Abstract
Bardet-Biedl syndrome (BBS) is a human genetic disorder resulting in obesity, retinal degeneration, polydactyly, and nephropathy. Recent studies indicate that trafficking defects to the ciliary membrane are involved in this syndrome. Here, we show that a novel complex composed of three chaperonin-like BBS proteins (BBS6, BBS10, and BBS12) and CCT/TRiC family chaperonins mediates BBSome assembly, which transports vesicles to the cilia. Chaperonin-like BBS proteins interact with a subset of BBSome subunits and promote their association with CCT chaperonins. CCT activity is essential for BBSome assembly, and knockdown of CCT chaperonins in zebrafish results in BBS phenotypes. Many disease-causing mutations found in BBS6, BBS10, and BBS12 disrupt interactions among these BBS proteins. Our data demonstrate that BBS6, BBS10, and BBS12 are necessary for BBSome assembly, and that impaired BBSome assembly contributes to the etiology of BBS phenotypes associated with the loss of function of these three BBS genes.
Keywords: Bardet-Biedl Syndrome, ciliopathy, molecular chaperone, protein trafficking
The primary cilium is a microtubule-based subcellular organelle that projects from the surface of the cell. It plays an essential role in the transduction of extracellular signals (1, 2). In vertebrates, loss of cilia or ciliary dysfunction leads to various defects such as situs inversus, polydactyly, neural tube defects, and obesity (2 –4). Ciliary dysfunction is also involved in several human genetic syndromes (2, 3). Bardet-Biedl syndrome (BBS) is one of the most studied human genetic disorders associated with ciliary dysfunction. Individuals with BBS display retinal degeneration, obesity, polydactyly, hypertension, hypogonadism, renal anomalies, and cognitive impairment (5 –7). BBS displays autosomal recessive inheritance with extensive genetic heterogeneity.
BBS proteins are required for the maintenance of ciliary structure and function. Mutation of BBS genes in mice results in absence of flagella in spermatozoa (8 –10) and abnormalities in cilia in brain ependymal cells, airway epithelial cells (11, 12) and olfactory neurons (13). At the molecular level, BBS proteins are involved in protein/vesicle trafficking along microtubules. For example, knockdown of BBS genes in zebrafish results in delay in retrograde melanosome transport, which is mediated by dynein motor proteins along the microtubule (14, 15). In C. elegans, mutations in bbs1, bbs7, or bbs8 cause defects in the movement of the intraflagellar transport (IFT) subcomplexes inside the cilium (16). During the last decade, twelve BBS genes (BBS1–12) have been identified (17 –26). More recently, hypomorphic mutations in two additional genes (MKS1 and CEP290) were reported to be associated with BBS, representing BBS13 and BBS14, respectively (27). Null mutations in MKS1 and CEP290 cause Meckel-Gruber syndrome, a related but more severe disorder (28 –30). Seven of the known BBS proteins (BBS1-2, BBS4-5, BBS7-9) have been shown to form a stable complex, the BBSome, and this complex is proposed to function in vesicle trafficking to the ciliary membrane through a Rab8-mediated mechanism (31). Consistent with this, several G-protein–coupled receptors failed to localize to neuronal cilia in Bbs2 and Bbs4 null brains (32). In addition, we have recently demonstrated that one component of the BBSome, BBS1, directly interacts with the leptin receptor and that leptin signaling is attenuated in BBS gene knockout mice, implicating BBS function in a broad range of membrane receptor signaling (33).
Three of the remaining BBS proteins (BBS6, BBS10, and BBS12) have sequence homology to the CCT (also known as TRiC) family of group II chaperonins (17, 24, 25). CCT proteins form an ≈900 kDa hetero-oligomeric complex that mediates protein folding in an ATP-dependent manner (34, 35). The CCT complex consists of two stacked rings, each of which is composed of radially arranged eight subunits (CCT1–8). Although mutations in BBS6, BBS10, and BBS12 account for ≈30% of the mutational load in BBS, the roles of these proteins have not been well characterized. More specifically, it is unknown (i) whether these chaperonin-like BBS proteins interact with each other and form a multisubunit complex or function individually in a common, linear pathway, (ii) whether they indeed have molecular chaperone function, and (iii) if so, which proteins/processes are regulated by these proteins. In addition, although chaperonin-like BBS proteins are not components of the BBSome, the phenotypes of Bbs6 null mice and human patients with mutations in BBS6, BBS10, or BBS12 genes are similar to mice and human patients with mutations in BBSome components (6, 8–10). The overlap in phenotypes implies that the functions of chaperonin-like BBS proteins are closely related to those of other BBS proteins. However, the details of the functional relationship and mechanism of action are unclear.
Here we address the functional relationship among chaperonin-like BBS proteins and BBSome proteins. We describe a complex composed of three chaperonin-like BBS proteins and six CCT chaperonins. This complex associates with a subset of the BBSome subunits and mediates BBSome assembly. We also explore the role of CCT with respect to BBS.
Results
Identification of the BBS/CCT Complex.
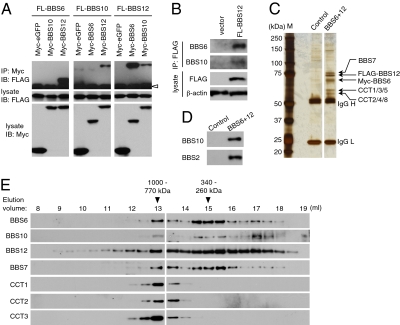
We tested the hypothesis that three chaperonin-like BBS proteins interact with each other and form a multisubunit complex. In transient transfection and coimmunoprecipitation (IP) assays, we found that BBS12 interacts with BBS6 and BBS10 (Fig. 1A). Myc-tagged green fluorescent protein (GFP) was used as a negative control and did not interact with these proteins. To test whether these interactions occur at physiological conditions, we transfected low levels of FLAG-BBS12 and probed its association with endogenous BBS6 and BBS10. As shown in Fig. 1B , endogenous BBS6 and BBS10 proteins were coimmunoprecipitated with BBS12, indicating that BBS12 associates with BBS6 and BBS10 in vivo.
Fig. 1.
Identification of the BBS/CCT complex. (A) Interactions between BBS6, BBS10, and BBS12. Myc- or FLAG-tagged BBS6, BBS10, and BBS12 expression constructs were transfected into HEK293T cells as indicated and lysates were subjected to co-IP assay. (Middle and Bottom) Expression of each component in the lysate. (Top) Amounts of FLAG-tagged proteins coimmunoprecipitated by anti-Myc antibody. IP, immunoprecipitation; IB, immunoblotting. Open arrowhead indicates IgG heavy chain. (B) BBS6, BBS10, and BBS12 associate in vivo. HEK293T cells were transfected with 1 μg of FLAG-BBS12 in one 10-cm dish and immunoprecipitated with anti-FLAG antibody. Co-IP of endogenous BBS6 and BBS10 was probed by Western blotting using antibodies against BBS6 and BBS10. β-Actin was used for normalization of the input. (C) Purification of BBS6 and BBS12 containing protein complexes. Proteins from Myc-BBS6– and FLAG-BBS12–expressing cells (BBS6+12) were purified by sequential affinity purification and analyzed by SDS/PAGE and silver staining. Parental cells were used as a control. Size markers (M) are shown in the left and IgG heavy chain (H) and light chain (L) are marked. (D) Copurification of BBS10 and BBS2 as minor interacting proteins of the BBS/CCT complex. Proteins isolated by sequential purification were resolved in SDS/PAGE and immunoblotted for BBS10 and BBS2. (E) Chaperonin-like BBS proteins form a multisubunit complex with CCT chaperonins. Proteins partially purified by anti-FLAG affinity gel were fractionated by Superose-6 size exclusion chromatography. Elution volumes and approximate molecular weights of fractions where BBS proteins were found were denoted at Top. Void volume was ∼7.6 mL. Fractions were immunoblotted for Myc (BBS6), FLAG (BBS12), BBS10, BBS7, CCT1, CCT2, and CCT3.
Next, we sought to isolate chaperonin-like BBS protein complexes with their associated proteins. To this end, we generated a stable cell line expressing both Myc-BBS6 and FLAG-BBS12. Protein complexes containing both BBS6 and BBS12 were isolated from this stable cell line by sequential purification using anti-FLAG and anti-Myc affinity gels. Isolated proteins were analyzed by SDS/PAGE and mass spectrometry (MS) (Fig. 1C). In addition to BBS6 and BBS12, three additional protein bands were copurified. Analysis by MS revealed that two of the bands contain six CCT chaperonin proteins (CCT1, CCT2, CCT3, CCT4, CCT5, and CCT8), and that the remaining band is BBS7 (Table S1). Although we could not detect BBS10 in our MS analysis, its presence in this complex was confirmed by immunoblotting (Fig. 1D), suggesting that BBS10 exists at a substoichiometric ratio or only transiently associates with the complex.
To determine the supramolecular organization of these proteins, we loaded partially purified BBS12-containing protein complexes onto a Superose-6 column and performed size exclusion chromatography (Fig. 1E). BBS12 was found in a wide range of fractions presumably because of overexpression. However, copurified BBS6, BBS7, BBS10, and CCT proteins showed more distinct elution profiles. They were all eluted in the same fraction with an estimated molecular weight of 770–1,000 kDa. This observation suggests that the three chaperonin-like BBS proteins, BBS7, and six CCT proteins form a multisubunit complex (the BBS/CCT complex). BBS6, BBS10, and BBS7 were also found in additional fractions with molecular weight of 260–340 kDa and a large portion of BBS10 was eluted as monomeric and dimeric forms. The latter pool may represent BBS10 dissociated from BBS12 after purification. Unlike BBS6 and BBS10, CCT proteins were found only in the larger protein complex. These data are consistent with the three chaperonin-like BBS proteins along with BBS7 forming a subcomplex and this subcomplex forming a larger BBS/CCT complex together with six CCT family chaperonins.
To test whether BBS6, BBS10, and BBS12 form the BBS/CCT complex in vivo, we performed size exclusion chromatography analysis with extracts from mouse testis and eye. Protein extracts were loaded onto a Superose-6 gel filtration column and elution profiles of BBS6 and BBS10 were analyzed by immunoblotting (Fig. S1). Consistent with the results from cultured cells, BBS6 was eluted as a complex with an estimated molecular weight of 770–1,000 kDa, and a subset of BBS10 was found in the same fractions in the testis. The majority of BBS10 was found as a monomeric form or in relatively small complexes with molecular weight ranging from 75 to 110 kDa. These data are in good agreement with the data obtained from the cell line (Fig. 1E) and indicate that the BBS/CCT complex indeed exists in vivo. In contrast, the 260–340 kDa subcomplex containing BBS6, BBS7, BBS10, and BBS12 was not detected in the testis or eye extracts, suggesting that only small amounts of such subcomplexes exist in vivo and/or that the equilibrium was biased toward the subcomplex because of over-expression of BBS6 and BBS12 in the cell line. Because CCT chaperonins also constitute the CCT complex, which does not contain chaperonin-like BBS proteins and is likely to be more abundant than the BBS/CCT complex, the elution profiles of CCT1, CCT2, and CCT3 in Fig. S1 are likely to reflect more of the CCT complex than the BBS/CCT complex. However, the elution profile of the BBS/CCT complex (e.g., BBS6 and BBS12-associated CCT chaperonins in Fig. 1E) is identical to that of CCT, implying that the BBS/CCT complex may have a similar structure to that of CCT, a two-tier ring configuration (35, 36).
Requirement of Chaperonin-Like BBS Proteins for BBSome Assembly.
BBS7 was previously found as a subunit of the BBSome. Interestingly, BBS7 was also isolated as a component of the BBS/CCT complex (Fig. 1 C and E). This observation raised the possibility that chaperonin-like BBS proteins may interact with some of the BBSome subunits and assist in their folding and/or in BBSome assembly. Indeed, when BBS7 was tandem affinity purified, components of the BBS/CCT complex were copurified in addition to BBSome subunits (Fig. S2A). In contrast, tandem affinity purification of BBS5, also a component of the BBSome, resulted in isolation of only BBSome subunits. These findings are consistent with the idea that the BBS/CCT complex selectively and transiently associates with BBS7 and dissociates before complete assembly of the BBSome. It also should be noted that like BBS10, the presence of BBS2 in the BBS/CCT complex was demonstrated by immunoblotting (Fig. 1D). To systematically investigate the interactions between chaperonin-like BBS proteins and BBSome subunits, we transfected BBSome subunits (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9) with each of the chaperonin-like BBS proteins and conducted co-IP experiments. In these analyses, we found a physical interaction between BBS2 and BBS6 (Fig. S2B). We also found that BBS10 interacts with BBS7 and BBS9 and that BBS12 interacts with BBS1, BBS2, BBS4, BBS7, and BBS9 (Fig. S2 C and D). These results indicate that chaperonin-like BBS proteins interact with a subset of the BBSome subunits.
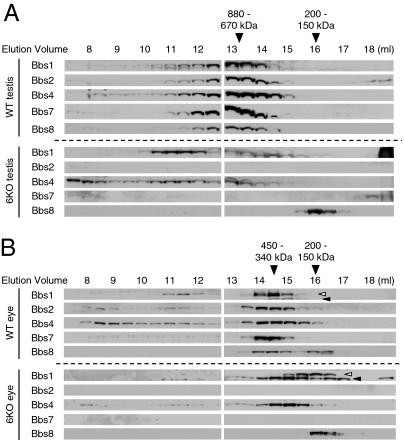
To test whether chaperonin-like BBS proteins are required for BBSome assembly, we investigated the BBSome assembly in Bbs6 null mouse tissues (9). Mouse testis and eye extracts were applied to a Superose-6 gel filtration column, and elution profiles of the BBSome subunits were examined (Fig. 2). In wild-type tissues, all BBSome subunits examined (Bbs1, Bbs2, Bbs4, Bbs7, and Bbs8) were eluted with the same elution profiles, indicating that they are associated with each other. However, in Bbs6 null testis and eye, Bbs1, Bbs4, and Bbs8 were eluted with distinct elution profiles. In the testis, the elution profiles of Bbs1 and Bbs4 were more widely spread over many fractions. A subset of Bbs1 and Bbs4 appeared to be aggregated or “misassociated” with unknown proteins. The elution of Bbs8 was shifted to later fractions (smaller molecular weights). In the Bbs6 null eye, elution profiles of Bbs1, Bbs4, and Bbs8 were all shifted to smaller but nonoverlapping molecular weight fractions. These data demonstrate that Bbs1, Bbs4, and Bbs8 are not associated with each other in Bbs6 deficient cells. In addition, Bbs2 and Bbs7 protein levels were severely reduced in Bbs6 null testis and eye (Fig. 2 and Fig. S3A). Although the amounts of Bbs1 and Bbs4 were not as severely reduced as Bbs2 and Bbs7, these proteins also showed a notable reduction in Bbs6 null eye and testis (Fig. S3A). To investigate whether BBS gene expression is decreased in Bbs6 null mice, we compared the mRNA levels of BBS genes (Bbs1, Bbs2, Bbs4, and Bbs7) between wild-type and Bbs6 null eye and testis. Quantitative PCR results indicated that there are no significant differences in mRNA levels of BBS genes in Bbs6 null mice (Fig. S3B). These data show that Bbs6 is required for the stability of Bbs2 and Bbs7, possibly by mediating their association with other BBSome subunits. Combined, our data indicate that Bbs6 is required for BBSome assembly and stable expression of Bbs2 and Bbs7.
Fig. 2.
BBSome is not formed in Bbs6 null mouse tissues. (A) Wild-type (WT) and Bbs6 null (6KO) mouse testis extracts were fractionated by size exclusion chromatography. Elution fractions were subjected to SDS/PAGE and immunoblotting. Membranes were probed with antibodies against Bbs1, Bbs2, Bbs4, Bbs7, and Bbs8. (B) Extracts from wild-type and Bbs6 null eye were analyzed as in A. Open arrowhead indicates Bbs1; closed arrowhead indicates a cross-reacting protein. It should be noted that Bbs1 blot from Bbs6 null eye was exposed longer than that of wild-type.
Intriguingly, the elution profiles of the BBSome subunits were significantly different between the testis and the eye even in wild-type mice (compare Fig. 2 A and B), whereas that of Bbs6 and CCT chaperonins was identical in these tissues (Fig. S1). The BBSome in the testis (670–880 kDa) was notably larger than the BBSome in the eye (340–450 kDa), where the estimated molecular weight of the BBSome is in good agreement with the calculated molecular weight of the BBSome (∼470 kDa). In addition, the elution profiles of Bbs1 and Bbs4 from Bbs6 null eye and testis were also different. These observations suggest that the conformation and/or composition of the BBSome vary in different tissues and that the fates of the BBSome subunits in the absence of Bbs6 also differ depending on the cell type.
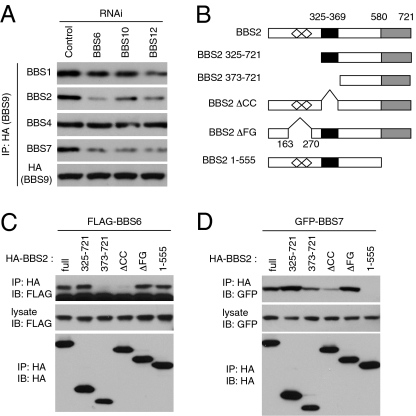
The requirement of BBS10 and BBS12 for BBSome assembly was determined in HEK293T cells depleted of these proteins using RNA interference (RNAi). The efficiency and specificity of gene knockdown was confirmed by quantitative PCR and immunoblotting (Fig. S4). In these and control cells, HA-BBS9 was transfected and its association with other BBSome subunits was analyzed by IP (Fig. 3A). In accordance with the previous study (31), BBSome subunits BBS1, BBS2, BBS4, and BBS7 were coprecipitated with BBS9 in control cells. In contrast, association of BBS2 and BBS7 with BBS9 was greatly reduced in BBS6-, BBS10-, or BBS12-depleted cells. Association of BBS1 with BBS9 was moderately or only slightly reduced, and interaction between BBS4 and BBS9 was not affected in these cells.
Fig. 3.
Chaperonin-like BBS proteins are required for BBSome assembly. (A) BBSome assembly in BBS6-, BBS10-, and BBS12-depleted cells. HA-BBS9 was transfected into control, BBS6-, BBS10-, and BBS12-depleted HEK293T cells, and BBSome assembly was assessed by measuring BBSome subunits associated with BBS9. (B) BBS2 deletion mutant constructs used to map BBS6- and BBS7-interacting domains. Open diamonds represent FG-GAP motifs (FG); black box indicates the coiled-coil (CC) domain; and gray box indicates the C-terminal α-helix–rich domain. Numbers represent amino acid residues. (C) Coiled-coil domain in BBS2 interacts with BBS6. HA-tagged BBS2 deletion mutants were cotransfected with FLAG-BBS6 into HEK293T cells, and lysates were immunoprecipitated with anti-HA antibodies. (D) C-terminal domain in BBS2 interacts with BBS7, but BBS6-interacting domain is required for efficient binding with BBS7. GFP-BBS7 was cotransfected with BBS2 deletion mutants.
Interaction domain–mapping experiments revealed that BBS2 interacts with BBS6 through its central coiled-coil domain (Fig. 3 B and C), whereas the C-terminal α-helix–rich domain of BBS2 is essential for interaction with BBS7 (Fig. 3 B and D). Consistent with the requirement of BBS6 for BBS2-BBS7 interaction, BBS2 coiled-coil domain deletion mutants, which are not able to interact with BBS6, showed a markedly reduced ability to interact with BBS7 (Fig. 3D). Combined, these data demonstrate that BBS6, BBS10, and BBS12 are required for complete assembly of the BBSome.
CCT Chaperonins Localize to Centrosomes and CCT Activity Is Essential for BBSome Assembly.
As many BBS proteins including BBS6, BBS10, and BBS12 have been found in centrosomes (21, 31, 37–39) (Fig. S5A), we examined whether CCT proteins also localize to centrosomes. With indirect immunofluorescence microscopy, all CCT proteins tested (CCT1, CCT2, CCT4, CCT5 and CCT8) showed abundant expression in the cytoplasm (Fig. S5B). Of note, with the exception of CCT2, we detected enrichment of each CCT protein in the centrosome. These data indicate that a subset of CCT proteins localize to the centrosome. Interestingly, BBS7 and CCT1, but not other BBS or CCT proteins, localize specifically to the “mother” centriole in ciliated cells, suggesting that BBS7 and CCT1 may have independent roles in the basal body (Fig. S6). Immunoblotting results indicated that each of the CCT antibodies recognizes a single protein with predicted molecular weight in ARPE-19 cells (Fig. S5C).
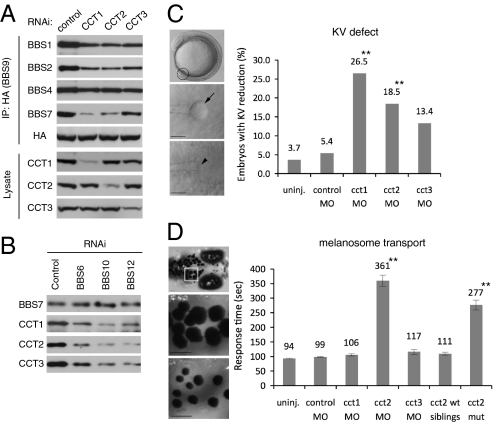
Next, we examined whether CCT chaperonins are required for BBSome assembly. To this end, we blocked the expression of CCT1, CCT2, and CCT3 by transfecting siRNAs into HEK293T cells and measured BBSome assembly as described above. Compared to control cells, association of BBS2 and BBS7 with BBS9 was markedly decreased in CCT1-, CCT2-, and CCT3-depleted cells (Fig. 4A). Similar to the results from cells depleted of chaperonin-like BBS proteins, BBS9-associated BBS1 and BBS4 protein levels were moderately or only slightly reduced in CCT1-, CCT2-, and CCT3-depleted cells. BBSome assembly was relatively weakly affected in CCT3-depleted cells, presumably because of less effective knockdown of CCT3 expression compared with CCT1 and CCT2.
Fig. 4.
Requirement of CCT activity for BBSome assembly, (A) CCT chaperonins are required for BBSome assembly. HA-BBS9 was cotransfected with siRNAs against CCT1, CCT2, CCT3, or control in HEK293T cells, and BBS9-associated BBSome subunits were probed by immunoprecipitation followed by immunoblotting. (B) Chaperonin-like BBS proteins are required for the interaction between BBS7 and CCT chaperonins. Expression of BBS6, BBS10, and BBS12 was blocked by RNAi in HEK293T cells, and endogenous BBS7 was immunoprecipitated in control, BBS6-, BBS10-, and BBS12-depleted cells. Association of CCT chaperonins with BBS7 was measured by immunoblotting. (C) MO-mediated knockdown of CCT chaperonins results in Kupffer’s vesicle (KV) defects in zebrafish. Representative images of zebrafish embryos and KVs are shown at Left. (Top) Lateral view of a 10-somite stage embryo with the location of the KV circled, dorsal views; (Middle and Bottom) of a normal KV (arrow) and a reduced KV (arrowhead) at the 10-somite stage. Embryos were injected with 15 ng of indicated MOs, except for cct1, for which a 5-ng dose was used because of severe developmental defects and lethality. The percentage of embryos with reduced or absent KVs are represented for each MO treatment. Sample size and P values are in Table S2. **P < 0.01 determined by Fisher’s exact test compared with control MO-injected group. (D) Retrograde melanosome transport is delayed in cct2 morphants and genetic mutants. Melanosome transport is assessed by monitoring melanosome trafficking in the head region of zebrafish (Top). Below are representative images of the boxed area with dispersed melanosomes of dark-adapted zebrafish before epinephrine treatment (Middle) and contracted melanosomes after epinephrine induced transport (Bottom). Rate of melanosome transport to the endpoint (perinuclear distribution) is noted in seconds on y-axis. Data are represented as mean ± SEM. **P < 0.01, determined by one-way ANOVA and Tukey test. (Scale bar, 100 μm.)
Recent proteomic interaction studies determined a group of proteins as potential CCT substrates, and many of them contained β-propeller domains (40 –42). Because BBS7 contains a β-propeller domain and is copurified with the BBS/CCT complex, we tested whether BBS7 associates with CCT independently of chaperonin-like BBS proteins. Indeed, CCT chaperonins were coprecipitated with endogenous BBS7 (Fig. 4B). However, in BBS6-, BBS10-, and BBS12-depleted cells, CCT chaperonins associated with BBS7 were markedly decreased. These findings indicate that CCT activity is essential for BBSome assembly and that chaperonin-like BBS proteins mediate the association of BBS7 with CCT chaperonins.
Having established a role for CCT1, CCT2 and CCT3 in BBSome assembly in cultured cells, we next examined the in vivo roles of these proteins in zebrafish. We previously demonstrated that knockdown of BBS genes results in the reduction or loss of Kupffer's vesicle (KV) and delays in melanosome transport. These phenotypes are shared among all BBS genes tested (14, 15, 31). To test whether knockdown of cct genes in zebrafish results in these BBS phenotypes, we designed anti-sense morpholino oligonucleotides (MOs) directed against the cct1, cct2 and cct3 transcription start sites. KV size at the 8–10 somite stage (10–12 h postfertilization) and melanosome transport at 5 days postfertilization (dpf) was assessed in MO-injected embryos (morphants). Knockdown of cct1 and cct2 generated KV defects and cct2 morphants displayed melanosome transport delay (Fig. 4 C and D and Table S2). Specificity of gene knockdown was confirmed with a second cct2 MO (splice-block MO) that also generated KV defects and melanosome transport delay (Table S2). Furthermore, we observed melanosome transport delay in homozygous genetic mutants for cct2 (43). Because of severe defects induced by cct1 knockdown, a low MO dose was used, and cct1 gene knockdown may not be efficient at day 5 to disrupt melanosome transport. cct3 Morphant phenotypes resembled that of cct3 genetic mutant (Fig. S7) (44); yet, cct3 morphants did not display statistically significant KV or transport defects. Of note, knockdown of cct1, cct2, and cct3 leads to additional, overlapping phenotypes in the zebrafish embryos, including brain abnormalities, small eyes, and cardiac edema (Fig. S7) which are also shared by cct genetic mutants (43, 44). Together these results indicate that at least a subset of cct proteins interact with the BBS pathway in vivo.
Disease-Causing Mutations Found in BBS6, BBS10, and BBS12 Disrupt Interactions Among Their Encoded Proteins.
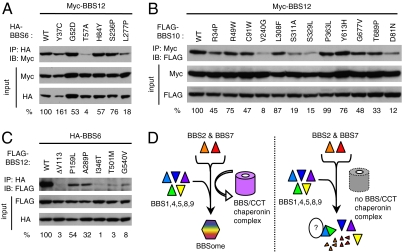
We investigated the impact of human mutations found in BBS6, BBS10, and BBS12 genes. We generated missense variants of BBS6, BBS10, and BBS12 (Fig. S8A) and examined their ability to interact with other BBS proteins. Of the mutations we tested in BBS6, the L277P mutation moderately affected interaction of BBS6 with BBS2 (decreased to 42% of wild-type; Fig. S8B). The T57A and L277P mutations greatly reduced the ability of BBS6 to interact with BBS12 (4% and 18% of wild-type, respectively; Fig. 5A). Many BBS10 variants tested had moderately reduced ability to interact with BBS7 and BBS9 compared with wild-type (Fig. S8C). However, V240G, S311A, and S329L mutations in BBS10 severely reduced the interaction of BBS10 with BBS12 (8%, 19%, and 15% of wild-type, respectively: Fig. 5B). Although mutations at amino acid D81 have not been reported in human BBS patients, this residue is part of the ATP binding pocket of CCT family chaperonins and is partly conserved in BBS10 (24, 36). In addition, mutations in this residue in GroEL group I chaperonin convert it to a dominant negative form, which is able to bind nonnative forms of substrates but not release them (45). Thus, we introduced a point mutation in this residue of BBS10 and tested its effect on protein–protein interactions. The D81N mutation greatly decreased all interactions of BBS10 with BBS7, BBS9, and BBS12 (Fig. 5B and Fig. S8C), indicating that the D81 residue may be required for overall BBS10 protein conformation rather than required for ATP binding and substrate folding. In BBS12, all mutations tested either abolished or significantly reduced the interaction between BBS6 and BBS12 (Fig. 5C). Most BBS12 mutant variants (except P159L) also showed significantly decreased interaction with BBS7 (Fig. S8D). The interaction between BBS10 and BBS12 was not affected by these mutations. In general, disease-causing mutations in BBS6, BBS10, and BBS12 have a more severe impact on interactions among chaperonin-like BBS proteins, whereas interactions with BBSome subunits are relatively mildly affected. These results suggest that many human mutations found in BBS6, BBS10, and BBS12 genes disrupt physical interactions among them and that formation of the multimeric complex of BBS6, BBS10, and BBS12 is critical for their function.
Fig. 5.
Many disease-causing missense mutations found in BBS6, BBS10, and BBS12 disrupt interactions among these proteins. (A) Interactions of BBS6 missense mutants with BBS12. HEK293T cells were transfected with Myc-BBS12 together with HA-BBS6 variants and lysates were subject to coimmunoprecipitation (IP) and immunoblotting (IB). Numbers at bottom represent ratio of coprecipitated proteins compared with wild-type protein after normalization with input. (B) Interactions of BBS10 missense mutants with BBS12. (C) Interactions of BBS12 missense mutants with BBS6. (D) Model for BBS/CCT complex function. BBS/CCT complex initially binds to BBS7 and potentially to BBS2, and mediates their association with other BBSome subunits (BBS1,4,5,8,9) to assemble BBSome. When chaperonin-like BBS genes are inactivated, at least two BBSome subunits (BBS2 and BBS7) are degraded, and the remaining BBSome subunits exist in monomeric form or aggregates with unidentified proteins.
Discussion
The BBSome is an ≈470 kDa protein complex composed of seven BBS proteins (31). The BBSome has been proposed as the functional unit that mediates vesicle trafficking to the ciliary membrane. Loss of any proteins in this complex abolishes the functionality of the BBSome, as shown by the similar phenotypes of BBS patients and BBS animal models with inactivation of different BBS genes. Although BBS6, BBS10, and BBS12 are not components of the BBSome, mutations in these genes also lead to the same phenotype as loss of BBSome subunits. However, the functional relationship between these two groups of BBS genes has been elusive. In this study, we demonstrate that chaperonin-like BBS proteins (BBS6, BBS10, and BBS12) are required for BBSome assembly (Fig. 5D). BBS6, BBS10, and BBS12 form a higher-order complex in conjunction with six CCT chaperonin proteins. These BBS proteins also interact with a subset of BBSome subunits and mediate their association. In murine tissues, inactivation of Bbs6 causes reduction of Bbs2 and Bbs7 and failure of association of the remaining BBSome subunits. Depletion of BBS6, BBS10, and BBS12 in cultured cells also leads to failure of BBSome assembly. Our study indicates that the lack of a functional BBSome in the absence of chaperonin-like BBS proteins is the cause of BBS in patients with mutations in these genes.
Our study also demonstrates previously unrecognized in vivo roles of CCT in ciliary function. First, six CCT proteins form a complex with chaperonin-like BBS proteins, and this complex mediates BBSome assembly. Second, although CCT proteins are relatively abundant cytosolic proteins, a subpopulation of at least four CCT proteins localizes to the centrosome. Indeed, centrosomal localization of CCT chaperonins has been previously reported in both immunological and biochemical studies (46, 47). Finally, knockdown of cct1 and cct2 in zebrafish leads to BBS-like phenotypes. Combined, these findings indicate that CCT chaperonins are essential for BBSome assembly and normal ciliary function.
Although BBS6, BBS10, and BBS12 form a complex with CCT chaperonins, they do not appear to be conventional chaperonins that promote protein folding with ATPase activity. For example, the ATP binding motif, which is essential to induce changes in protein folding and highly conserved in all CCT proteins (36), is either not conserved (BBS6 and BBS12) or only partially conserved (BBS10) (17, 24, 25). Mutations in this motif in BBS10 do not trap BBS7 in the BBS10-associated state. These findings suggest that BBS6, BBS10, and BBS12 are not likely to have ATPase activity. Instead, BBS6, BBS10, and BBS12 mediate the association of BBS7 with CCT chaperonins, thus acting as substrate binding subunits of the chaperone complex. The protein folding activity is likely to be accomplished by CCT proteins.
Interestingly, BBS6, BBS10, and BBS12 are vertebrate-specific BBS genes, and their homologs are not found in invertebrates. In contrast, BBSome constituting BBS proteins are conserved in all ciliated organisms including Caenorhabditis elegans and Chlamydomonas reinhardtii. BBS6, BBS10, and BBS12 might have evolved in vertebrate to mediate the association of CCT chaperonins with β-propeller domain containing BBSome subunits such as BBS2 and BBS7. It is an interesting question as to whether, in invertebrates, CCT chaperonins do not need BBS6, BBS10, and BBS12 to interact with BBSome subunits or the BBSome assembles without molecular chaperone functions. In addition, the composition of the BBSome appears to vary depending on tissues. How the BBSome is assembled and whether there are tissue-specific variations in BBSome with functional ramifications are important remaining questions.
Materials and Methods
HEK293T cells were cultured in DMEM (Invitrogen) with 10% FBS (Invitrogen). Small interfering RNAs (siRNAs) were purchased from Dharmacon. Antibodies against BBS1, BBS2, BBS4, and BBS7 were described previously (31). Sources of other antibodies are in SI Methods. For protein-protein interaction studies, cells were lysed in the lysis buffer (20 mM hepes pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100) supplemented with Complete Protease Inhibitor Mixture (Roche Applied Science). Lysates were subjected to immunoprecipitation with indicated antibodies for 2 h or overnight at 4°C. SDS/PAGE and immunoblotting was performed following standard protocols. Size exclusion chromatography was performed using Superose-6 10/300 GL column (GE Healthcare). MO injection and analysis of KV and melanosome transport in zebrafish was performed as previously described (14, 15). For details, please see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Lokesh Gakhar and Yalan Li for assistance in size exclusion chromatography and mass spectrometry, respectively. We also thank Kevin Bugge, Charles C. Searby, and Gun-Hee Kim for their technical assistance, and Dr. Maxence V. Nachury for discussion and critical reading of the manuscript. This work was supported by US National Institutes of Health grants REY110298 and REY017168 (to V.C.S.) and R01CA112369 (to D.C.S.) and by an Iowa Cardiovascular Center Institutional Research Fellowship (to L.M.B). V.C.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910268107/DCSupplemental.
References
- 1.Singla V, Reiter JF. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 2.Marshall WF, Nonaka S. Cilia: Tuning in to the cell's antenna. Curr Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 4.Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- 5.Blacque OE, Leroux MR. Bardet-Biedl syndrome: An emerging pathomechanism of intracellular transport. Cell Mol Life Sci. 2006;63:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore SJ, et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: A 22-year prospective, population-based, cohort study. Am J Med Genet A. 2005;132:352–360. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheffield VC, Heon E, Stone EM, Carmi R. In: Inborn Errors of Development: The Molecular Basis of Clinical Disorders of Morphogenesis. Epstein CJ, Erickson RP, Wynshaw-Boris A, editors. New York: Oxford University Press; 2004. pp. 1044–1049. [Google Scholar]
- 8.Mykytyn K, et al. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA. 2004;101:8664–8669. doi: 10.1073/pnas.0402354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fath MA, et al. Mkks-null mice have a phenotype resembling Bardet-Biedl syndrome. Hum Mol Genet. 2005;14:1109–1118. doi: 10.1093/hmg/ddi123. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura DY, et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc Natl Acad Sci USA. 2004;101:16588–16593. doi: 10.1073/pnas.0405496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah AS, et al. Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0712327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis RE, et al. A knockin mouse model of the Bardet-Biedl syndrome 1 M390R mutation has cilia defects, ventriculomegaly, retinopathy, and obesity. Proc Natl Acad Sci USA. 2007;104:19422–19427. doi: 10.1073/pnas.0708571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulaga HM, et al. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- 14.Yen HJ, et al. Bardet-Biedl syndrome genes are important in retrograde intracellular trafficking and Kupffer’s vesicle cilia function. Hum Mol Genet. 2006;15:667–677. doi: 10.1093/hmg/ddi468. [DOI] [PubMed] [Google Scholar]
- 15.Tayeh MK, et al. Genetic interaction between Bardet-Biedl syndrome genes and implications for limb patterning. Hum Mol Genet. 2008;17:1956–1967. doi: 10.1093/hmg/ddn093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 17.Katsanis N, et al. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 18.Mykytyn K, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura DY, et al. Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2) Hum Mol Genet. 2001;10:865–874. doi: 10.1093/hmg/10.8.865. [DOI] [PubMed] [Google Scholar]
- 20.Mykytyn K, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- 21.Ansley SJ, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 22.Chiang AP, et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimura DY, et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoetzel C, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- 25.Stoetzel C, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slavotinek AM, et al. Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet. 2000;26:15–16. doi: 10.1038/79116. [DOI] [PubMed] [Google Scholar]
- 27.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 28.Kyttälä M, et al. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- 29.Baala L, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank V, et al. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum Mutat. 2008;29:45–52. doi: 10.1002/humu.20614. [DOI] [PubMed] [Google Scholar]
- 31.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 32.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo S, et al. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1). Multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biochem. 1995;230:3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. [DOI] [PubMed] [Google Scholar]
- 35.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ditzel L, et al. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 37.Kim JC, et al. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci. 2005;118:1007–1020. doi: 10.1242/jcs.01676. [DOI] [PubMed] [Google Scholar]
- 38.Hirayama S, et al. MKKS is a centrosome-shuttling protein degraded by disease-causing mutations via CHIP-mediated ubiquitination. Mol Biol Cell. 2008;19:899–911. doi: 10.1091/mbc.E07-07-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marion V, et al. Transient ciliogenesis involving Bardet-Biedl syndrome proteins is a fundamental characteristic of adipogenic differentiation. Proc Natl Acad Sci USA. 2009;106:1820–1825. doi: 10.1073/pnas.0812518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 41.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 42.Valpuesta JM, Martín-Benito J, Gómez-Puertas P, Carrascosa JL, Willison KR. Structure and function of a protein folding machine: The eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529:11–16. doi: 10.1016/s0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- 43.Amsterdam A, et al. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda N, Mishina M. Identification of chaperonin CCT gamma subunit as a determinant of retinotectal development by whole-genome subtraction cloning from zebrafish no tectal neuron mutant. Development. 2004;131:1913–1925. doi: 10.1242/dev.01085. [DOI] [PubMed] [Google Scholar]
- 45.Farr GW, Scharl EC, Schumacher RJ, Sondek S, Horwich AL. Chaperonin-mediated folding in the eukaryotic cytosol proceeds through rounds of release of native and nonnative forms. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 46.Liu Q, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown CR, Doxsey SJ, Hong-Brown LQ, Martin RL, Welch WJ. Molecular chaperones and the centrosome. A role for TCP-1 in microtubule nucleation. J Biol Chem. 1996;271:824–832. doi: 10.1074/jbc.271.2.824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.