Abstract
p53 is a master regulatory, sequence-specific transcription factor that directly controls expression of over 100 genes in response to various stress signals. Transactivation is generally considered to occur through p53 binding to a consensus response element (RE) composed of two 5′-RRRCWWGYYY-3′ decamers. Recently, studying the human angiogenesis-related gene FLT1 we discovered that p53 can mediate limited transactivation at a noncanonical 1/2 site and could synergize with the estrogen receptor (ER) acting in cis at a nearby ER 1/2 site. To address the generality of concerted transactivation by p53 and ER, the 1/2 site in the FLT1 promoter was replaced with a variety of 1/2 sites, as well as canonical weak and strong p53 REs of human target genes. The p53 transactivation of all tested sequences was greatly enhanced by ligand-activated ER acting in cis. Furthermore, enhanced transactivation extends to several cancer-associated p53 mutants with altered function, suggesting ER-dependent mutant p53 activity for at least some REs. The enhanced transactivation was also found with p63 and p73. We propose a general synergistic relationship between p53 family and ER master regulators in transactivation of p53 target canonical and noncanonical REs, which might be poorly responsive to p53 on their own. This relationship greatly expands the transcriptional master network regulated by p53 in terms of genes affected and levels of expression and has implications for the appearance and possible treatments of cancer.
Keywords: FLT1, noncanonical response element, half-site, transcriptional synergy
The tumor suppressor protein p53 is a master regulator that plays a major role in genome stability, cell propagation, and apoptosis. The sequence-specific DNA binding of p53 is critical to its tumor suppressor activity in response to cellular and environmental stresses. The canonical target sequence consists of two decameric motifs, originally defined by the consensus RRRCWWGYYY separated by 0–13 bp (1), although the separation is considered to be typically less than or equal to two bases (2 –5). Although the interactions of p53 bound to some DNAs are established, the mechanisms by which WT or mutant p53 can transactivate to different extents from the many variants of the consensus motif are still not well-understood.
Different strategies merging bioinformatic analysis along with experimental approaches including microarrays, ChIP, ChIP-on-chip, or ChIP followed by cloning and DNA sequencing, have been used to reveal potential p53 sequence-specific gene targets in the human genome (5, 6). This has refined the p53 consensus sequence and also revealed many more p53 potential binding sites in the genome. However, not all expected target sites based on prior knowledge of p53 target genes have been retrieved with these methodologies, suggesting that experimental conditions such as cell type or p53-inducing stimuli could influence p53 occupancy across the genome. Also, technical limitations of ChIP approaches could introduce a bias toward more accessible genomic regions or against weak affinity p53 binding sites. Furthermore, the dynamic modulation of p53 occupancy at a target regulatory region and the correlation between occupancy levels and transcriptional rates of the nearby genes are complex and likely influenced by other sequence-specific transcription factors and cofactors. In addition, there are regions where p53 has been found to bind that do not conform to p53 consensus (7, 8). This is consistent with protein–protein interactions as well as protein–DNA interactions contributing to p53 recruitment at genomic regions. Also, there may be alternative modes of p53 binding to target DNA sequences that strongly deviate from the canonical consensus in terms of sequence and/or spatial arrangements of the 1/4 sites. For example, alternative arrangements of 1/4 sites (i.e., head-to-tail vs. head-to-head) or a specific separation between 1/2 sites result in p53 driving transcriptional repression instead of activation (1, 9). Also, combinatorial interactions of p53 with other sequence-specific transcription factors may influence target gene modulation. There is still a gap in what constitutes a functional cis-acting target DNA element for p53 transcriptional control of a gene, especially in the context of the overall promoter, the chromatin landscape and the potential crosstalk with other cis-acting TFs (10).
Recently, we examined 1/2 sites that could bind p53 and lead to transactivation (3). Specifically, a C to T single nucleotide polymorphism SNP in the promoter of the human angiogenesis-related gene FLT1 generated a consensus 1/2 site that incorporated this gene into the p53 transcriptional network (11). More recently, we also established that a 1/2 site could function in transactivation of the DNA repair gene RAP80 (12). An additional complexity in the p53 network was revealed wherein p53 could act in cis at the FLT1 promoter with other transcription factors, the estrogen receptors (ERs). The interaction occurred through an ER 1/2 site located approximately 250 nt upstream from the p53 1/2 site (13) and resulted in a synergistic increase in FLT1 transcription. We have examined the FLT1 promoter motif to address the generality of ER being able to act in cis with p53 at canonical, as well as noncanonical, p53 sequences (i.e., 1/2 sites) to enhance transactivation. In addition we investigated whether this synergy applies to cancer-associated p53 mutants as well as to the p53-related p63 and p73 proteins.
Results
p53 Sequence-Dependent Transactivation from Noncanonical 1/2 Site Motifs.
To assess further the ability of noncanonical sequences to support p53 transactivation in human cells, a panel of 15 p53 1/2 site response elements (REs) were cloned upstream of the luciferase reporter in the pGL4.26 vector (i.e., 1/2RE::Luc). Although they completely match consensus sequence (RRRCWWGYYY), each differs by one or more bases at different positions of the RE. Transactivation capacity toward the 1/2 sites was evaluated in p53 null SaOS2 cells transiently cotransfected with a WT p53 expression vector. As shown in Fig. 1A, p53 dependent transactivation is strongly influenced by RE sequence. (The levels of p53 were comparable in the transfected cells as described in Fig. S1). The 1/2 site from the FLT1 promoter (GGACATGCTC) is only modestly responsive to p53. For the CWWG core, the CATG combination was the most responsive, although changes in the purines (R) and pyrimidines (Y) had different effects, even lowering the responsiveness to nearly background levels. For positive controls, we included the REs from two well-known p53 targets p53-regulated apoptosis-inducing protein 1 (AIP) and p21 5′RE. Several of the 1/2 sites supported transactivation to levels comparable to the AIP weak canonical RE.
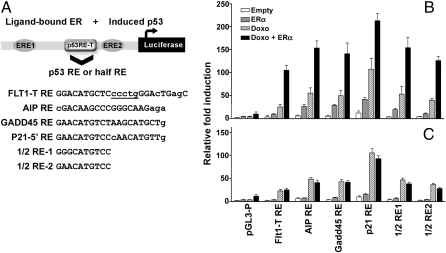
Fig. 1.
p53 transactivation from noncanonical 1/2 sites in human cells. Sequence dependence of p53 transactivation from various 1/2 site p53 REs. Luciferase assays were performed 48 h after transfection of SaOS2 cells with the reporter constructs along with plasmids expressing WT p53 (A) or different p53 mutants (B). The original p53 1/2 site associated with FLT1 gene is indicated by the *. For evaluation of mutant p53 transactivation, the constructs designated 1/2 RE1 and RE2 were used as well as the full site p21 5′RE. Results are reported as average-fold luciferase induction relative to the empty pGL4.26 vector ± SD (n = 3).
Nearly 1/2 of human cancers have associated p53 mutations, most of which are missense mutations in the DNA-binding domain (DBD). These can lead to nuclear accumulation of mutant protein and loss of transcriptional activation of target genes. However, the functional consequences of only approximately 20% of the nearly 1,100 amino acid changes toward established p53 REs have been examined in detail in mammalian cells. Very few have been tested for transactivation at noncanonical p53 sequences. We, therefore, determined the transactivation capacity of a panel of 12 cancer-associated altered-function p53 mutants (described in Table S1 and Fig. S2) toward two of the most responsive p53 1/2 site REs identified in Fig. 1A (GGGCATGTCC corresponding to 1/2 RE1 and GAACATGTCC corresponding to 1/2 RE2 derived from p21 5′ RE). A full p53RE (p21 5′RE) was included as a positive control. (Transfected p53 constructs were equally expressed as shown in the Fig. S3). As shown in Fig. 1B and summarized in Table 1, the loss-of-function hotspot cancer mutants R175H, G245S, and G279E lacked function toward both 1/2 sites, as well as the full REs, although Y220C, M237I, and E285K were weakly active toward at least one 1/2 site RE and the full p21-5′RE. Surprisingly, several of the altered function p53 mutants could support transactivation of 1/2 site REs, as well as a full p53RE including A138V and R337H, that have been found associated with several sporadic and familial cancers, respectively. As observed for WT p53, differences in the 1/2 site sequences also affect the transactivation potential of some p53 mutants (S121F, T125R, A138V, and E285K). In addition, we also tested p53 mutants that have lost the ability to adopt tetrameric or dimeric conformation (3). Interestingly, mutants that form dimers but not tetramers (L344A and N345S) retain some functionality toward the 1/2 REs, unlike mutants that cannot form dimers (i.e., only form monomers: L330H, Q331stop, and L344P). Finally, a deletion mutant (Δ368) that removes the C-terminal-tail of p53 required for structure–specific and nonsequence specific binding also was able to transactivate 1/2 REs (Fig. S4).
Table 1.
Transactivation by WT and mutant p53 at 1/2 and full sites in the FLT1-T motif, alone and at endogenous FLT1-T
| Full and 1/2 site REs within1 Kb Flt1 motif::LUC | 1/2 site::LUC | Endogenous FLT1-T | ||||||||||||
| Full sites | 1/2 sites | |||||||||||||
| AIP | GADD45 | p21 | FLT1-T | 1/2 RE1 | 1/2 RE1 | 1/2 RE2 | ||||||||
| p53 mutation | −ERα | +ERα | −ERα | +ERα | −ERα | +ERα | −ERα | +ERα | −ERα | +ERα | −ERα | −ERα | −ERβ | +ERβ |
| WT | Yes | YES** | Yes | YES** | Yes | YES** | Yes | YES** | Yes | YES** | Yes | Yes | Low | YES** |
| S121F | Yes | YES** | Yes | YES** | Slight | Slight | Yes | YES** | Yes | YES** | Yes | Yes | Slight | YES** |
| A138V | Slight | YES** | Yes | YES** | Yes | YES** | No | YES** | Yes | YES** | Yes | Yes | No | YES** |
| G279R | No | No | Slight | YES** | Yes | YES** | No | YES** | Slight | YES** | Yes | Yes | Slight | YES** |
| R337H | Slight | YES** | Yes | YES** | Yes | YES** | No | YES** | Yes | YES** | Yes | Yes | Low | YES** |
| E285K | No | No | Yes | YES** | No | No | No | No | Slight | Slight | Yes | No | No | No |
| R337C | No | No | Slight | YES** | Yes | YES** | No | No | No | No | No | No | No | No |
| T125R | No | No | Yes | Yes | Slight | Yes | No | No | Yes | Slight | Yes | No | No | No |
| Y220C | No | No | No | No | No | No | No | Slight | No | Slight | Slight | No | No | Low |
| M237I | No | No | No | Yes | No | No | No | No | Slight | Slight | Slight | No | Slight | No |
YES**, transactivation by WT or mutant p53 is enhanced by ERα or ERβ.
*R175H, G245 and G279E were also examined (Fig. 1B) but exhibited no transactivation under any condition.
Impact of p53 RE Sequence on Transcriptional Cooperation Between p53 and Estrogen Receptors.
Based on the in cis transcriptional cooperation between p53 and ERα or ERβ in transactivation at a 1/2 site p53 RE (13), we explored further the generality of this transcriptional cooperation. We used the FLT1 promoter motif and developed 5 constructs, described in Fig. 2A, where the original 1/2 site (FLT1-T RE) was replaced by other p53 1/2 sites or by canonical p53REs derived from known human target genes that had previously been shown to exhibit various transactivation potentials in a yeast-based assay (2, 14, 15): AIP (weak), GADD45 (moderate) and p21-5′ (strong). As a control for p53 responsiveness, independent from the FLT1 motif, we used the isolated 5′ RE from the p21 gene. The ability of these constructs to support p53 transactivation was examined using a luciferase reporter (downstream from the constructs) assay in U2OS cells that are WT p53 but ER deficient (Fig. 2B). Activation of p53 by doxorubicin alone induced transcription of all FLT1 promoter constructs to varying levels, including those containing the two noncanonical 1/2 sites. Relative to the original 1/2 site p53RE-T, the other 1/2 sites were more responsive to p53, consistent with the negative impact of the “CTC” sequence motif in the native FLT1 1/2 site (Fig. 1A) (see also ref. 3). Except for AIP which was somewhat higher, the canonical REs maintained their expected relative level of responsiveness when placed into the FLT1 motif. Over-expression of ERα alone resulted in weak to modest transactivation depending on the constructs, probably reflecting the cooperation with the basal levels of p53 (Fig. 2B). Ectopic expression of ERα and concomitant activation of p53 by doxorubicin treatment resulted in a dramatic transcriptional synergy that was dependent on RE sequence. Remarkably, the level of induction for the p53 1/2 site REs was comparable to that of the full site human p21 RE.
Fig. 2.
The p53 RE sequence determines the level of synergy between p53 and ERα in the FLT1 promoter motif. (A) A schematic description of the FLT1-T promoter showing the relative position of the Flt1-T RE flanked by the two identified EREs. The original 1/2 site Flt1-T RE was replaced by other 1/2 sites (previously tested in Fig. 1B) or by well-established canonical p53 REs as indicated in A. Small letters denote deviations from the consensus, although the underlined letter refers to the spacer between p53 1/2 sites. (B) The ability of these constructs to support p53 transactivation was examined using a luciferase reporter assay in U2OS cells following p53 activation by doxorubicin in the presence or absence of transfected ERα. When indicated, cells were treated with doxorubicin (0.3μg/mL) for 16 h before cell lysis. Luciferase activity was measured 48 h posttransfection and normalized for transfection efficiency. (C) Site-directed mutagenesis of the EREs in the FLT1 promoter constructs abolished the synergistic transactivation between p53 and ERα. Presented are the average-fold luciferase induction relative to the empty pGL4.26 vector ± SD (n = 3).
The 1 kb promoter region of FLT1-T contains two 1/2 estrogen response elements (EREs). One site, which is located 225nt upstream of the p53 RE (GGTCAgagTcACt; mismatches from a consensus full-site ERE are underlined), was required for p53-induced transactivation in three different cancer cell lines derived from colon or bone tissue (11, 13). A second putative ERE 1/2 site (GGTCAggagcggC), is located 145nt downstream to the p53 RE. Given that ER and p53 proteins were reported to physically interact (16, 17), we changed the two EREs by site-directed mutagenesis to confirm that the synergistic transactivation observed with the variant FLT1 constructs require ER sequence-specific DNA binding (changes: aaTCAgagTcACt for ERE1 and aaTCAggagcggC for ERE2; the double mutant construct is referred to as ere1,2 in Fig. 2C for U2OS cells and Fig. S5 for MCF7 cells). The removal of the EREs resulted in the complete loss of the ERα synergistic impact for every construct (Fig. 2C).
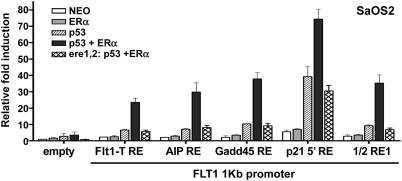
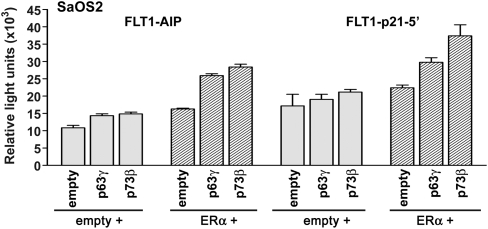
To further confirm the role of p53, we repeated the luciferase reporter assays in the p53 null SaOS2 cell line which is ERβ negative and weakly expresses ERα (Fig. 3). The transient restoration of WT p53 expression by transfection alone was sufficient to induce the transcription of all of the FLT1 promoter constructs tested although the expression of only the ERα transcription factor in the absence of p53 failed to induce significant transactivation of the reporter gene. As observed previously in U2OS cells, the presence of both TFs resulted in the synergistic transactivation of all FLT1 constructs, confirming the cooperativity between these two master regulators. The level of luciferase activity relative to the mock condition (pGL3P empty promoter) was driven by the strength of the p53 sequence introduced to the FLT1 promoter. Unlike p53, ectopic expression of the p53 family members p63γ and p73β did not lead to transactivation of the FLT1-AIP RE and FLT1-p21 RE constructs (Fig. 4). However, concomitant ectopic expression ERα resulted in a small but significant increase in reporter activity, suggesting that p63γ and p73β can also cooperate with ERα.
Fig. 3.
Transcriptional cooperation between ERα and p53 enhances the sequence-dependent transcriptional potential in the FLT1 promoter motif. FLT1 motif constructs were created containing noncanonical and canonical p53 REs upstream of a luciferase reporter as described in Fig. 2. The constructs contained EREs as present in the FLT1 motif or mutated EREs (referred to as ere1,2). These constructs were cotransfected with WT p53 and/or ERα expression vectors into SaOS2 cells and the luciferase promoter activity was compared to mock transfected cells. Luciferase activity was measured 48 h posttransfection and normalized for transfection efficiency. Results are shown as the average-fold luciferase induction relative to the empty vector ± SD (n = 3).
Fig. 4.
The p53-related p63γ and p73β proteins can cooperate with ERα to transactivate canonical p53 REs in the FLT1 promoter. A derivative of the 1-kb FLT1 promoter construct containing the p53REs from AIP or p21 genes was cotransfected into SaOS2 cells in the presence of p73β and p63γ alone or in combination with an ERα expression vector. Luciferase activity was measured 48 h posttransfection. Empty + represents a vector (pCMV-Neo) used to keep constant the amount of total DNA transfected. Presented is the average-fold luciferase induction relative to the empty reporter vector ± SD (n = 3).
The various FLT1 reporter constructs were also examined in MCF7 cells that are p53 and ER positive for ERα and ERβ. Cells were treated with doxorubicin to activate p53 and cultured in normal medium containing the estradiol ligand. We noticed that in MCF7 cells the empty pGL3 vector was doxo responsive; nevertheless we confirmed that the responsiveness of the FLT1 derived constructs in untreated and doxorubicin treated cells was proportional to the intrinsic transactivation potential of the cloned p53 REs (Fig. S5). Transactivation was greater for the 1/2 site RE1 in the FLT1 motif as compared to the native p53 RE-T. For the full site REs, the FLT1-p21 was highly responsive with FLT1-AIP and FLT1-GADD45 being much less, similar to findings in U2OS cells. Also consistent with the U2OS results, inactivation of the two ERE sites (ere1,2) reduced p53 transactivation of all REs in the FLT1-motif. The FLT1-p21 construct was less impacted by the deletion of the EREs compared to the results in U2OS cells (Fig. 2C).
Transcriptional Synergy Between Mutant p53 and ERs.
Having established that some p53 mutants retain transactivation at noncanonical 1/2 sites, we addressed the potential interaction between ten p53 mutants with different transactivation capacities in combination with the ERα. As shown in Fig. S6 and summarized in Table 1 each of the altered function p53 mutants had its own transactivation spectrum toward the various full and 1/2 sites in the FLT1-derived luciferase constructs when expressed alone or in the combination with ERα. As expected, the p53 mutant G279E was not functional toward any of the REs.
For some mutants (e.g., S121F, A138V, G279R and R337H) transcriptional cooperation with ERα was evident with most of the reporter constructs evaluated including the 1/2 site from FLT1 and 1/2 RE1, resulting in comparable or even higher transactivation than with WT p53 for some sites. Thus, ERα could in some cases actually lead to reactivation of the mutants toward at least some REs. As with WT p53, the cooperation with ER was dependent on the presence of the EREs, with the possible exception of T125R and M237I. The other mutants, T125R, Y220C, M237I, E285K and R337C, were inactive toward noncanonical 1/2 REs placed within the FLT1 promoter. However, ERα stimulated E285K and R337C at the p21 (for R337C) and GADD45 REs.
Effect of p53 Mutants on Endogenous FLT1 Gene Expression.
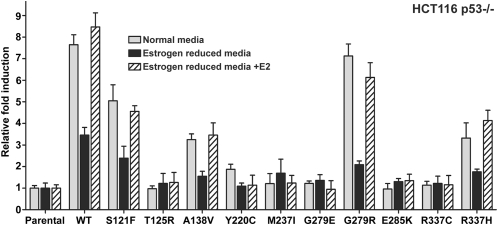
Previously, we showed that WT p53 activation by genotoxic stress could result in allele-specific up-regulation of the endogenous FLT1 gene in cancer cell lines that are heterozygous for the p53 RE SNP in the promoter region of this gene (11, 13). Having established here that some p53 mutants can cooperate with ERα to transactivate FLT1-T and derived promoter constructs, we sought to evaluate the potential for mutant p53 to activate the endogenous FLT1 gene. We used the HCT116 p53−/− ERβ+ cells that are heterozygous for the C/T SNP at the p53 RE in FLT1. Cells were cultured in normal as well as estrogen-reduced media (Materials and Methods) and transfected with p53 mutant expression vectors. As shown in Fig. S7, nuclear levels of WT and p53 mutants after transfection were comparable; however, the Y220C mutant protein was predominantly expressed in the cytoplasm, as previously reported (18). The FLT1 mRNA levels were evaluated approximately 24 h after transient transfection. As shown in Fig. 5 and summarized in Table 1, the expression of WT p53 in p53−/− cells resulted in approximately eightfold induction of FLT1, as expected. Consistent with the above results (Fig. S2 and Table 1), p53 mutants S121F, A138V, G279R, R337H, and to a limited extent Y220C were able to induce the expression of FLT1 in vivo, whereas mutants T125R, M237I, and R337C had no effect on the expression. Induction was greatly reduced when cells were cultured in estrogen-reduced conditions, strongly implicating a role for ERβ in the FLT1 transcription. This reduction was reversed when transfected cells were cultured in medium containing the ER ligand 17β-estradiol (E2), resulting in synergy between ERβ and p53 for the responsive mutants as well as for the WT. Thus, ERβ bound to a ligand and acting in cis is required or can stimulate the function of selected p53 mutants toward at least some 1/2 and full site REs and also an endogenous gene target.
Fig. 5.
ERβ cooperates with mutant p53 to induce transcription from the endogenous FLT1 gene. HCT116 p53−/− cells cultured in normal media (gray bars), in estrogen-reduced media alone (black bars), or in the presence of 10−9 M of E2 (strip bars) were transiently transfected with WT or mutant p53 expression vector. After 24 h, the relative expression of the FLT1 mRNA was measured by real time PCR. The HCT116 cells are heterozygous for the p53 target 1/2 site in the FLT1 where p53 induction leads to specific induction of the T allele (11). Results are reported as average-fold ± SD (n = 3) of induction relative to the housekeeping gene β2 microglobulin.
Discussion
Identification of genes regulated by p53 is critical to a full understanding of the p53 transcriptional network and how modulations of the network relate to various possible biological outcomes elicited by p53 activation and cancer prevention. The establishment of p53 transcriptional targets that are regulated by sequences differing from the commonly accepted p53 consensus RE greatly expands the p53 transcriptional network. These noncanonical REs include 1/2 and 3/4 sites with deviations in the most conserved domain of the p53 RE, the CWWG core (12, 13, 19). A recent review listed 129 validated p53 transcriptional targets (7). We have reevaluated these validated targets and concluded that approximately 30% should be considered as noncanonical (20).
Here, using luciferase reporter transactivation assays in human cancer cells, we developed functionality rules regarding WT p53 transactivation at 1/2 site REs, similar to our previous approaches with full sites (20), and identified cancer-associated mutant p53s that retained function at 1/2 sites. Moreover, we establish the general ability of ERα acting in cis to amplify transcription by WT and mutant p53s at a variety of noncanonical and canonical p53 REs.
Sequence-Dependent WT and Mutant p53 Activity at Noncanonical p53 REs.
p53 transactivation of noncanonical 1/2 site REs in human cells is highly sequence-specific (Fig. 1) in agreement with initial observations using a yeast-based system (3). The requirements for functionality of 1/2 sites overlap with those for the canonical p53 REs containing two decamers (2, 20, 21). The CWWG core has the greatest influence on p53 transactivation from 1/2 sites in human SaOS2 cells. Although the overall transactivation levels were modest, CATG was dramatically more effective than the other 3 possibilities, with the palindrome GGGCATGCCC being the most active decamer followed by GGGCATGTCC, suggesting that mechanisms governing p53-DNA interactions, such as flexibility (22 –24), are also relevant to 1/2 sites. Notably, these two REs showed transactivation levels similar to the weak responding full-site p53RE from the AIP gene. The same trend had been seen with canonical REs. These findings with 1/2 sites are likely relevant to previously described full-sites where decamers that are separated by more than a few bases appear to act independently (3).
Changes in purines and pyrimidines flanking the core RE sequence also affect responsiveness of 1/2 site p53 REs. Bases flanking the CWWG consisting of GGG and C/TCC enhanced transactivation, as also reported for full site REs (25). In agreement with results in a yeast-based system (3) and the p53 1/2 site RE in the FLT1 gene (11), a “CTY” motif following CATG reduces the functionality of a 1/2 site.
Nearly one-third of cancer-associated mutants retain transactivation capability (15) and many exhibit a change-in-spectrum in terms of transactivation from various REs. Among 12 cancer-associated mutants examined (Table 1 and Fig. S6), four were transcriptionally active at full site REs (S121F, A138V, G279R and R337H) and also retained activity at the two 1/2 sites. Of the remaining five that were active at full sites, T125R, Y220C, M237I, and E285K exhibited limited transactivation at 1/2 sites. The p53 cancer hotspot mutants (R175H, G245S, and G245S) that were transcriptionally null toward full site REs, behaved similarly toward 1/2 sites.
Transcriptional Cooperation Between Estrogen Receptors and WT or Mutant p53.
Having established transcriptional cooperation in cis between p53 and ER (each binding to their respective 1/2 sites), we asked about the generality of this cooperation especially because the corresponding motifs are expected to be common in the genome. Using a variety of p53 full and 1/2 site REs, we demonstrated that p53 and ER could greatly increase transactivation in a manner that was dependent on a functional ERE in cis. Remarkably, not only was there ER/p53 synergy in the responsiveness to p53, but the greatest relative increases were obtained with the p53 1/2 sites and the weaker REs (up to eightfold). Furthermore, the p53-related transcription factors p73β and p63γ could also cooperate with ER to induce transcription from the p21 and AIP REs, although to a lesser degree than p53. Because some p53 target genes can also be transactivated by p63 and p73, it is possible that the mechanism of cooperativity with the ERs is similar to that for p53 and ERs. These results suggest the possibility that ER, and potentially other hormone nuclear receptors that act on ERE related sequences, could broadly influence p53 as well as p63γ and p73β responses, especially because ER pentamer 1/2 sites are common across the genome.
The ability of ER to influence p53 mediated transcription in cis extends to functional p53 mutants. Among the nine mutants examined that retain at least some transactivation (Table 1), six exhibited increases with ER for at least one full site RE (GADD45). Among these six, there were four (S121F, A138V, G279R, and R337H) that yielded strong, synergistic interactions with 1/2 sites (Fig. S6). These were the same mutant p53s that responded well with different 1/2 site REs independent of ER. Although the familial cancer mutants R337C and R337H interact with ER to drive transcription from full site REs, only the R337H can transactivate from 1/2 sites and also exhibit synergy with ER at a 1/2 site. Both these mutants are associated with familial cancer predispositions; however, R337H is uniquely associated with the risk of developing adrenocortical carcinomas (ACC) (26, 27).
Not only was there synergy between ERα and WT p53 toward transfected FLT1-T constructs, there was also synergy with endogenous ERβ toward the endogenous FLT1-T allele in the HCT116 p53−/− colon cancer cell line. As previously observed for the WT p53 (13), the synergy was clearly dependent on ligand because there was little ER responsiveness for cells grown in medium greatly reduced for estrogen content; the p53-ER synergy was restored when the ligand for ERs was added back. Similar results were obtained with S121F, A138V, G279R, and R337H mutants.
As for many other transcription factors, p53 regulation of gene expression can take place both by direct binding to target gene promoters and through the interaction with other TFs that can impact negatively or positively on p53-mediated transactivation (8, 20). Our results with the various FLT1 motif constructs that contain mutated ERE sequences demonstrate that the transcriptional cooperation between p53 and ERs is mediated by their cognate response elements in cis rather than through physical interactions between p53 and ER proteins (16, 17). Previously, we reported that ER binding at FLT1 promoter is dependent on p53 binding and the recruitment of cofactors including the TRAP/Mediator complex (13).
Like the WT protein, mutant p53 proteins can also interact with several sequence-specific transcription factors and cooperatively modulate transactivation characteristics (28 –30). The possibility that mutant p53 proteins can bind DNA with different/altered specificity from the WT p53 and induce the recruitment of chromatin-modifying cofactors distinct from those targeted by WT p53 has also been suggested (31, 32). It is clear that further analyses of how mutant p53 interacts with genomic sequences in vivo, the possible protein partners interacting with mutant p53 and the chromatin modifications associated will provide more insights on the mechanism of how mutant p53 controls gene expression in vivo.
Conclusions
Because 1/2 sites respond weakly to p53, they may provide opportunities for regulation at high levels of p53 expression as found for several REs associated with apoptosis genes. Importantly, we found that among validated p53 REs conserved between rodents and humans, one third (20/58) were comprised of 1/2 or of 3/4 sites containing a consensus 1/2 site (RRRCATGYYY) (20). This is consistent with our previous findings of conservation of weak sites (21).
Because several mutations have been identified that affect the ability to form p53 oligomers, we expanded our previous analysis of tetramerization domain mutants (3) to their ability to transactivate from strong half-sites. Based on results presented in Fig. S4, we conclude that p53 dimers, but not monomers, can bind and transactivate at strong 1/2 sites. This agrees with in vitro results where WT p53 and the dimer mutant L344A can bind the same 1/2 site corresponding to our 1/2 RE1 (GGGCATGTCC) (33), although there is little if any binding to weak MDM2 half-sites that do not contain a CATG cores (34). We also found that the R337H and R337C mutants, which can affect tetramer stability differed in their ability to transactivate from the 1/2 sites. Interestingly, deletion of the C-terminal nonspecific p53 DNA binding domain appeared to have only a twofold effect on transactivation of both full and half sites relative to WT p53.
An important factor to consider is the level of p53 proteins needed for transactivation at 1/2 and full sites. For WT p53, we reported that unlike for full sites, the 1/2 sites are transactivated only when p53 protein levels are high (3). Furthermore, in the case of p53 mutants that form dimers, there was a strong dependency on high protein levels for 1/2 as well as full site REs. Collectively, these results suggest that p53 tetramers can be formed by two p53 dimers binding independently at adjacent 1/2 sites or even, although less efficiently, when only a 1/2 site RE is present with a significant contribution from nonspecific DNA binding.
Our results demonstrate the potential for ER to interact in cis with p53 to generate transcriptional cooperation at poorly responsive p53 1/2 and full site RE targets that normally might be unrecognizable as responsive to p53 alone. These findings raise many questions, including the generality of p53/ER synergy across the genome and requirements for synergy and the distance be tween the ER and p53 targets, variation in target sequence, as well as the type of stress activating p53 and the type of ligands activating the ERs. Given the key role that p53 and ER have in tumor development and cancer progression along with the high frequency of p53 and ERE 1/2 sites, the molecular rules for in cis cooperation warrant further detailed investigation. These observations also set the stage for investigating other transcriptional factors that may play a role in p53-targeted gene expression. Because the weak RE sites are unlikely to have been identified in previous ChIP experiments, approaches (currently underway) will need to be modified to accommodate the ability to detect them in the genome with or without the presence of ER or other TFs.
The inclusion of the weak 1/2 sites, along with transcriptional amplification, adds another dimension to the “piano model” used to describe the variations in transcription of p53 (or other) target genes where the REs are the keys and the p53 is the hand. Because of variation in REs (and, therefore, level of response), the “sound” of the resulting chord (i.e., the biological response) can be quite varied. In this model, the ER acts to strengthen (or simply include) the sound of the individual keys in the final chord. In a more general sense, determining the matrix of relationships between p53 expression levels and responsiveness at canonical and noncanonical target sequences, along with in cis (direct physical) interactions, is important to understanding the overall impact of the p53 tumor suppressor. These concepts also apply to mutant p53. In fact, we established that several p53 mutants that retain function can cooperate with ER in transactivation at strong as well as weak p53 target REs, including 1/2 sites. The potential for partial-function p53 alleles to interact with other families of sequence-specific transcription factors, could be a relevant parameter to consider when correlating p53 mutant status and clinical variables in breast as well as other cancers. In addition, tumors that are ER positive may have a better opportunity for turning on or reactivating p53-related functions toward some genes even if the levels of p53 are reduced or there are altered function p53 mutants.
Materials and Methods
Cell Lines and Drug Treatments.
Human colon carcinoma HCT116 and p53-null derivatives cells were supplied by Dr. B. Vogelstein (The Johns Hopkins Kimmel Cancer Center, Baltimore, MD; ref 35), although the human osteosarcoma cell lines SaOS2 (HTB-85,) and U2OS (HTB-96) were obtained from ATCC. The human breast adenocarcinoma-derived MCF-7 cell line was obtained from the InterLab Cell Line Collection bank (ICLC) Repository (Genoa, Italy). More information about cell culture conditions, protein levels determination, DNA damage stresses, and reagents used for the present study is given in SI Materials and Methods.
Plasmids, Reporter Constructs, and Luciferase Assays.
Information about mammalian expression plasmids used in this study, as well as for reporter constructs, transfection, and luciferase assays are provided in SI Materials and Methods.
Gene Expression by Real-Time RT-PCR.
To determine the FLT1 mRNA levels, real-time PCR was carried out using the ABI Prism 7000 sequence detection system. More information about the protocol is presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We appreciate O. Bandele and A. M Jetten for critical comments on the manuscript. This work was supported by the Intramural Research Program of the National Institute on Environmental Health Sciences (Grant Z01 ES065079 to D.M. and M.A.R.) and partially supported by the Italian Association for Cancer Research, Associazione Italiana per la Ricerca sul Cancro (to A.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909129107/DCSupplemental.
References
- 1.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 2.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol. 2002;22:8612–8625. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan JJ, et al. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veprintsev DB, Fersht AR. Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res. 2008;36:1589–1598. doi: 10.1093/nar/gkm1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 6.Smeenk L, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 8.Vousden KH, Prives C. Blinded by the light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 9.Johnson RA, Ince TA, Scotto KW. Transcriptional repression by p53 through direct binding to a novel DNA element. J Biol Chem. 2001;276:27716–27720. doi: 10.1074/jbc.C100121200. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–4023. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menendez D, et al. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci USA. 2006;103:1406–1411. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Menendez D, Yang XP, Resnick MA, Jetten AM. A regulatory loop composed of RAP80-HDM2-p53 provides RAP80-enhanced p53 degradation by HDM2 in response to DNA damage. J Biol Chem. 2009;284:19280–19289. doi: 10.1074/jbc.M109.013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menendez D, et al. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol Cell Biol. 2007;27:2590–2600. doi: 10.1128/MCB.01742-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inga A, Monti P, Fronza G, Darden T, Resnick MA. p53 mutants exhibiting enhanced transcriptional activation and altered promoter selectivity are revealed using a sensitive, yeast-based functional assay. Oncogene. 2001;20:501–513. doi: 10.1038/sj.onc.1204116. [DOI] [PubMed] [Google Scholar]
- 15.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci USA. 2003;100:9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Schwartz JA, Brooks SC. Estrogen receptor protects p53 from deactivation by human double minute-2. Cancer Res. 2000;60:1810–1814. [PubMed] [Google Scholar]
- 17.Liu W, et al. Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J Biol Chem. 2006;281:9837–9840. doi: 10.1074/jbc.C600001200. [DOI] [PubMed] [Google Scholar]
- 18.Dearth LR, et al. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis. 2007;28:289–298. doi: 10.1093/carcin/bgl132. [DOI] [PubMed] [Google Scholar]
- 19.Cai BH, et al. Functional four-base A/T gap core sequence CATTAG of P53 response elements specifically bound tetrameric P53 differently than two-base A/T gap core sequence CATG bound both dimeric and tetrameric P53. Nucleic Acids Res. 2009;37:1984–1990. doi: 10.1093/nar/gkp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 21.Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. Proc Natl Acad Sci USA. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 23.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 24.Nagaich AK, Bhattacharyya D, Brahmachari SK, Bansal M. CA/TG sequence at the 5′ end of oligo(A)-tracts strongly modulates DNA curvature. J Biol Chem. 1994;269:7824–7833. [PubMed] [Google Scholar]
- 25.Halazonetis TD, Kandil AN. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueiredo BC, et al. Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation. J Med Genet. 2006;43:91–96. doi: 10.1136/jmg.2004.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro RC, et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc Natl Acad Sci USA. 2001;98:9330–9335. doi: 10.1073/pnas.161479898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E, Deppert W. The versatile interactions of p53 with DNA: When flexibility serves specificity. Cell Death Differ. 2006;13:885–889. doi: 10.1038/sj.cdd.4401909. [DOI] [PubMed] [Google Scholar]
- 29.Mazur SJ, et al. Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J Mol Biol. 1999;292:241–249. doi: 10.1006/jmbi.1999.3064. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, et al. p53 domains: Identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7(12B, 12B):2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, Deppert W. Interactions of mutant p53 with DNA: Guilt by association. Oncogene. 2007;26:2185–2190. doi: 10.1038/sj.onc.1210312. [DOI] [PubMed] [Google Scholar]
- 32.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol Cell Biol. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterman JL, Shenk JL, Halazonetis TD. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14:512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 35.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.