Thioalkalivibrio jannaschii sp. nov., novel species of haloalkaliphilic, obligately chemolithoautotrophic sulfur-oxidizing bacteria from hypersaline alkaline Mono Lake (California) Int J Syst Evol Microbiol. 2002;52:913–920. doi: 10.1099/00207713-52-3-913. [DOI] [PubMed] [Google Scholar]
- 29.Knittel K, et al. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int J Syst Evol Microbiol. 2005;55:781–786. doi: 10.1099/ijs.0.63362-0. [DOI] [PubMed] [Google Scholar]
- 30.Dhillon A, et al. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol. 2005;71:4592–4601. doi: 10.1128/AEM.71.8.4592-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ Microbiol. 2005;7:118–132. doi: 10.1111/j.1462-2920.2004.00672.x. [DOI] [PubMed] [Google Scholar]
- 32.Bradley AS, Hayes JM, Summons RE. Extraordinary 13C enrichment of diether lipids at the Lost City Hydrothermal Field indicates a carbon-limited ecosystem. Geochim Cosmochim Acta. 2009;73:102–118. [Google Scholar]
- 33.Ludwig KA, Shen C, Kelley DS, Cheng H, Edwards R. U-Th isotopic systematics and ages of carbonate chimneys at the Lost City Hydrothermal Field. Eos Trans AGU. 2009 90(Suppl):Abstract V31D-2007. [Google Scholar]
- 34.Grove TL, Parman SW. Thermal evolution of the Earth as recorded by komatiites. Earth Planet Sci Lett. 2004;219:173–187. [Google Scholar]
- 35.Nesbet EG, Sleep NH. The habitat and nature of early life. Nature. 2001;409:1082–1091. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]
- 36.Olson JM. Photosynthesis in the Archean era. Photosynth Res. 2006;88:109–117. doi: 10.1007/s11120-006-9040-5. [DOI] [PubMed] [Google Scholar]
- 37.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quince C, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 41.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: Pyrosequencing errors lead to artificial inflation of diversity estimates. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.02051.x. 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 42.Schloss PD, Handelsman J. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl Environ Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All V6 sequence data are available at the VAMPS database (http://vamps.mbl.edu), under dataset names ICM_LCY_Av6 and ICM_LCY_Bv6 and in the NCBI Short Read Archive under submission number SRP000912. See Supplementary Information for sample names in the databases.