Abstract
The pleckstrin homology domain leucine-rich repeat protein phosphatase 1 (PHLPP1) differentially attenuates Akt, PKC, and ERK1/2 signaling, thereby controlling the duration and amplitude of responses evoked by these kinases. PHLPP1 is expressed in the mammalian central clock, the suprachiasmatic nucleus, where it oscillates in a circadian fashion. To explore the role of PHLPP1 in vivo, we have generated mice with a targeted deletion of the PHLPP1 gene. Here we show that PHLPP1-null mice, although displaying normal circadian rhythmicity, have a drastically impaired capacity to stabilize the circadian period after light-induced resetting, producing a large phase shift after light resetting. Our findings reveal that PHLPP1 exerts a previously unappreciated role in circadian control, governing the consolidation of circadian periodicity after resetting.
Keywords: Per, period, after-effect, suprachiasmatic nucleus, phase shift
Essential properties of the mammalian circadian clock are a self-sustained oscillation under constant conditions and the dynamic resetting induced by light. These properties of the clock are necessary for humans and other mammals where they control cyclical activities as disparate as the sleep/wake cycle, metabolism, and cardiac function (1 –3). The fine tuning of the endogenous clock by exogenous cues (zeitgebers) can be especially apparent when one travels across time zones or is transferred to a night shift working schedule (4). Circadian rhythms in mammals are driven by the central pacemaker, which is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (5). Light administration during the subjective night activates neurons of the retino-hypothalamic tract that target the SCN, and thereby resets behavioral rhythmicity. If mice are exposed to a light stimulus in the early night the clock delays, whereas it advances if light is given at late night (5, 6), underscoring the adaptability of the clock to changing lighting environment. Although there have been remarkable advances in deciphering the molecular organization of the circadian clock (1, 7), understanding of the molecular pathways involved in light-induced clock resetting is still incomplete (5).
PHLPP1 and PHLPP2 (pleckstrin homology domain leucine-rich repeats protein phosphatases 1 and 2) are Ser/Thr protein phosphatases that have been implicated in the regulation of various signaling pathways. These phosphatases negatively regulate Akt and protein kinase C (PKC) by direct dephosphorylation (8 –10). In addition, a splice variant isoform of PHLPP1 (PHLPP1β, also known as suprachiasmatic nucleus circadian oscillatory protein, SCOP, see ref. 11) has been implicated in cognitive processes and its degradation appears to be required for long-term memory formation (12). PHLPP1 is thought to be involved in long-term memory formation by inhibiting ERK1/2 via direct binding to and inhibition of K-Ras, a small GTPase operating upstream of ERK1/2 (13). Interestingly, PHLPP1 (SCOP) is expressed in a circadian manner in the SCN, peaking in the subjective night (11). Its role in circadian function, however, remains obscure.
Here, we report the generation of PHLPP1-deficient mice by homologous recombination. These PHLPP1−/− mice display a delayed shortening of circadian period length (tau) after light-induced phase shift. The delayed shortenings result in a large phase shift in PHLPP1−/− mice following light resetting tasks. Considering the previously published role of PHLPP1 in cognitive function (12), the results presented here implicate PHLPP1 in SCN neuronal plasticity, where it appears to fine tune the clock, adapting it specifically to changes in external cues. Our results indicate that the phosphatase PHLPP1 plays a critical role in the consolidation of circadian periodicity after resetting.
Results
PHLPP1 −/− Mice Show a Normal Free-Running tau.
To elucidate the physiological functions of PHLPP1, mice were generated in which the PHLPP1 gene was ablated by homologous recombination (Fig. 1 A–C). The mutant mice show no detectable levels of the PHLPP1 mRNA and protein (Fig. 1 D and E). Importantly, PHLPP2 expression levels in PHLPP1 −/− versus the PHLPP1 +/+ mice were equivalent (Fig. 1F), a notion that confirmed previous PHLPP1 interference experiments (10). These animals develop normally and show no gross anatomical defects (Fig. S1 A and B). Daily activities (running wheel revolutions under light–dark cycle) do not differ between in PHLPP1 −/− and PHLPP1 +/+ mice (Fig. S1C). The circadian expression of PHLPP1 in the SCN (11) prompted us to study the circadian behavior of the mutant mice. To determine whether PHLPP1 −/− mice had normal circadian rhythmicity, we first analyzed tau under constant conditions. After entrainment on a LD 12:12 cycle for more than 3 weeks, PHLPP1 −/− and wild-type (PHLPP1 +/+) littermate mice were moved into constant darkness (DD, free running) starting at the time of lights “off,” zeitgeber time (ZT) 12. This day was defined as day 1. To measure tau, two different detection systems were used, passive (pyroelectric) infrared sensors (Fig. 2, Top) and running wheels (Fig. S1D), obtaining equivalent results. Tau length was measured from day 2 to day 21. In both systems, there was no difference in tau between genotypes (Fig. S1E) (P > 0.05 Welch’s t test).
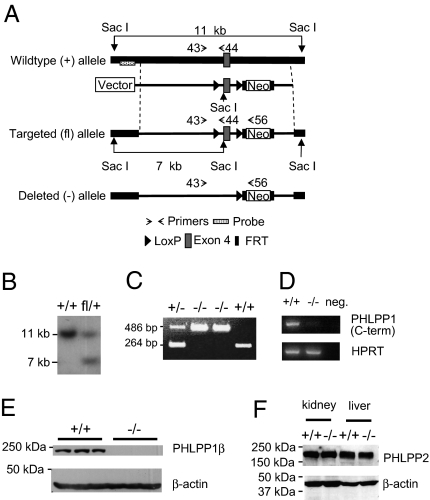
Fig. 1.
Generation of PHLPP1 −/− mice. (A) Targeting strategy to disrupt the PHLPP1 gene. Shown are the wild-type allele, the targeting vector, the targeted (floxed) allele, and the deleted allele generated by Cre-mediated recombination of the floxed allele. The DNA probe used for screening the Southern blots is marked by a dotted box, and the PCR primers used for genotyping are indicated by small arrows. (B) Southern blot analysis of SacI-digested DNA from embryonic stem cells used to generate chimeric mice. The probe labels an 11-kb fragment in the wild-type cells and a 7-kb fragment in cells in which homologous recombination has occurred. (C) Genotyping PCR results from a cross between heterozygous PHLPP1 mutant mice. The PHLPP1 WT (+) allele gives a 264-bp PCR product whereas the deleted allele (−) produces a 486-bp fragment. (D) Lack of PHLPP1 transcript expression in PHLPP1−/− mice. Total RNA isolated from PHLPP1 +/+ and PHLPP1−/− mouse brain was used as templates for the RT-PCR analysis. The PHLPP1-specific primers used were located in exons 16 and 17, respectively. The hypoxanthine phosphoribosyltransferase 1 (HPRT) primers were used as controls for RT-PCR reactions. For a negative control, the cDNA template was omitted in the reaction (the lane labeled “neg”). (E) Western blot of PHLPP1 protein in brain lysates derived from PHLPP1+/+ and PHLPP1−/− mice. β-Actin serves as a loading control. (F) Western blot of PHLPP2 protein in liver and kidney whole cell lysates derived from PHLPP1+/+ and PHLPP1−/− mice. β-Actin serves as a loading control.
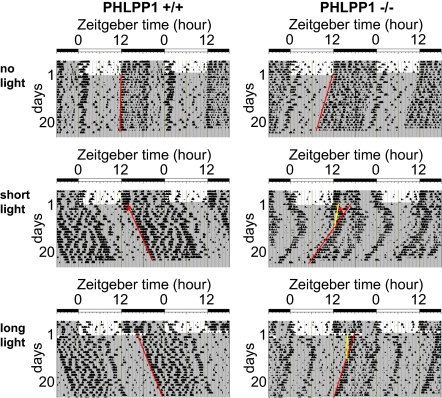
Fig. 2.
Differential light-induced phase shifts in PHLPP1-null mice. Double-plotted activity records of wild-type mice (PHLPP1+/+) and PHLPP1-deficient mice (PHLPP1−/−). Mice were entrained in 12:12 light–dark (LD) cycles and then placed in constant darkness (DD) from the light off (ZT12), on day 1. After 50 h in DD (CT14, day 3), no light pulse (no light; Top), or a 30-min light pulse (short light; Middle) was administered to PHLPP1+/+ and PHLPP1−/− mice. A separate group of mice were entrained and moved to constant darkness following 8 h of light prolongation (long light; Bottom) on the last day (day 1) of LD cycle. Locomotor activities were monitored by infrared sensors and are expressed in the histogram. Periods of darkness are indicated by gray backgrounds. Short-light pulses are denoted by asterisks. Red regression lines estimate the phase shift from days 10 to 21. Yellow regression lines estimate the phase shifts occurring several days after light pulses (short-light task = days 4–10; long-light task = days 2–10; lines are shown for phase shifted PHLPP1−/− only).
Late tau Shortening Produces a Large Phase Resetting in PHLPP1-Null Mice.
To compare the ability of PHLPP1-null mice versus wild-type littermates to adapt to light-induced resetting of the clock phase, we administered a short-light pulse to the animals during the subjective night. Mice were exposed to a 30-min light pulse of ∼200 lx during the second early subjective night (CT14) in DD (50 h following entry into DD following entrainment) (Fig. 2, Middle). Our results demonstrate that several cycles after the resetting task, the circadian period shortens spontaneously in the PHLPP1 −/− mice (Fig. 2, Middle Right). Two regression lines were made, one for the first several activity cycles after light manipulation [early phase (E), day 4 to day 10] and one for the following activity cycles [late phase (L), day 10 to day 21].
Tau values were compared from early-phase and late-phase days in each lighting task (i.e., “no light” and “short light”). When no light was administered (no-light group), the estimated phases are located around light offset (Fig. S2A). No statistical difference between the two genotypes was observed (repeated measure ANOVA; P > 0.05) (Fig. S2A, Upper). Interestingly, in the short-light exposed group, only the value from late-phase days (L) of PHLPP1 −/− mice showed a larger phase angle to light off (repeated measure ANOVA; P < 0.005) (Fig. S2A, Lower). This data suggests that the delayed phase change was solely responsible for the larger phase delays in short-light-exposed PHLPP1 −/−mice.
Behavioral rhythms from early-phase days (E) and late-phase days (L) were also tested for differences in the phase shifts. Calculations from the early-phase groups, showed no difference in the phase shift between PHLPP1 +/+ and PHLPP1 −/− mice (two-way ANOVA; P > 0.05) (Fig. 3A, Upper). However, by late-phase days, PHLPP1 −/− mice phase delayed more than wild-type animals in response to the short-light tasks (two-way ANOVA, P < 0.05). The short-light task induced a more pronounced phase delay (∼3 h) in PHLPP −/− compared to PHLPP1 +/+ mice (Fisher’s PLSD P < 0.005) (Fig. 3A, Lower). These results indicate that PHLPP1 contributes to light-induced clock resetting. During the first several days, there was no phase difference between PHLPP1 +/+ and PHLPP1 −/− mice, but PHLPP1 −/− mice ultimately showed a larger phase shift than PHLPP1 +/+ mice after 21 days of consecutive recording due exclusively to the tau shortening during the late-phase days.
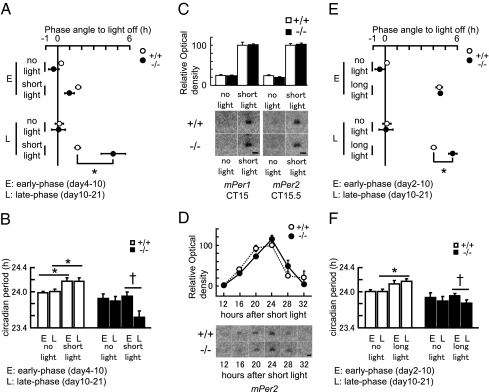
Fig. 3.
Light task effects on the phase and circadian period of PHLPP1−/− mice. (A) Quantification of short-light task-induced changes in activity rhythm phases [no light; +/+ (n = 6): −/− (n = 6), short light; +/+ (n = 8]): −/− (n = 8)] from days 4 to 10 (E, early phase) and days 10 to 21 (L, late phase). Extrapolated activity onsets of early phase and late phase are indicated by open circles (PHLPP1+/+) and filled circles (PHLPP1−/−) (mean ± SEM). *P < 0.005 (Fisher’s PLSD). (B) Circadian period during days 4–10 (E, early phase) and from days 10 to 21 (L, late phase) are indicated by open bars (PHLPP1+/+) and filled bars (PHLPP1−/−) (mean ± SEM). *P < 0.05 (Welch’s t test), †P < 0.01 (paired t test). (C) Acute expression of mPer1 and mPer2 in the SCN by short-light tasks. Upper: quantified values of mPer1 [+/+ (n = 3), −/− (n = 3)] and mPer2 [+/+ (n = 4), −/− (n = 4)]. Short-light induced RNA levels of PHLPP1+/+ are adjusted to 100. Normalized RNA levels are indicated by open bars (PHLPP1+/+) and filled bars (PHLPP1−/−) (mean ± SEM). After short-light exposure (CT14–CT14.5, 200 lx) mice were returned to DD and killed 30 min later (CT15) for mPer1 and 60 min later (CT15.5) for mPer2. (Lower) representative films (Scale bar, 0.5 mm.) (D) mPer2 expression rhythms in the SCN after short-light tasks. Upper: quantified values of mPer2 [+/+ (n = 3), −/− (n = 3)]. Peak level of RNA in PHLPP1+/+ mice is adjusted to 100. Normalized RNA levels are indicated by open circles (PHLPP1+/+) and filled circles (PHLPP1−/−) (mean ± SEM). After short-light exposure (CT14–CT14.5, 200 lx) mice were returned to DD and killed 12–32 h later from the short-light onset. (Lower) representative films (Scale bar, 0.5 mm.) (E) Quantification of long-light task-induced changes in activity rhythm phases [no light; +/+ (n = 6): −/− (n = 6), long light; +/+ (n = 14): −/− (n = 14)] days 2–10 (E, early phase) and days 10–21 (L, late phase). Extrapolated activity onsets of the early phase and late phase are indicated by open circles (PHLPP1+/+) and filled circles (PHLPP1−/−) (mean ± SEM). *P < 0.0005 (Fisher’s PLSD). (F) Circadian periods of activities days 2–10 (E, early phase) and from days 10 to 21 (L, late phase) are indicated by open bars (PHLPP1+/+) and filled bars (PHLPP1−/−) (mean ± SEM). *P < 0.05 (Welch’s t test), †P < 0.005 (paired t test).
A large period change several days after resetting induced a drastic phase change in PHLPP1 −/− mice (Fig. 2, Middle Right), even though the initial phase shift was equivalent in both genotypes (Fig. 3A, Upper). To address this further, we next estimated the tau length during early-phase days and late-phase days. The tau of PHLPP1 +/+ mice was extended by the phase delay (Fig. 3B, Left; P < 0.05 Welch’s t test). However, no differences between the early-phase days and the late-phase days in both no-light and short-light groups were observed. The unchanged tau is reflected by the lack of change in phase delay (Fig. 3A).
In contrast, there was no tau extension induced by short-light exposure in PHLPP1 −/− mice (Fig. 3B, Right). Furthermore, the tau shortened in the late-phase period (compare E and L, Fig. 3B, Right; P < 0.01 paired t test). This drastic and late tau change in PHLPP1 −/− mice is reflected in the final phase delay (Fig. 3A). Quantified tau analysis revealed a twofold effect caused by the PHLPP1 mutation. Finally, there is a loss of acute tau extension by phase delay lighting conditions but, there is a “late effect” revealed by a shortening of tau after several days in DD, which induced the large phase resetting in PHLPP1 −/− mice.
Intact Response of the SCN to Resetting Light in PHLPP1 −/− Mice.
Due to the normal initial behavioral shifts of PHLPP1 −/− mice, we analyzed light responsiveness of their SCN. The response to light of the SCN appeared to be normal in the mutant mice. Specifically, we analyzed the expression of the light-inducible clock genes, mPer1 and mPer2 (14, 15) and found the kinetics of induction and the final expression levels to be equivalent in wild-type and mutant mice. All mice were exposed to a 30-min light pulse (short light, ∼200 lx, CT14–14.5). This treatment induced a robust induction of mPer1 at CT15 in the SCN, 30 min following the light pulse. mPer2 was also induced, peaking at CT15.5, 60 min after the light pulse (Fig. 3C).
Next, we examined the circadian mPer2 expression in the SCN during the first cycle after resetting light. Mice were exposed to short light (∼200 lx) at CT14–14.5 (30 min) and sampled every 4 h from 12 h to 32 h after the beginning of light pulse. Using this approach, mPer2 expression was found to be lower at 12-h and 32-h time points and higher at 20-h and 24-h time points in both PHLPP1 +/+ and PHLPP1 −/− mice. Although PHLPP1 −/− mice showed a larger phase delay (∼3 h), there was no phase difference between PHLPP1 +/+ and PHLPP1 −/− animals (Fig. 3D; two-way ANOVA P > 0.05). The magnitude and profile of light-induced gene expression and subsequent gene expression rhythms in the SCN were equivalent in both genotypes.
Long-Light Treatment Induces Large Phase Delays in PHLPP1 −/− Mice by Changing the Period Length.
Because two distinct light tasks, short light (30 min) and long light (∼8–12 h), play differential roles in clock resetting (16 –18), we tested whether long-light tasks similarly affected the period length of PHLPP1−/− mice. Mice were exposed to 8 h of light (∼200 lx) from the last day of LD 12-h cycle (ZT12 of day 1) (Fig. 2, Lower) (16). Our results show that several cycles after the resetting task, the circadian period again shortened spontaneously in the PHLPP1 −/− mice similar to the tau shortening after the short-light task (Fig. 2, Lower Right). Two regression lines were made (early phase, day 2 to day 10, late phase, day 10 to day 21) and the values from early-phase and late-phase days were compared for each task (no light and long light). Without light exposure (no-light group) there was no statistical difference in estimated phases (repeated measure ANOVA; P > 0.05) (Fig. S2B, Upper). In the long-light exposed group, only the tau from the late-phase days (days 10–21) of PHLPP1 −/− mice behavior showed a larger phase angle to lights off (repeated measure ANOVA; P < 0.005) (Fig. S2B, Lower). This suggests that similarly to the short-light exposure, a delayed change in tau after long-light exposure produced the ultimate phase delay observed in PHLPP1 −/− animals.
The tau values from the early-phase and late-phase days were subsequently examined. During the early-phase, there was no difference in the phase shift between PHLPP1 +/+ and PHLPP1 −/− mice (two-way ANOVA; P > 0.05) (Fig. 3E, Upper), although long-light exposure delayed activity phase extensively. During the late-phase period, PHLPP1 −/− mice showed a delayed phase compared to wild-type animals (two-way ANOVA, P < 0.05). The long-light task induced a more pronounced phase delay (∼1.5 h) in PHLPP −/− mice when compared to their wild-type controls (Fisher’s PLSD, P < 0.005) (Fig. 3E, Lower). These results indicate that PHLPP1 contributes not only to short light but also to long-light-induced clock resetting.
We next examined the effect of long-light exposure on tau (Fig. 3F). The tau of PHLPP1 +/+ mice was extended by long light significantly during the late phase (Fig. 3F, Left; P < 0.05 Welch’s t test). There was no difference, however, between early-phase and late-phase tau, with or without long light in these animals (Fig. 3F, Left). In contrast, long-light exposure did not induce tau extension in PHLPP1 −/− mice (Fig. 3F, Right), similar to the effect elicited by the short-light task. In addition, the tau of PHLPP1 −/− mice in the long-light group shortened in the early-phase (compare E and L, Fig. 3F, Right; P < 0.005 paired t test). This late but drastic tau shortening is reflected in the final phase shift in PHLPP1 −/− mice (Fig. 3E).
Finally, it is noteworthy that tau length in PHLPP1 −/− mice was shorter after short-light exposure than after long-light exposure (repeated measure ANOVA, P < 0.05; Fisher’s PLSD, P < 0.05) (Fig. S3). Interestingly, this indicates that the length of the light stimulus may have an impact on the molecular determinants of tau stability.
Impairment of Shortening of Circadian Period by Phase Advancing Light in PHLPP1 −/− Mice.
Although phase delaying light extends the circadian period, phase advancing light typically shortens the circadian period (19). We next examined the circadian period after phase advance in PHLPP1 −/− mice. Mice were exposed to 4 h of light (∼200 lx) at the last day of LD 12-h cycle (ZT20 to ZT24 of day 1) (Fig. S4A). In contrast to phase delaying tasks, there was no difference in phase advance behavior between PHLPP1 −/− and PHLPP1 +/+ mice (Fig. S4B; P > 0.05 two-way ANOVA). Phase advancing tasks shorten tau in PHLPP1 +/+ animals both in early phase (day 2 to day 10) and late phase (day 10 to day 21) (Fig. S4C; P < 0.01 Welch’s t test). Interestingly, similar to phase delay experiments (Figs. 2 and 3), tau was not affected in PHLPP1 −/− mice (Fig. S4C; P > 0.05 Welch’s t test). Differences in the circadian period between early phase and late phase was not observed in PHLPP1 −/− mice (Fig. S4C; P > 0.05 Welch’s t test).
Discussion
A comprehensive understanding of the molecular pathways by which light is able to induce clock resetting is still lacking. In this study we show that the Ser-Thr phosphatase PHLPP1 is critical for the control of clock resetting. Our results indicate that light-induced acute phase shift is unchanged in PHLPP1 −/− mice, but that the tau length is drastically modified, a property that increases the final phase delay. This model shows that resetting light differentially regulates acute clock resetting, causing the subsequent period change (Fig. 4). Therefore, resetting light can directly (Fig. 4A) or indirectly (Fig. 4B) (via phase resetting) control tau. In this process, PHLPP1 appears to act as fine regulator of tau after resetting, ascribing to PHLPP1 an in vivo role unappreciated to date (Fig. 4). Importantly, PHLPP1 has been implicated in memory formation (12), via the attenuation of MAPK signaling. PHLPP1 also contributes to the duration and amplitude of responses of other kinases implicated in neuronal plasticity, including PKC and Akt (20, 21). It is tempting to speculate that the circadian phenotype of the PHLPP1-null mice described here could be parallel to the function of PHLPP1 in memory formation. This intriguing possibility is supported by the fact that PKC and MAPKs are important for circadian function (22, 23) and that PHLPP1 itself oscillates in the SCN (11). Our findings support a scenario in which PHLPP1 may operate in controlling the consolidation of circadian periodicity after resetting. Whereas this particular function may be conceptually different from a classical view of “memory,” our results indicate that some common molecular pathways may contribute to the plasticity required for circadian tau maintenance.
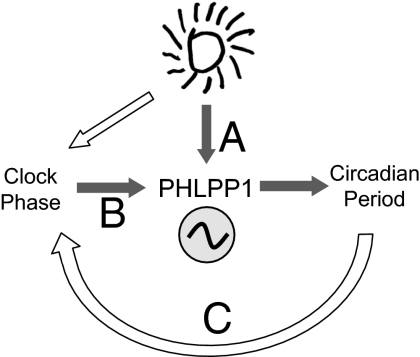
Fig. 4.
Model depicting how PHLPP1 affects the circadian system. Light pulse-induced phase shift of circadian rhythms and period change. Light pulse directly (A) or via clock resetting (B) controls circadian period after the phase shift. Irregular and delayed circadian period change in PHLPP1−/− affects the magnitude of resetting (C).
Our study reveals that ablation of PHLPP1 in the mouse has dual effects on tau length. One effect is indicated as an “acute effect,” wherein there is absence of tau extension (shortening) after phase delay (advance)-inducing light exposure. Another effect is the “late effect,” wherein there is a delayed shortening of tau, which subsequently is expressed as a large phase delay in PHLPP1 −/− mice during the late-phase period after light exposure (Fig. 4C).
After light-induced phase shifting, tau length can change in subsequent days. This plasticity of tau has been referred to as the “after-effect” (19). Light pulses that cause a phase delay in the clock are typically followed by a lengthening of tau whereas light that triggers a phase advance subsequently shortens the tau. Congenital factors that define tau include species differences (19), different substrains within species (i.e., specific mouse strains) (24) and clock gene mutations (7). The determining factors responsible for after effects have remained elusive. In our studies, the lengths of tau that accompany phase delay (advance) shifts are longer (shorter) than when they are not accompanied by phase shifts in PHLPP1 +/+ mice (Fig. 3 B and F and Fig. S4). This response to light in PHLPP1 +/+ mice is compatible with canonical “after effects of phase shifts” that occur under free running conditions (19). Typically there is extension of tau by phase delay. Conversely, there is a shortening of tau by phase advance. The loss of acute tau extension and shortening in PHLPP1 −/− mice (Fig. 3 B and F and Fig. S4) indicates a specific contribution of PHLPP1 to the after effect. Lighting cycles that deviate from 24 h (T cycles) also induce after effects. Shorter (<24 h) and longer (>24 h) lighting cycles shorten and lengthen behavioral tau, respectively. Interestingly, these lighting tasks have reversed effects on the SCN. For example, SCN isolated from animals entrained to short lighting cycle shows long period and vice versa (25, 26). Although the molecular basis for this discrepancy are still undetermined, PHLPP1 appears to operate as an after effect controlling gene, and thereby is expected to play a central role in this complex phenomenon.
Interestingly, the late-phase tau shortening may indicate that the function of PHLPP1 is to stabilize the period after phase shift. Thus, the PHLPP1 −/− mice show a very unique response which is demonstrated by a delayed change in tau after resetting. This phenotype is different (although maybe related to) the “delayed disappearance” of rhythmicity in Clock (27 –29) and Per2 (30) mutant mice that occurs after ∼2 weeks in DD. Perhaps PHLPP1 may also regulate the phosphorylation state of some circadian regulators, including CLOCK and PER2, the phosphorylation of which has been shown to be critical for normal circadian function (31, 32). Furthermore, PKC and MAPK, (PHLPP1-regulated kinases) have been shown to be important for clock resetting (22, 23). Future studies will establish whether PHLPP1 physiologically dephosphorylates any components of the clock machinery. Interestingly, evidence of multiple targets of PHLPP1 as a phosphatase exists in vivo (8–10, 13). Involvement of these and/or other novel pathways requiring PHLPP1 function may clarify how PHLPP1 contributes to tau stability.
In the context of this study it is notable that short-light (30 min) and long-light (∼8–12 h) treatments play different roles in clock resetting (18). For example, mice with mutations in the clock gene, Per1, or in the inhibitor of DNA-binding 2 (Id2) gene show phase delays that are larger than wild-type littermates after long-light tasks, an effect that is absent when animals are exposed to short light (16, 17, 33). In the present study, short light and long light had differential impact on the behavioral response in PHLPP1 −/− mice. Interestingly, short light shortened tau more than long light in PHLPP1 −/− mice (Fig. S3), a situation opposite to what is observed in the Per1 or Id2 mutant animals. Although the after effects were different in these mouse models, collectively, they demonstrate that the stimulus produced by short light versus long light has different effects on tau stability. That PHLPP1 also plays a role in the differential response to short and long lights implicates it in SCN neuronal plasticity.
When activity rhythm is reset by a light pulse, changes in the SCN program of gene expression typically occur within the first hour or two after the light pulse (34). Experiments using in vitro real time monitoring systems have shown that activation of NMDA receptors induces this rapid SCN phase resetting as demonstrated by the change in circadian Per1 promoter activity (35). However, what maintains tau length in free-running conditions? Although the molecular feedback loops of the circadian clock are necessary for tau maintenance in free-running conditions (1, 7), how these contribute to the plasticity required for such maintenance is yet unclear. Our data indicate that PHLPP1 may occupy a privileged position in the mechanism by which rhythmicity is stabilized in the SCN in the absence of zeitgeber cues. As PHLPP1 has already been implicated in neuronal plasticity (12), the data presented here ushers in a unique perspective on the role of this phosphatase in long-lasting plasticity in the circadian system. Its role in tau maintenance indicates that it may be central to the fine tuning necessary in the central pacemaker for long-term synaptic plasticity, especially in response to changing stimuli.
Materials and Methods
Gene Targeting and Generation of Homozygous Null Mice.
The targeting exon (exon 4) of PHLPP1 was subcloned in between two LoxP sites on the pFlox-FRT vector. A SacI restriction site was introduced into the targeting vector for detection of homologous recombination events by Southern blot analysis. For negative selection, sequences encoding diphtheria toxin (DT) were amplified using PCR and subcloned into the vector. The neomycin selection cassette (neor) is flanked by two FRT sites, which allow deletion of the neor gene via Flp-mediated recombination in mice. The targeting vector was electroporated into 129/Sv ES cells and the cells were subjected to positive and negative selection on the basis of neomycin and DT sensitivity (Fig. 1A). Genomic DNA was isolated from the ES cells and digested with SacI, and relevant products were detected by Southern blot using the probe as marked on the diagram. The wild-type allele generates an 11-kb fragment whereas the targeted knockout allele gives a 7-kb fragment. ES cells with a recombinant allele were injected into C57BL/6 blastocysts and transplanted into pseudopregnant C57BL/6 mice to generate chimeric pups. PHLPP1 fl/+ chimeras were bred with Protamine-Cre mice (129 background; 129-Tg(Prm-cre)58Og/J; The Jackson Laboratory) to generate fl/+, Protamine-Cre/+ males (Fig. 1B). Breeding of these mice to wild-type mice resulted in recombination of the LoxP sites and deletion of exon 4 in the male gametes, yielding PHLPP1+/− mice. Genotyping PCR was performed using the following primers: FP43 (forward primer): 5′-TAG GAG AGA CTA GTG ACA TC-3′, RP44 (reverse primer 1): 5′-TGA GCT TAT ACG CTG TGA TGC-3′, and RP56 (reverse primer 2): 5′-AGC CGA TTG TCT GTT GTG C-3′ (Fig. 1A). Primer pair FP43/RP44 generates a 264-bp product from the wild-type allele and a 336-bp product from the floxed allele. Primer pair FP43/RP56 generates a 486-bp product from the deleted allele (Fig. 1C). RNA from mouse brain samples was isolated using a Qiagen RNEasy lipid tissue kit, and RT-PCR was performed using a Qiagen OneStep RT-PCR kit and the following primers: (forward) 5′-TCT GTC GAA ATG GGA AGC CAC TGT C-3′ and (reverse) 5′-TGT ACC ACC ACA GCA CTG ATG C-3′. Protein extracts from PHLPP1+/+ and PHLPP1−/− brains, livers, and kidneys were subjected to Western blot analysis using the PHLPP antibodies from Bethyl Laboratories (PHLPP1; cat. no. A300-659A, PHLPP2; cat. no. A300-661A) and the β-actin antibody from Sigma (cat. no. A2228).
Animals and Behavioral Rhythm Monitoring.
Animal protocols used in this study were reviewed and approved by the Institutional Animal Care and Use Committee of the the University of California, Irvine and University of California, San Diego.
PHLPP1+/+ and PHLPP1−/− mice were bred and housed under 12-h light (fluorescent, Sylvania 25 w no. 4100K, ∼200 lx) /dark (LD) cycles. Locomotor activity was detected using running wheels (33) and passive (pyroelectric) infrared sensors (PU-2201; EK Japan). Running wheels and passive infrared sensors were used for tau analysis and passive infrared sensors were used for resetting analysis. Locomotion data were collected using the VitalView data acquisition system (Mini-Mitter) using a sampling interval of 5 min. Actograms were acquired using Actiview Biological Rhythm Analysis software (Mini-Mitter). Circadian period and phase shift of the activity rhythms were analyzed by Clocklab software (Actimetrics).
In Situ Hybridization.
In situ hybridization histochemistry using free-floating sections was performed according to methods detailed previously (14). We used 33P-radiolabeled cRNA probes for mPer1 and mPer2 for the in situ hybridization studies. mPer1 (75-411 NM_011065) fragment was obtained by RT-PCR and subcloned into pCR-BluntII-TOPO vector (Invitrogen). mPer1 and mPer2 (33) cDNA containing vectors were linearized with restriction enzymes and used as templates for standard 33P-labeled cRNA synthesis (14).
Statistical Analysis.
The effects of genotypes, lighting tasks, and number of days after the light task on the behavioral rhythms were tested by two-way or repeated measure ANOVA. A post hoc Fisher test was used for the comparison between the values. The effects of light tasks to tau were tested by Welch’s or paired t test.
Supplementary Material
Acknowledgments
We thank Yasukazu Nakahata, Jun Hirayama, Yiling Chen, Yasufumi Shigeyoshi, Mamoru Nagano, Qun-Yong Zhou, and Jia-Da Li and all members of the Newton and Sassone-Corsi laboratories for help, reagents, and support. S.M. was supported by the Uehara Memorial Foundation Research Fellowship for study abroad and by the Mochida Memorial Foundation for Medical and Pharmaceutical Research Fellowship. K.E.-M. is supported by National Institutes of Health (NIH) (CA 113265, T32). This work was supported by grants from Institut National de la Sante et de la Recherche Medicale, France (P.S.-C.), and NIH GM 67946 (to A.C.N.); R01-GM081634-01; and R21 AG033888 (to P.S.-C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910292107/DCSupplemental.
References
- 1.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis AM, et al. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci USA. 2007;104:3450–3455. doi: 10.1073/pnas.0611680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barion A, Zee PC. A clinical approach to circadian rhythm sleep disorders. Sleep Med. 2007;8:566–577. doi: 10.1016/j.sleep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijer JH, Schwartz WJ. In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J Biol Rhythms. 2003;18:235–249. doi: 10.1177/0748730403018003006. [DOI] [PubMed] [Google Scholar]
- 6.Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in nocturnal rodents II. The variability of phase response curves. J Comp Physiol. 1976;106:253–266. [Google Scholar]
- 7.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 8.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem. 2008;283:6300–6311. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 10.Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu K, Okada M, Takano A, Nagai K. SCOP, a novel gene product expressed in a circadian manner in rat suprachiasmatic nucleus. FEBS Lett. 1999;458:363–369. doi: 10.1016/s0014-5793(99)01190-4. [DOI] [PubMed] [Google Scholar]
- 12.Shimizu K, Phan T, Mansuy IM, Storm DR. Proteolytic degradation of SCOP in the hippocampus contributes to activation of MAP kinase and memory. Cell. 2007;128:1219–1229. doi: 10.1016/j.cell.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J Biol Chem. 2003;278:14920–14925. doi: 10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- 14.Shigeyoshi Y, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 15.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 16.Masubuchi S, Kataoka N, Sassone-Corsi P, Okamura H. Mouse Period1 (mPER1) acts as a circadian adaptor to entrain the oscillator to environmental light/dark cycles by regulating mPER2 protein. J Neurosci. 2005;25:4719–4724. doi: 10.1523/JNEUROSCI.4761-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffield GE, et al. A role for Id2 in regulating photic entrainment of the mammalian circadian system. Curr Biol. 2009;19:297–304. doi: 10.1016/j.cub.2008.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masubuchi S, Sassone-Corsi P. Circadian biology: an unexpected invitee to new time zones. Curr Biol. 2009;19:R298–R300. doi: 10.1016/j.cub.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: I. The stability and lability of spontaneous frequency. J Comp Physiol. 1976;106:223–252. [Google Scholar]
- 20.Colley PA, Sheu FS, Routtenberg A. Inhibition of protein kinase C blocks two components of LTP persistence, leaving initial potentiation intact. J Neurosci. 1990;10:3353–3360. doi: 10.1523/JNEUROSCI.10-10-03353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CH, et al. A role for the PI-3 kinase signaling pathway in fear conditioning and synaptic plasticity in the amygdala. Neuron. 2001;31:841–851. doi: 10.1016/s0896-6273(01)00433-0. [DOI] [PubMed] [Google Scholar]
- 22.Jakubcakova V, et al. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007;54:831–843. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 23.Akashi M, Hayasaka N, Yamazaki S, Node K. Mitogen-activated protein kinase is a functional component of the autonomous circadian system in the suprachiasmatic nucleus. J Neurosci. 2008;28:4619–4623. doi: 10.1523/JNEUROSCI.3410-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwartz WJ, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED. Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythms. 2004;19:198–207. doi: 10.1177/0748730404264156. [DOI] [PubMed] [Google Scholar]
- 26.Molyneux PC, Dahlgren MK, Harrington ME. Circadian entrainment aftereffects in suprachiasmatic nuclei and peripheral tissues in vitro. Brain Res. 2008;1228:127–134. doi: 10.1016/j.brainres.2008.05.091. [DOI] [PubMed] [Google Scholar]
- 27.Vitaterna MH, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antoch MP, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng B, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 31.Yoshitane H, et al. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toh KL, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 33.Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best JD, Maywood ES, Smith KL, Hastings MH. Rapid resetting of the mammalian circadian clock. J Neurosci. 1999;19:828–835. doi: 10.1523/JNEUROSCI.19-02-00828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asai M, et al. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr Biol. 2001;11:1524–1527. doi: 10.1016/s0960-9822(01)00445-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.