Abstract
Hybrid incompatibility in F1 hybrids or later generations is often observed as sterility or inviability. This incompatibility acts as postzygotic reproductive isolation, which results in the irreversible divergence of species. Here, we show that the reciprocal loss of duplicated genes encoding mitochondrial ribosomal protein L27 causes hybrid pollen sterility in F1 hybrids of the cultivated rice Oryza sativa and its wild relative O. glumaepatula. Functional analysis revealed that this gene is essential for the later stage of pollen development, and distribution analysis suggests that the gene duplication occurred before the divergence of the AA genome species. On the basis of these results, we discuss the possible contribution of the “founder effect” in establishing this reproductive barrier.
Keywords: F1 pollen sterility, gene duplication, BDM incompatibility, mitochondrial ribosomal protein
In On the Origin of Species, Darwin argued the importance of natural selection in the evolution of species (1). Through natural selection, mutations increasing fitness will become fixed in a population, whereas those reducing fitness will be removed. The sterility and inviability observed in hybrids between two species are obvious traits that decrease fitness. Therefore, the existence of these traits has been pointed out as a contradiction of natural selection. Bateson, Dobzhansky, and Muller explained this contradiction using a genetic model in which hybrid sterility and inviability are caused by sets of interacting “complementary genes” (2, 3). This model is widely accepted and called the Bateson–Dobzhansky–Muller (BDM) incompatibility model. A molecular study of Drosophila revealed that the interaction of protein products from multiloci (4 –7) and gene transposition (8) could lead the BDM incompatibility in hybrids. In plants, hybrid incompatibility conferring inviability, sterility, and necrosis (or weakness) in the F1 and F2 generation has been frequently found (9 –11). Epistatic interaction between dominant or recessive alleles at multiple loci triggers zygotic (or sporophytic) BDM incompatibility in hybrids in various species. Recent gene cloning studies contribute to an understanding of the mechanism of the epsitatic interaction between the loci in plants (12, 13). One study of “hybrid necrosis,” one of the reproductive barriers widely distributed in plant species (11), demonstrated that one dominant allele encoding nucleotide binding site–leucine-rich repeat (NBS-LRR) genes causes characteristic autoimmune reactions in F1, which are similar to phenotypes associated with response to environmental stress, such as pathogen attack in Arabidopsis thaliana (12). Another recent study demonstrated that interaction of recessive alleles between two loci, which emerged from gene duplication and its reciprocal loss in each diverged population, cause hybrid inviability in the F2 generation in A. thaliana (13). On the other hand, cloning of the gene for gametic (or gametophytic) BDM incompatibility is limited, except as cited in one seminal work for the Sa locus identified in an inter-subspecific cross in rice (14). The Sa locus had been recognized as a single Mendelian factor, but this locus contained two adjacent genes interacting with each other, and divergence of the genes in each subspecies caused gametophytic pollen sterility in their hybrid.
In rice, pollen sterility in F1 hybrids is a major reproductive isolation barrier between cultivated rice Oryza sativa and its wild relatives (15, 16). To explain the genetic mechanism of such F1 sterility, two models have been proposed according to the mode of Menderian inheritance: the one-locus allelic interaction and the two-locus epistatic interaction (16). Recently, two cases of the gametophytic F1 pollen and embryo sac sterility fitting the one-locus allelic interaction model were analyzed at the molecular level (14, 17). However, the molecular mechanism of gametophytic F1 sterility fitting the two-locus model could not be understood although the possible cause of sterility was estimated to be due to duplicated genes, which are essential for gamete development in the classic genetic analysis in rice (18).
In this study, we demonstrated that reciprocal loss of duplicated genes causes hybrid sterility in F1 between O. sativa and O. glumaepatula. These genes encode mitochondrial ribosomal protein L27, which is essential for the late developmental stage of pollen. Furthermore, we analyzed the distribution of the duplicated locus in the rice AA genome species, and its evolutional trajectory was discussed.
Results
Epistatic Interaction Between S27 and S28.
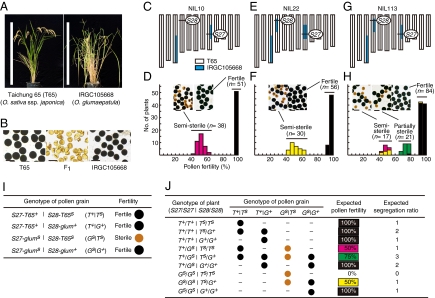
O. glumaepatula is a wild rice distributed in the Amazon basin (19). The F1 hybrid between the cultivated variety Taichung 65 [O. sativa ssp. japonica (T65)] and the O. glumaepatula accession IRGC105668 exhibits complete pollen sterility (Fig. 1 A and B), but its pistils can accept fertile pollen and produce mature seeds. On the basis of these characteristics, we developed near-isogenic lines (NILs) by pollinating the F1 hybrids with T65 pollen. Genetic studies using these NILs revealed that the F1 pollen sterility is controlled by several loci, including S27 and S28 (20 –23). On the basis of a genetic analysis of three NILs (Fig. 1 C, E, and G) using two tightly linked markers, S27_ssr1 for S27 and S28_10 for S28, we found an epistatic interaction between S27 on chromosome 8 and S28 on chromosome 4.
Fig. 1.
Epistasis between S27 and S28. (A) T65 and IRGC105668 plants. Scale bars: 1 m. (B) I2–KI-stained pollen of T65, IRGC105668, and their F1 hybrid. Scale bars: 50 μm. (C and D) Genetic analysis of S27 using NIL10. (E and F) Genetic analysis of S28 using NIL22. (G and H) Genetic analysis of the epistatic interaction between S27 and S28 using NIL113. (C, E, and G) The graphical genotypes of the NILs. (D, F, and H) Frequency distribution of pollen fertility in the progeny of each NIL. Pink, yellow, green, and black in the histograms indicate the genotypes of individuals represented in J. (I) Gametophytic epistatic model of hybrid pollen sterility. (J) Expected classes of genotypes and pollen fertility in self-pollinated progeny of plants heterozygous at both S27 and S28.
In the self-pollinated progeny of NIL10 (S27 heterozygous and S28-T65 homozygous) (Fig. 1C), S27 heterozygotes were semisterile and S27-T65 homozygotes were completely fertile, and no S27-glum homozygote was obtained (Fig. 1D). This result indicates that the S27-glum allele causes pollen sterility in this genetic background. In the self-pollinated progeny of NIL22 (S27-glum homozygous and S28 heterozygous) (Fig. 1E), S28 heterozygotes were semisterile and S28-glum homozygotes were completely fertile, and no S28-T65 homozygote was obtained (Fig. 1F). Again, this indicates that the S28-T65 allele causes pollen sterility in this genetic background. Taken together, we hypothesized that S27 and S28 interact epistatically in a gametophytic manner and that pollen grains carrying S27-glum and S28-T65 would be sterile.
To confirm this hypothesis, the self-pollinated progeny of NIL113 (S27 and S28 heterozygous) were tested (Fig. 1 G and H). According to the hypothesis, the fertility of a single pollen grain is determined by the genotype of the pollen grain at the S27 and S28 loci (Fig. 1I); i.e., S27-glum and S28-T65 alleles are sterile, whereas S27-T65 and S28-glum are fertile alleles (hereafter, sterile alleles are indicated by a superscript s and fertile ones by a superscript “+”). Consequently, pollen grains carrying a set of sterile alleles, S27-glums and S28-T65s, should be sterile whereas pollen grains carrying at least one fertile allele, S27-T65+ or S28-glum+, should be fertile (Fig. 1I). In this context, the pollen fertility of plants heterozygous at both the S27 and S28 loci should become 75%, and self-pollinated plants will show 100%, 75%, and 50% pollen fertility with a segregation ratio of 7:3:2, respectively (Fig. 1J). Indeed, the progeny of NIL113 were divided into three classes (Fig. 1H): semisterile, partially sterile, and fertile with an average pollen fertility of 48.9%, 74.1%, and 94.8%, respectively, and the segregation of pollen fertility in the progeny fit our hypothesis (Table S1). Spikelet fertility of each pollen fertility class was normal, indicating that S27 and S28 did not affect female gamete fertility.
Map-Based Cloning.
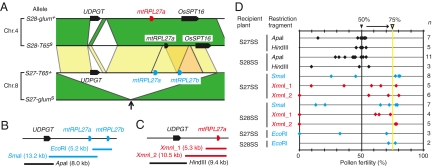
We performed map-based cloning of S27 and S28. High-resolution mapping revealed that S27 and S28 are located in 134.9-kb and 1.68-Mb regions on chromosomes 8 and 4, respectively (Fig. S1). Ueda et al. (24) reported ≈30-kb duplicated segments in our candidate regions for S27 and S28 in the Nipponbare genome (Fig. S1, yellow boxes). Therefore, we determined the sequences around these duplicated segments of T65 and O. glumaepatula and found similar duplicated segments in the S27-T65+, S28-T65s, and S28-glum+ alleles, but not in the S27-glums allele (Fig. 2A, arrow). We speculated that genes located in these duplicated segments are associated with the epistatic interaction between S27 and S28, whereas S27-glums may be a loss-of-function allele due to the absence of the duplicated segment. The gene prediction by the Rice Genome Automated Annotation System (25) suggested that the corresponding region of S27-T65+ contains the UDP-glycosyltransferase gene (UDPGT) and two tandem mitochondrial ribosomal protein L27 genes (mtRPL27a and mtRPL27b), whose amino acid sequences are identical (Fig. 2A and Fig. S2). In the S28 region, one UDPGT and one mtRPL27 are predicted in S28-glum+, and one mtRPL27 and no UDPGT are predicted in S28-T65s. On the basis of the genomic sequence similarity, including the promoter region of the genes, the mtRPL27 genes predicted in S28-T65s and S28-glum+ appeared to correspond to mtRPL27a in S27-T65+. The S28-T65s and S28-glum+ regions also contain another gene (OsSPT16), which encodes a homolog of yeast spt16 (24) and is not seen in the S27-T65+ region (Fig. 2A).
Fig. 2.
Map-based cloning of S27 and S28. (A) The genomic sequences of T65 and O. glumaepatula in the duplicated segments at S27 and S28. The conserved genomic regions between homologous chromosomes and between nonhomologous chromosomes are connected with green and yellow, respectively. The tandem genomic duplicate containing mtRPL27b in the S27-T65+ allele is indicated by orange. Pentagons represent the predicted genes. The segmental duplicated segments did not exist in S27-glums allele (arrow). (B and C) Locations of the restriction fragments of S27-T65+ (B) and S28-glum + (C) used in the complementation test. (D) Pollen fertility of T0 plants transformed with the restriction fragments in B and C. S27SS and S28SS represent the S27 semisterile and S28 semisterile plants, respectively (see text for their genotypes). The expected pollen fertility of the recipient plants is 50%. The rescued plants are expected to show 75% pollen fertility.
Next, we transformed the genomic fragments containing various genome sequences derived from the fertile alleles S27-T65+ and S28-glum+ (Fig. 2 B and C) into the pollen-semisterile plants carrying S27-T65+/S27-glums|S28-T65s/S28-T65s (S27 semisterile plants) or plants carrying S27-glums/S27-glums|S28-T65s/S28-glum+ (S28 semisterile plants). Theoretically, when the transformed genomic fragment has the ability to rescue pollen sterility and is introduced at one locus of the host plants, pollen fertility is rescued from 50% to 75% in T0 plants (the rightmost line in Fig. 2D). When the genomic fragment containing mtRPL27a (the SmaI fragment from T65 in Fig. 2B and the XmnI_1 or XmnI_2 fragment from O. glumaepatula in Fig. 2C) or mtRPL27b (the EcoRI fragment in Fig. 2B) was transformed, transgenic plants exhibited ≈75% pollen fertility, whereas the introduction of UDPGT (the ApaI fragment in Fig. 2B or the HindIII fragment in Fig. 2C) did not affect pollen fertility (Fig. 2D). These results demonstrate that the duplicated genes encoding the mtRPL27 protein at S27 and S28 are involved in this pollen sterility, whereas mtRPL27a on S28-T65s is a loss-of-function allele caused by failure of expression (see below).
Mechanism of Pollen Sterility.
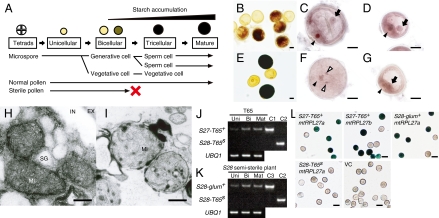
To unravel the mechanism of pollen sterility, we observed the pollen of S28 semisterile plants at each developmental stage (Fig. 3A). No phenotypic abnormality was observed at the tetrad or unicellular stages (Fig. S3). At the late bicellular stage, we detected generative and vegetative cells in all pollen grains (Fig. 3 C and D), but half of them failed to initiate starch accumulation (Fig. 3B). At the mature stage, two sperm cells and one vegetative cell were detected in half of the pollen grains (Fig. 3F), and the remainder were still arrested at the bicellular stage without starch accumulation (Fig. 3 E and G). Furthermore, because the causal genes of S27 and S28 encode mtRPL27 protein, which is localized in mitochondria (24), we observed mitochondria in pollen grains at the mature stage using transmission electron microscopy (Fig. 3 H and I). In the mitochondria of the fertile pollen, a developed matrix structure and intermembrane space were observed (Fig. 3H), whereas no such complex structure was observed in the mitochondria of sterile pollen grains (Fig. 3I). In bacteria, ribosomes lacking RPL27 protein are defective in protein synthesis (26). This suggests that a defect of mtRPL27 inhibits protein synthesis in mitochondria, and consequently its respiratory activity is defective and it produces sterile pollen.
Fig. 3.
Mechanism of pollen sterility. (A) Schematic of postmeiotic pollen development. (B–G) Light-microscopic observation of the pollen phenotype in bicellular (B–D) and mature (E–G) stages in terms of starch accumulation (B and E) and nuclei (C, D, F, and G). (C, D, F, and G) Normal (C and F) and abnormal (D and G) pollen grains. Nuclei of vegetative cells, generative cells, and sperm cells are indicated by black arrowheads, black arrows, and white arrowheads, respectively. Scale bars: 10 μm. (H and I) The ultrastructure of normal (H) and abnormal (I) pollen grains. MI, mitochondria; EX, exine; IN, intine; SG, starch grain. Scale bars: 0.2 μm. (J and K) Allele-specific semiquantitative RT–PCR of each mtRPL27 allele in anthers during the unicellular (Uni), bicellular (Bi), and mature pollen stages (Mat) in T65 (J) and in the S28 semisterile plants (K). C1, C2, and C3 indicate the plasmid DNA carrying mtRPL27 of the S27-T65+, S28-T65s, and S27-glum+ alleles as controls, respectively. (L) Analysis of GUS activity under the control of promoters of each mtRPL27 gene in pollen. VC indicates the control. Scale bars: 50 μm.
An expression analysis of mtRPL27 was performed. Transcription of mtRPL27 was observed in various plant tissues, including leaf, leaf sheath, root, and flower (Fig. S4). The expression of mtRPL27 in anther during pollen development was analyzed by quantitative RT–PCR (Fig. S4). About twofold greater expression was observed in the bicellular and mature stages relative to the unicellular stage. This matches the microscopic observations and supports the importance of the mtRPL27 protein for pollen development from the bicellular to the mature stage.
Next, we examined the expression of mtRPL27a at S28-T65s in T65. Although several amino acid substitutions were observed (Fig. S2), we speculated that the loss-of-function at S28-T65S is caused by failure of expression because a loss of promoter activity of mtRPL27 at this locus has been reported in Nipponbare (24), a close relative of T65. When we analyzed the DNA sequence of the RT–PCR products from T65 anthers, we detected only the mtRPL27a or mtRPL27b sequence of S27-T65+, and not mtRPL27a of S28-T65s, which suggests that mtRPL27a at the S28-T65s locus is not transcribed. Furthermore, we confirmed the loss of expression of S28-T65s by using allele-specific semiquantitative RT–PCR analysis with locked nucleic acid (LNA) primers (27). In the T65 anther, increased expression of mtRPL27a or mtRPL27b of S27-T65+ was detected (a similar result is shown in Fig. S4B), whereas no PCR product was found using the S28-T65s allele-specific primers (Fig. 3J). When we used anthers from S28 semisterile plants, the expression of S28-glum+ was observed in a pattern similar to S27-T65+, but again, no PCR product for S28-T65s was detected (Fig. 3K). To confirm the expression of mtRPL27 in pollen grains, we analyzed the β-glucuronidase (GUS) activity under the control of the promoters of S27-T65+ mtRPL27a, S27-T65+ mtRPL27b, S28-T65s mtRPL27a, and S28-glum+ mtRPL27a (Fig. 3L). GUS activity was observed under the control of the promoters of the three fertile alleles (+), but not S28-T65s (Fig. 3L). All these results clearly demonstrate that S28-T65s contains a loss-of-function allele of mtRPL27a with failure of expression.
Origin and Distribution of the S27-glums Allele During AA Genome Divergence.
On the basis of a comparison of genomic sequences, Ueda et al. (24) postulated that the duplicated segment on chromosome 8 (i.e., S27) was derived from chromosome 4 (i.e., S28). We investigated the distribution of the S27-glums alleles among rice AA genome species using two PCR markers, S27D and S27ND, which determine the presence or absence of the duplicated segment at S27 (Fig. S5). Among the 172 accessions of the eight AA genome species, the accessions carrying the duplicated segment at S27 (S27-T65+ allele) were widely distributed in all seven species, except for O. glumaepatula (Table 1, Fig. S6). This indicates that the segmental genomic duplication carrying mtRPL27 at S27 occurred before the divergence of the rice AA genome species and that the duplicated segments at S27 spread in each species, except O. glumaepatula, which is the sole South American species possessing the AA genome and is thought to have an African origin (29). Indeed, the O. glumaepatula accessions used in this study were closely related to the O. barthii and O. glaberrima complex (28, 30, 31) (also see Fig. S6). This evidence led us to hypothesize that the ancestral population of O. glumaepatula and O. barthii contained both duplicated and nonduplicated genomes at S27, and a population with a nonduplicated genome dispersed from Africa to South America and evolved into O. glumaepatula in the Amazon basin. In other words, the frequency of the S27-glums allele might have been more prominent in O. glumaepatula due to the founder effect, which is a special case of random genetic drift in a population derived from a small number of individuals geographically isolated from the ancestral population. In population genetics, the frequency of genes, including neutral genes of duplicated loci, sometimes changes rapidly through a random sampling effect in a small founder population. These data suggest that geographical isolation played a central role in establishing reproductive isolation between the divergent species despite the selectively neutral allele.
Table 1.
Distribution of the duplicated segment at S27 among rice AA genome species
| No. of accessions | |||
| Duplicated segment at S27 | |||
| Species* | Present | Absent | Total |
| O. sativa ssp. japonica group | 31 | 0 | 31 |
| O. sativa ssp. indica group | 70 | 0 | 70 |
| O. rufipogon group | 23 | 0 | 23 |
| O. nivara group | 12 | 0 | 12 |
| O. glaberrima group | 8 | 0 | 8 |
| O. barthii group | 5 | 3 | 8 |
| O. glumaepatula group | 0 | 7 | 7 |
| O. longistaminata group | 1 | 1 | 2 |
| O. meridionalis group | 11 | 0 | 11 |
Discussion
Reproductive barriers are thought to play an important role in plant speciation, as in animal speciation (10, 32). The analysis of morphometric data from more than 200 plant genera revealed significant relationships between species and postmating reproductive barriers, especially postzygotic barriers (10, 32). Therefore, a study on the mechanism of postzygotic barriers is essential to understanding the mechanism of plant species diversification. A large fraction of plant genomes consists of duplicated loci due to the frequent occurrence of segmental genome duplication or polyploidization (33, 34), which has led various authors to propose that the loss-of-function allele of duplicated genes maintained in the genome is a potential source of postzygotic isolation (35, 36). However, only one example of this was reported very recently in an intraspecific hybrid within A. thaliana (13). Our study provides experimental evidence that the duplication and subsequent reciprocal loss of the gene, which is expressed and essential for the male gametophyte, also play an important role in hybrid sterility in the F1 generation.
Materials and Methods
Plant Materials.
The Asian cultivated rice T65 (O. sativa L. ssp. japonica cv. Taichung 65) was crossed with pollen of the South American wild rice species O. glumaepatula Steud. (accession: IRGC105668, Brazil) to produce F1 plants with T65 cytoplasm. Three NILs were developed by continuous backcrossing with T65 and screened at the BC4 F1 generation: NIL10 (S27 heterozygous and S28-T65 homozygous), NIL22 (S28 heterozygous and S27-glum homozygous), and NIL113 (both S27 and S28 heterozygous). The genotypes of each NIL across the entire genomic region were determined using restriction fragment length polymorphism markers. The progeny of NIL10 and NIL22 were used for high-resolution mapping of S27 and S28, respectively. To analyze the epistatic interaction, the progeny of NIL113 were used.
Evaluation of Pollen Fertility.
Panicles at the flowering stage were fixed and stored in 70% (wt/wt) ethanol. Six anthers from one spikelet collected just before flowering were squashed on a glass slide in 1% (wt/vol) iodine-potassium iodide (I2–KI) solution. More than 200 pollen grains were evaluated for pollen fertility under an Axioplan light microscope (Zeiss). Pollen grains that were morphologically the same as grains of T65 were scored as normal. Aborted, unstained, and small grains were scored as sterile.
Map-Based Cloning.
For the high-resolution mapping of S27 and S28, we used simple sequence repeat, cleaved amplified polymorphic sequence, and single nucleotide polymorphism markers (Table S2). Genomic DNA fragments containing the candidate genes were cloned in the Ti-plasmid binary vector pPZP2H-lac (37). Using Agrobacterium-mediated transformation (38), these genomic fragments were introduced into the pollen-semisterile plants with the genotype S27-T65+/S27-glums| S28-T65s/S28-T65s and into the other pollen-semisterile plants with the genotype S27-glums|S27-glums/S28-T65s/S28-glum+.
Observation of Pollen Development.
Panicles in the meiotic-to-mature stages were collected continuously to monitor pollen development from the unicellular-to-mature stage. Panicles were placed in fixative solution containing 4% (wt/vol) paraformaldehyde, 0.25% (wt/vol) glutaraldehyde, 0.02% (vol/vol) Triton X-100, and 100 mM sodium phosphate (pH 7.5) for 24 h at 4°C and then rinsed in 100 mM sodium phosphate buffer. The hematoxylin staining procedure used followed that of Chang and Neuffer (39) with minor modifications.
Ultrastructural Analysis.
Anthers at the mature stage were vacuum-infiltrated three times for 10 min with fixative (4% paraformaldehyde and 1% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.4). The procedures for the ultrastructural studies were essentially the same as those described by Hara-Nishimura et al. (40).
Expression Analysis.
Total RNA was extracted using the RNeasy plant kit (Qiagen). The sequences of the primer pairs were 5′-CCG TGA CTC TAA CCC TAA G-3′ and 5′-ATG GAA GCG GGT TCC TCT T-3′ for mtRPL27 and 5′-CTG TCA ACT GCC GCA AGA AG-3′ and 5′-GGC GAG TGA CGC TCT AGT TC-3′ for UBQ1 (Os03g0234200). Following Itoh et al. (41), we sampled 1–2 mm of the anthers at the unicellular stage, 2 mm of the anthers at the bicellular stage, and the anthers at the mature stage. First-strand cDNA was synthesized from ∼1 μg of total RNA with an oligo(dT) primer and the OmniScript RT kit (Qiagen). Expression of mtRPL27 at the various stages was quantified with real-time PCR analysis using 10% of the resulting cDNA as template. Real-time PCR was performed with the LightCycler system (Roche) and a QuantiTect SYBR Green PCR kit (Qiagen). For this analysis, a linear standard curve and threshold cycle number versus log (designated transcript level) were constructed using a series of dilutions of each PCR product (10−17, 10−18, 10−19, and 10−20 M); the transcript levels in all unknown samples were determined using the standard curve. We used UBQ1 as an internal standard for normalizing the cDNA concentrations. Data represent the average of three replicates of the first-strand cDNA. Allele-specific semiquantitative RT–PCR was performed using LNA primers (27). The primer sequences are listed in Table S3. The PCR was conducted using AmpliTaq Gold DNA polymerase (Applied Biosystems) and consisted of a 5-min Taq activation step followed by 33 cycles at 94°C for 30 s, 68.4°C for 20 s, and 72°C for 20 s.
Promoter GUS Assay.
To analyze the GUS activity in the pollen, the promoter sequence for each mtRPL27 gene was digested using the appropriate restriction enzyme or amplified with PCR using a genomic clone as the template. When PCR was used to clone the promoter sequence, the resulting product was cloned into pCRII (Invitrogen) and was sequenced to confirm that no base substitution had occurred during PCR. The promoter sequences were introduced at the front of the GUS reporter gene of pBI-Hm (42) to produce a fusion with the GUS reporter gene. The constructs were introduced using Agrobacterium-mediated transformation (38) into T65. The regenerated plants containing only one copy of the insert were screened by Southern blot analysis using a GUS DNA fragment as the probe. Pollen of regenerated plants was sampled, and the GUS activity was analyzed as described previously by Matsuoka and Sanada (43).
Distribution Analysis.
The presence and absence of the duplicated segment at S27 was investigated in the AA genome rice species with the markers S27D and S27ND, respectively, using KOD plus DNA polymerase (TOYOBO). The primer pair used for the S27D marker was 5′-TTG AAT TCC GAG AGT CTG GC-3′ and 5′-GGT CGT CGG AGT GGT AGA CGA AG-3′ and that used for the S27ND marker was 5′-GGT CGT CGG AGT GGT AGA CGA AG-3′ and 5′-TGT TCA CCA AAT GAC TCC AAA TCG-3′. The PCR comprised an initial denaturation at 96°C for 2 min, followed by 30 cycles at 96°C for 35 s, 60°C for 30 s, and 68°C for 2 min.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Integrated Research Project for Plants, Insects, and Animals Using Genome Technology GD-2003 to A. Y, and K. D., and Genomics for Agricultural Innovation QTL-5002 to A.Y. and IPG-0015 to K.D.) and a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (18075006 to M.M.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Date deposition: The sequences reported in this paper have been deposited in the DNA Data Base in Japan [accession nos. AB496673 (mtRPL27a S27-T65+), AB496674 (mtRPL27b S27-T65+), and AB496675 (mtRPL27a S28-glum+)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0908283107/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: J. Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 3.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- 4.Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila . Proc Natl Acad Sci USA. 2003;100:5302–5307. doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brideau NJ, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila . Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 6.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila . Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 7.Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masly JP, Jones CD, Noor MAF, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila . Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 9.Stebbins GL. The inviability, weakness, and sterility of interspecific hybrids. Adv Genet. 1958;9:147–215. doi: 10.1016/s0065-2660(08)60162-5. [DOI] [PubMed] [Google Scholar]
- 10.Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bomblies K, Weigel D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- 12.Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5 doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana . Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- 14.Long Y, et al. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci USA. 2008;105:18871–18876. doi: 10.1073/pnas.0810108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu YE, Morishima H, Oka HI. Reproductive barriers distributed in cultivated rice species and their wild relatives. Jpn J Genet. 1969;44:207–223. [Google Scholar]
- 16.Oka HI. Origin of Cultivated Rice. Tokyo: JSSP/Elsevier; 1998. [Google Scholar]
- 17.Chen J, et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA. 2008;105:11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka HI. Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics. 1974;77:521–534. doi: 10.1093/genetics/77.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akimoto M, Shimamoto Y, Morishima H. Population genetic structure of wild rice Oryza glumaepatula distributed in the Amazon flood area influenced by its life-history traits. Mol Ecol. 1998;7:1371–1381. [Google Scholar]
- 20.Sobrizal, Matsuzaki Y, Sanchez PL, Ikeda K, Yoshimura A. Identification of a gene for male gamete abortion in backcross progeny of Oryza sativa and Oryza glumaepatula . Rice Genet Newsl. 2000;17:59–61. [Google Scholar]
- 21.Sobrizal, Matsuzaki Y, Sanchez PL, Ikeda K, Yoshimura A. Mapping of F1 pollen semi-sterility gene found in backcross progeny of Oryza sativa L. and Oryza glumaepatula Steud. Rice Genet Newsl. 2000;17:61–63. [Google Scholar]
- 22.Sobrizal, Matsuzaki Y, Yoshimura A. Mapping of a gene for pollen semi-sterility on chromosome 8 of rice. Rice Genet Newsl. 2001;18:59–61. [Google Scholar]
- 23.Sobrizal, Matsuzaki Y, Yoshimura A. Mapping of pollen semi-sterility gene, S28(t), on rice chromosome 4. Rice Genet Newsl. 2002;19:80–82. [Google Scholar]
- 24.Ueda M, et al. Promoter shuffling at a nuclear gene for mitochondrial RPL27. Involvement of interchromosome and subsequent intrachromosome recombinations. Plant Physiol. 2006;141:702–710. doi: 10.1104/pp.105.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakata K, et al. RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 2002;30:98–102. doi: 10.1093/nar/30.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wower IK, Wower J, Zimmermann RA. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J Biol Chem. 1998;273:19847–19852. doi: 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 27.Latorra D, Arar K. Design considerations and effects of LNA in PCR primers. Mol Cell Probes. 2003;17:253–259. doi: 10.1016/s0890-8508(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 28.Doi K, Nonomura MN, Yoshimura A, Iwata N, Vaughan DA. RFLP relationships of A-genome species in the genus Oryza . J. Fac. Agr. Kyushu Univ. 2000;45:83–98. [Google Scholar]
- 29.Vaughan DA, Kadowaki K, Kaga A, Tomooka N. On the phylogeny and biogeography of the genus Oryza . Breed Sci. 2005;55:113–122. [Google Scholar]
- 30.Wang ZY, Second G, Tanksley SD. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet. 1992;83:565–581. doi: 10.1007/BF00226900. [DOI] [PubMed] [Google Scholar]
- 31.Cheng CY, Tsuchimoto S, Ohtsubo H, Ohtsubo E. Evolutionary relationships among rice species with AA genome based on SINE insertion analysis. Genes Genet Syst. 2002;77:323–334. doi: 10.1266/ggs.77.323. [DOI] [PubMed] [Google Scholar]
- 32.Rieseberg LH, Wood TE, Baack EJ. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 34.Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Werth CR, Windham MD. A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate-gene expression. Am Nat. 1991;137:515–526. [Google Scholar]
- 36.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 37.Fuse T, Sasaki T, Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 2001;18:219–222. [Google Scholar]
- 38.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 39.Chang MT, Neuffer MG. Maize microsporogenesis. Genome. 1989;32:232–244. [Google Scholar]
- 40.Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni . Plant Cell. 1993;5:1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh J, et al. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- 42.Nomura M, et al. The promoter of rbcS in a C3 plant (rice) directs organ-specific, light-dependent expression in a C4 plant (maize), but does not confer bundle sheath cell-specific expression. Plant Mol Biol. 2000;44:99–106. doi: 10.1023/a:1006461812053. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka M, Sanada Y. Expression of photosynthetic genes from the C4 plant, maize, in tobacco. Mol Gen Genet. 1991;225:411–419. doi: 10.1007/BF00261681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.