Abstract
Assembling the tree of life is a major goal of biology, but progress has been hindered by the difficulty and expense of obtaining the orthologous DNA required for accurate and fully resolved phylogenies. Next-generation DNA sequencing technologies promise to accelerate progress, but sequencing the genomes of hundreds of thousands of eukaryotic species remains impractical. Eukaryotic transcriptomes, which are smaller than genomes and biased toward highly expressed genes that tend to be conserved, could potentially provide a rich set of phylogenetic characters. We sampled the transcriptomes of 10 mosquito species by assembling 36-bp sequence reads into phylogenomic data matrices containing hundreds of thousands of orthologous nucleotides from hundreds of genes. Analysis of these data matrices yielded robust phylogenetic inferences, even with data matrices constructed from surprisingly few sequence reads. This approach is more efficient, data-rich, and economical than traditional PCR-based and EST-based methods and provides a scalable strategy for generating phylogenomic data matrices to infer the branches and twigs of the tree of life.
Keywords: next-generation DNA sequencing, phylogenetics, transcriptome, Anopheles, orthology
Recent advances in statistical phylogenetics, information technology, and molecular biology make it feasible to assemble a complete tree of life. Efforts so far have largely sought wide taxonomic breadth, which is reflected in the GenBank records of DNA sequences from approximately 300,000 species (1). Although broad taxonomic coverage is necessary for assembly of the tree, the sequence records of the overwhelming majority of species in GenBank are quite small and phylogenetically uninformative (2). Furthermore, phylogenies reconstructed from small amounts of DNA can be error-prone (3 –7) and often fail to detect biological processes such as deep coalescence, introgression, and hybridization (8). Thus, full and accurate resolution of the tree of life will require a concomitant increase in the genomic depth of sequence sampling of each species (2, 3, 9–15).
The two most commonly used methodologies for capturing orthologous DNA sequence data—the PCR-based (16) and EST-based (9, 17–19) approaches—are costly, labor-intensive, error-prone (20 –22), and impractical for generating phylogenomic data matrices containing thousands of species. Next-generation DNA sequencing technologies dramatically increase sequencing throughput and efficiency (23, 24) and therefore offer an opportunity to increase genomic depth through whole-genome sequencing. Although this approach is a major source of data for phylogenomic studies (14, 15), de novo assembly of complete eukaryotic genomes is currently prohibitively expensive for most of the approximately 2 million described species.
Transcriptomes offer alternative sources of orthologous sequence that are easier to sample than genomes for three reasons. First, after RNA processing, transcriptomes are typically much smaller than the genome. For example, only 7% of the Anopheles gambiae genome codes for proteins (25). Second, transcriptomes contain few simple-sequence regions and repetitive elements (26, 27). This is important because assembly of such sequences from the short-read lengths produced by next-generation DNA sequencing technologies is challenging (28). Third, and most important, the grossly uneven abundance of transcripts [varying over five orders of magnitude (29)] means that even light sequence coverage should provide in-depth sampling of a few hundred loci simply by sequencing transcripts in proportion to their representation in the library (9, 17, 19, 30). Moreover, highly expressed genes are typically involved in housekeeping and energy functions and therefore tend to be well-conserved (29, 30), leading to the expectation that orthologous genes can be efficiently sampled across species (9, 17, 19, 30).
We tested the idea that RNA-Seq (31) of non-normalized transcriptomes could be a data-rich, accurate, and cost-effective source of orthologous sequence data for phylogenomic analysis. We sequenced the transcriptomes of 10 mosquito species and developed a novel methodology that enabled us to transform the 36-bp sequence reads into phylogenomic data matrices containing hundreds of thousands of orthologous nucleotides. These matrices were composed primarily of highly expressed genes and contained very few orthology assignment errors. Phylogenetic inferences made from these data matrices were robust and sensitive to biological processes such as introgression. The success of this approach suggests an efficient, robust, and cost-effective way of increasing the genomic depth of the tree of life.
Results
Large Phylogenetic Data Matrices Can Be Accurately Assembled from Short Transcript Sequences.
We obtained an average of 13 million 36-bp DNA sequence reads from non-normalized cDNA libraries of 10 mosquito species using Illumina’s Solexa next-generation DNA sequencing platform (32) (Tables S1 and S2). We assembled the sequence reads de novo (33) and retained all contigs ≥100 bp for further analysis (Table S2). Phylogenomic data matrices were constructed by mapping single contigs from each species to full transcripts of the nonredundant transcriptome of the outgroup species Aedes aegypti (deduced from its complete genome sequence), which we used as a reference sequence (“single-contig” strategy). All A. aegypti reference transcripts with reciprocal best-BLAST-hit matches to single contigs of all nine Anopheles species were retrieved, locally aligned, and stripped of all codons with gaps or missing data in the A. aegypti reference transcripts.
The data matrix constructed from all contigs ≥100 bp included sequences of 553 genes, yielding a total alignment length of 389,364 bp; a data matrix consisting only of contigs ≥300 bp included 69 genes in a total alignment of 72,564 bp (Table 1 and Table S3). The accuracy of ortholog assignment (evaluated by estimating the percentage of A. gambiae contigs accurately assigned to their A. aegypti reference transcript orthologs) was high in all data matrices, with matrices built from contigs ≥100 bp only marginally worse than matrices built from contigs ≥300 bp. For example, only one of the 69 loci contributing to the 300-bp data matrix was incorrect (99% accuracy), and only 27 of the 553 loci in the 100-bp data matrix were incorrect (95% accuracy) (Table 1 and Table S3).
Table 1.
Ortholog number, contig number, and ortholog assignment accuracy for data matrices constructed from ≥100 and ≥300 bp contigs using the single-contig and supercontig strategies
| Single-contig strategy | Supercontig strategy | |||||||||
| ≥100-bp contigs | ≥300-bp contigs | ≥100-bp contigs | ≥300-bp contigs | |||||||
| Data set | Orthologs* | Accuracy,† % | Orthologs | Accuracy, % | Orthologs | Contigs‡ | Accuracy, % | Orthologs | Contigs | Accuracy, % |
| 100,000 | 0 | NA | 0 | NA | 4 | 3 | 33 | 0 | 0 | NA |
| 250,000 | 1 | 0 | 0 | NA | 50 | 34 | 79 | 2 | 2 | 50 |
| 500,000 | 11 | 91 | 0 | NA | 124 | 128 | 86 | 10 | 6 | 67 |
| 1,000,000 | 72 | 96 | 0 | NA | 226 | 287 | 85 | 64 | 36 | 86 |
| 2,000,000 | 120 | 94 | 8 | 75 | 430 | 550 | 84 | 148 | 86 | 86 |
| 3,000,000 | 173 | 93 | 29 | 93 | 630 | 825 | 82 | 198 | 146 | 88 |
| 4,000,000 | 212 | 93 | 36 | 92 | 850 | 1,152 | 82 | 255 | 183 | 87 |
| 5,000,000 | 252 | 93 | 40 | 95 | 1,054 | 1,449 | 83 | 302 | 222 | 86 |
| ∼6,500,000 | 333 | 93 | 37 | 97 | 1,591 | 1,872 | 84 | 445 | 290 | 87 |
| ∼13,000,000 | 553 | 95 (89)§ | 69 | 99 (94)§ | 2,661 | 4,118 | 85 | 725 | 523 | 86 |
No. of A. aegypti orthologs in data matrix.
Percentage of A. gambiae contigs accurately assigned to their A. aegypti reference transcript orthologs in the data matrix.
No. of A. gambiae contigs assigned to A. aegypti reference transcripts in the data matrix.
Percentage of accurately inferred orthologs using a phylogeny-based assessment of orthology assignment.
To further verify that our single-contig strategy yielded accurate orthologous assignments, we visually examined the phylogenies of all 553 and 69 putative orthologs constructed from all contigs ≥100 and ≥300 bp, following inclusion of additional high-scoring, but not best-scoring, sequences retrieved by BLAST from all transcriptomes. Under this phylogeny-based assessment, all orthologs had been correctly included in 89% (491/553) and 94% (65/69) of data matrices built from contigs ≥100 and ≥300 bp, respectively (Table 1).
Constructed Data Matrices Yield Robust Phylogenies.
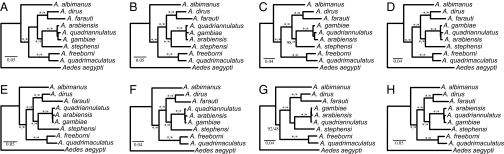
Maximum-likelihood (ML) (34) and Bayesian (35) phylogenetic analyses of the two data matrices yielded identical trees with strong clade support values (Fig. 1 A and E), with the exception of relationships between the three closely related species A. gambiae, Anopheles arabiensis, and Anopheles quadriannulatus, which we address below. The branching patterns recovered were mostly consistent with previous studies based on a few loci (36 –39). While previous analyses reported weak support for the sister grouping of Anopheles albimanus to the Anopheles freeborni–Anopheles quadrimaculatus clade (36) or failed to resolve the placement of A. albimanus (38, 39), our genome-scale analysis strongly supports the placement of A. albimanus outside all the other eight Anopheles species used in our study (approximately unbiased test (40): Δl = −41.5, p-value = 0.0001). Identical topologies were obtained when loci of ambiguous orthology were removed (Fig. 1 B and F and Fig. S1), when all columns with missing data were excluded from the analysis (Fig. 1 C and G), or when A. gambiae, an ingroup species, was used as the reference for data matrix assembly (Fig. 1 D and H).
Fig. 1.
Robust phylogenetic inference from short-read next-generation DNA sequencing. (A) ML phylogeny produced from data matrix constructed by considering all contigs ≥100 bp assembled from ∼13 million sequence reads per species using A. aegypti full transcripts as references under the single-contig strategy. (B) ML phylogeny of data matrix analyzed in A after exclusion of all loci of ambiguous orthology under either assessment strategy. (C) ML phylogeny of data matrix analyzed in A after exclusion of all sites with any missing data or gaps. (D) ML phylogeny produced from data matrix constructed by considering all contigs ≥100 bp assembled from ∼13 million sequence reads per species using A. gambiae full transcripts as references. (E–H) The same analyses as in A–D but on data matrices constructed by considering all contigs ≥300 bp. Clade support near internodes represents bootstrap support (ML) and posterior probability (Bayesian inference), respectively. Asterisks denote absolute support. Branch lengths represent estimated substitutions per site.
Three of the Anopheles species in this study, A. gambiae, A. arabiensis, and A. quadriannulatus, belong to the Anopheles gambiae species complex, a group of seven nearly indistinguishable sibling species (41). Several population genetic studies have provided evidence for introgression between species in this complex (41 –44). We tested whether our data also contained evidence for introgression by searching for single-gene alignments that significantly favored one of the three alternative topologies among A. gambiae, A. arabiensis, A. quadriannulatus, and Anopheles stephensi (45). Thirteen percent (32/237) of the single-gene alignments examined strongly supported one of the three alternative topologies and significantly rejected the two others (support was nearly equally distributed among the three alternative topologies). These data support and extend previous conclusions of introgression within the A. gambiae species complex (41 –44), although other processes, such as deep coalescence and hybridization (8), cannot be excluded.
Small Amounts of Input Data Are Sufficient for Phylogenetic Resolution.
The large size of our data matrices and the unequivocal support of the resulting phylogenies suggest that the amount of data that we collected may have been more than what is required for resolution of the clade. To evaluate how much data is necessary and sufficient to obtain phylogenetic resolution, we assembled phylogenetic data matrices from fewer sequence reads (Table S3). Even after substantial reduction in the amount of input data, the single-contig strategy was remarkably efficient at identifying large amounts of orthologous DNA and capable of resolving the clade (Fig. 2 A and B and Table S4). For example, reducing the input data from 13 to 3 million sequence reads per species still yielded large and information-rich data matrices (e.g., 91 kb from 173 loci for the 100-bp data matrix) that were sufficient to resolve all internodes of the clade (Fig. 2B). However, data matrices constructed from fewer than 2 million sequence reads were insufficient to resolve all internodes of the clade (Fig. 2B).
Fig. 2.
Constructed phylogenomic data matrices contain large amounts of orthologous DNA and are capable of yielding robust phylogenetic inferences even after substantial reductions in the amount of input data. (A) Numbers of total, variable, and parsimony-informative sites in data matrices constructed from different amounts of raw data using the single-contig strategy with contigs ≥100 bp. (B) Number of resolved internodes in data matrices constructed using the single-contig strategy. (C) Numbers of total, variable, and parsimony-informative sites in data matrices constructed from different amounts of raw data using the supercontig strategy with contigs ≥100 bp. (D) Number of resolved internodes in data matrices constructed using the supercontig strategy.
Because de novo transcriptome assemblies yield large numbers of short contigs (Table S2), the single-contig strategy likely fails to capture large amounts of potentially informative data because many genes are represented by two or three non-overlapping contigs, and the single-contig strategy uses only the “best” contig for a given gene in the data matrices. To improve data recovery and test the lower limit of raw sequence data required to recover accurate data matrices, we used a local alignment procedure to place all contigs from each Anopheles species that uniquely mapped to A. aegypti reference transcripts into supercontigs (“supercontig” strategy). This strategy dramatically increased data matrix size (e.g., from ∼390 to ∼971 kb for the full 100-bp data matrix), with only marginal decreases in ortholog assignment accuracy (Fig. 2C, Table 1, and Table S5). Importantly, the data matrices generated from supercontigs produced resolved phylogenies from even fewer sequence reads (Fig. 2 C and D and Table S6) than those constructed with the single-contig strategy. Indeed, the supercontig strategy generated a well-supported phylogeny from only 0.5 million sequence reads per taxon (Fig. 2D), one-fourth the lower limit with the single-contig strategy.
Constructed Data Matrices Are Composed Primarily of Highly Expressed Genes.
The recovery of so much usable data from so few sequence reads is remarkable because successful de novo assembly of short sequence reads requires deep coverage (33). For example, 0.5 million 36-bp sequence reads would cover only 0.06× –0.013× of the A. gambiae and A. aegypti genomes, and only 0.8–0.9× of their processed transcriptomes (25, 46), respectively. These estimates suggest that, even though light sequencing of non-normalized transcriptomes results in low average coverage, the coverage of highly expressed genes remains deep enough to enable their assembly.
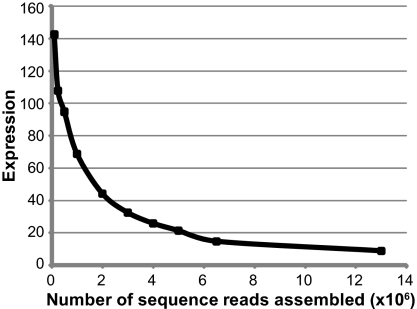
To evaluate the relative contribution of highly expressed genes to data matrix generation, we compared the average expression level of base pairs from our data matrices to the full transcriptome of A. aegypti. Base pairs contained in our data matrices have substantially enriched expression levels (Fig. 3), with the magnitude of this effect increasing dramatically (from 9- to 142-fold) in data matrices constructed from smaller amounts of data. Thus, light transcriptome short-read sequencing leverages the naturally skewed representation of transcripts to attain the deep coverage necessary to construct phylogenomic data matrices from short reads (Fig. 3).
Fig. 3.
Base pairs found in phylogenetic data matrices are derived from highly expressed transcripts, especially in data sets constructed from less input data. Expression is plotted against the number of sequence reads used. The average expression of a base pair included in a given supercontig data matrix (contigs ≥100 bp) was quantified from the A. aegypti data relative to the average expression of a base pair in the full A. aegypti transcriptome.
Discussion
It is widely recognized that successful reconstruction of the tree of life will require increases in both taxonomic breadth and genomic depth (2, 3, 9–15). However, constraints in taxon availability and sequence data acquisition have forced systematists to identify optimal experimental strategies for accurate resolution of major clades of the tree of life (47 –49). We have shown that highly informative phylogenomic data matrices can be constructed from a surprisingly small number of short sequence reads of non-normalized transcriptomes, suggesting that labor and cost should no longer prohibitively limit taxon selection.
We estimate that this clade could have been fully resolved with less than one-twentieth of the sequence data that we generated (Fig. 2). Thus, the cost of obtaining phylogenetically informative characters from dozens to hundreds of genes scattered across the genome is almost negligible compared to PCR-based and EST-based approaches. Of course, these approaches might still be more practical for some projects than light transcriptome short-read sequencing. For example, our approach currently requires large amounts of high-quality RNA, as well as considerable bioinformatics infrastructure and expertise. However, some third-generation sequencing technologies promise to provide direct sequencing of extraordinarily small RNA quantities (50), which should essentially eliminate some of the most labor- and resource-intensive steps and allow analysis of ever smaller and more precious tissue samples. Furthermore, the bioinformatics pipeline that we used was primarily driven by BLAST, PERL scripts, and standard phylogenetics software, and we anticipate its automation for large-scale phylogenomics projects in future software packages.
The accuracy, scalability, and sufficiency of light transcriptome short-read sequencing suggest that it is now feasible to generate genome-scale phylogenies of many challenging and speciose clades. Importantly, as new sequencing technologies are developed, sequencing efficiency will continue to improve, bringing more clades into focus (23, 24). Regardless of the specific future advances, leveraging skewed transcript abundance by RNA-Seq provides a robust and efficient way to increase the genomic depth and phylogenetic resolution of the vast diversity of life on earth.
Methods
Transcriptome Sequencing.
Eggs from all 10 species (Table S1) were obtained through the Malaria Research and Reference Reagent Resource Center (http://www.mr4.org/). Mosquito rearing, total RNA and poly(A+) RNA isolation, and cDNA synthesis, preparation, and sequencing were done following previously published protocols (30). Two lanes of massively parallel sequencing by synthesis were performed per species and processed into SCARF files containing millions of 36-bp sequencing reads with raw quality scores using the Solexa Genome Analyzers I and II and the Solexa Pipeline software, according to the manufacturer's instructions (Illumina) (32) (Table S2). Sequence data from A. aegypti and A. gambiae (30) were retrieved from the National Center for Biotechnology Information short read archive (study no. SRP001531 of submission no. SRA010234).
Transcriptome and Data Matrix Assembly.
We assembled varying amounts of sequence reads of each species de novo using the VELVET (version 0.7.23) software (33). VELVET generates assemblies by searching among sequence reads for identical matches of a certain length (referred to as k-mer length). To identify the optimal k-mer value (sensu ref. 33), we assembled our sequence reads using k-mer lengths of 17, 19, 21, 23, 25, 27, and 29, without imposing cutoffs for contig coverage or length. For data matrix construction, we used only those assemblies that yielded the greatest number of contigs ≥300 bp. Only contigs ≥100 bp were retained for further analysis. Single-contig data matrices were constructed by mapping single contigs from each species to full transcripts from the nonredundant complete transcriptome of the outgroup A. aegypti, which we used as a reference, using the reciprocal best-BLAST-hit criterion with a cutoff e-value of 10−6. All ortholog sets were locally aligned using DIALIGN2 (51) and stripped of all codons with gaps or missing data in the reference transcripts using custom PERL scripts.
Supercontig data matrices were constructed by relaxing the reciprocal best-BLAST-hit criterion to allow multiple nonconflicting VELVET contigs to be mapped to a single reference protein. For each species, all significant (e-value <10−5) TBLASTN (and the reciprocal BLASTX) hits between predicted A. aegypti proteins and VELVET contigs were considered for possible placements and conflicts in the order of TBLASTN scores for each reference protein. A VELVET contig was considered mapped to a reference protein if the contig's BLASTX score for the considered protein was the highest of any protein (same as the reciprocal best-BLAST-hit criterion) and the location of its proposed placement did not overlap with any placed contigs (local relaxation of the reciprocal best-BLAST-hit criterion). If any proposed contig placements conflicted, we first considered whether they might simply not have been joined by VELVET because the overlap was less than that of the k-mer and attempted to make joins on the basis of the exact matches of overhang base pairs from 3 to (k-mer − 1). If such a join could not be made, either because the length of the overlap was equal to or greater than the k-mer length or because the overlapping contigs’ overhangs were not identical, only the contig with the highest TBLASTN score was retained. Once all contigs had been considered for a given reference protein, 9–11 “N” base pairs were inserted between contigs to preserve the reading frame, as well as 0–20 “N” base pairs at the beginning. Once assemblies had been constructed for each species, all genes were locally aligned with DIALIGN2 (51) to their respective reference coding sequence and trimmed to retain only codons with data in the reference sequence that were unambiguously aligned in four or more taxa. All codons containing any base pairs considered unaligned by DIALIGN2 were recoded as “N” prior to phylogenetic analysis.
The data matrix used to examine introgression within the A. gambiae species complex was constructed by identifying all orthologs between A. gambiae reference transcripts and contigs ≥300 bp from A. arabiensis, A. quadriannulatus, and A. stephensi under the single-contig strategy.
Orthology Assignment Accuracy.
The accuracy of single-contig and supercontig putative orthology assignments was assessed for each placed A. gambiae VELVET contig by determining whether the putative source coding sequence in the annotated A. gambiae transcriptome was the reciprocal best-BLAST hit of the A. aegypti coding sequence to which it was mapped above. Those placements whose accuracy was not confirmed (below ∼15% for most data sets) contained an almost even mixture of likely errors (i.e., A. gambiae VELVET contigs placed to A. gambiae coding sequences that had no or a different putative ortholog among A. aegypti coding sequences) and candidates for unannotated A. gambiae transcripts (i.e., A. gambiae VELVET contigs that could not be reliably placed in an annotated A. gambiae coding sequence but had been placed in an annotated A. aegypti coding sequence).
The accuracy of single-contig putative orthology assignments was further assessed using a phylogeny-based strategy as follows. We first retrieved the 24 highest BLAST hits (if available) from a database that contained all assembled contigs of all 10 species and the A. aegypti and A. gambiae reference transcriptomes, using each A. aegypti reference transcript in the ortholog sets constructed from the ≥100- and ≥300-bp contigs as a query (only the portion of the transcript contained in the data matrix was used). We then merged the BLAST-retrieved sequences with the originally chosen putative orthologs, removed all duplicates, aligned each data matrix as above, and phylogenetically analyzed as below. Finally, we visually examined the topology of each data matrix to assess whether the originally chosen putative orthologs were correctly assigned.
Phylogenetic Analysis.
Phylogenetic reconstruction was performed using the optimality criteria of Maximum Likelihood (ML) and Bayesian inference (BI), as implemented in RAXML, version 7.0.4 (34) and MRBAYES, version 3.1.2 (35), respectively. For the ML analysis, robustness of inference was assessed by running 100 fast bootstrap replicates using the GTR + CAT approximation. The ML tree was calculated assuming a GTR + GAMMA model of sequence evolution. In all ML analyses, all free sequence model parameters were estimated by RAXML. The ML topology was compared to alternative topologies using the Shimodaira–Hasegawa test (45), as implemented in RAXML (34), using a p-value cutoff of 0.05. Alternative topologies were compared by the approximately unbiased test in CONSEL (version 0.1j) (40) after calculating the site-likelihood scores for each topology in RAXML (34). For the BI analysis, two independent analyses were run assuming a different GTR + GAMMA + I substitution model for each codon position. Each analysis was run using four chains (one cold and three hot) for 2 million generations. Trees were sampled every 1,000 generations and the first 2,000 sampled trees were discarded as burn-in, by which point stationarity had already been reached. When evaluating whether an internode was resolved (Fig. 2B and D), we required clade support values of at least 95% for both ML and BI.
Expression Analyses.
Expression was determined by mapping all A. aegypti sequence reads to the A. aegypti annotated transcriptome using the RMAPQ(version 0.45) software with three mismatches allowed and a quality filter of five (52). The length of each annotated transcript was then used to determine expression on a base-pair basis and to normalize expression of the average base pair in the transcriptome to a value of one. The relative enrichment of expression levels in the phylogenetic data matrices was determined as the weighted average of the expression of transcripts incorporating base pairs into the matrix (Fig. 3).
Supplementary Material
Acknowledgments
We thank Paul Howell and the MR4 facility at the Centers for Disease Control and Prevention for providing mosquito strains; Julian Hillyer, Larry Zwiebel, Jason Pitts, Jonas King, and Tania Estevez-Lao for providing advice, assistance, and access to mosquito-rearing facilities; and David Pollock for providing comments on an earlier draft. This work used resources of the Advanced Computing Center for Research and Education at Vanderbilt University. This work was supported by the James S. McDonnell Foundation (C.T.H. and M.J.), the Searle Scholars Program (A.R.), and the National Science Foundation (DEB-0844968) (A.R.). C.T.H. is the Maclyn McCarty Fellow of the Helen Hay Whitney Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910449107/DCSupplemental.
Data deposition: All sequence data have been deposited at the National Center for Biotechnology Information short read archive (www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) as study no. SRP001532 of submission no. SRA010237.
References
- 1.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37(Database issue):D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanderson MJ. Phylogenetic signal in the eukaryotic tree of life. Science. 2008;321:121–123. doi: 10.1126/science.1154449. [DOI] [PubMed] [Google Scholar]
- 3.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 4.Naylor GJP, Brown WM. Amphioxus mitochondrial DNA, chordate phylogeny, and the limits of inference based on comparisons of sequences. Syst Biol. 1998;47:61–76. doi: 10.1080/106351598261030. [DOI] [PubMed] [Google Scholar]
- 5.Cummings MP, Otto SP, Wakeley J. Sampling properties of DNA sequence data in phylogenetic analysis. Mol Biol Evol. 1995;12:814–822. doi: 10.1093/oxfordjournals.molbev.a040258. [DOI] [PubMed] [Google Scholar]
- 6.Rokas A, King N, Finnerty J, Carroll SB. Conflicting phylogenetic signals at the base of the metazoan tree. Evol Dev. 2003;5:346–359. doi: 10.1046/j.1525-142x.2003.03042.x. [DOI] [PubMed] [Google Scholar]
- 7.Castoe TA, et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci USA. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maddison WP. Gene trees in species trees. Syst Biol. 1997;46:523–536. [Google Scholar]
- 9.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 10.Regier JC, et al. Resolving arthropod phylogeny: Exploring phylogenetic signal within 41 kb of protein-coding nuclear gene sequence. Syst Biol. 2008;57:920–938. doi: 10.1080/10635150802570791. [DOI] [PubMed] [Google Scholar]
- 11.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 12.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 13.Hackett JD, et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of Rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 14.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 15.Miller W, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocher TD, et al. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. [Google Scholar]
- 17.Hughes J, et al. Dense taxonomic EST sampling and its applications for molecular systematics of the Coleoptera (beetles) Mol Biol Evol. 2006;23:268–278. doi: 10.1093/molbev/msj041. [DOI] [PubMed] [Google Scholar]
- 18.de la Torre JE, et al. ESTimating plant phylogeny: Lessons from partitioning. BMC Evol Biol. 2006;6:48. doi: 10.1186/1471-2148-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theodorides K, De Riva A, Gómez-Zurita J, Foster PG, Vogler AP. Comparison of EST libraries from seven beetle species: towards a framework for phylogenomics of the Coleoptera. Insect Mol Biol. 2002;11:467–475. doi: 10.1046/j.1365-2583.2002.00355.x. [DOI] [PubMed] [Google Scholar]
- 20.Bradley RD, Hillis DM. Recombinant DNA sequences generated by PCR amplification. Mol Biol Evol. 1997;14:592–593. doi: 10.1093/oxfordjournals.molbev.a025797. [DOI] [PubMed] [Google Scholar]
- 21.Sorek R, Safer HM. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 2003;31:1067–1074. doi: 10.1093/nar/gkg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark AG, Whittam TS. Sequencing errors and molecular evolutionary analysis. Mol Biol Evol. 1992;9:744–752. doi: 10.1093/oxfordjournals.molbev.a040756. [DOI] [PubMed] [Google Scholar]
- 23.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae . Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 26.Tóth G, Gáspári Z, Jurka J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lander ES, et al. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Hert DG, Fredlake CP, Barron AE. Advantages and limitations of next-generation sequencing technologies: A comparison of electrophoresis and non-electrophoresis methods. Electrophoresis. 2008;29:4618–4626. doi: 10.1002/elps.200800456. [DOI] [PubMed] [Google Scholar]
- 29.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 30.Gibbons JG, et al. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Sallum MAM, et al. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Syst Entomol. 2002;27:361–382. [Google Scholar]
- 37.Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: From subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–139. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- 38.Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: Insights from nuclear single-copy genes and the weight of evidence. Syst Biol. 2001;50:540–556. [PubMed] [Google Scholar]
- 39.Krzywinski J, Wilkerson RC, Besansky NJ. Evolution of mitochondrial and ribosomal gene sequences in Anophelinae (Diptera: Culicidae): Implications for phylogeny reconstruction. Mol Phylogenet Evol. 2001;18:479–487. doi: 10.1006/mpev.2000.0894. [DOI] [PubMed] [Google Scholar]
- 40.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 41.Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 42.Besansky NJ, et al. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc Natl Acad Sci USA. 1994;91:6885–6888. doi: 10.1073/pnas.91.15.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang-Sattler R, et al. Mosaic genome architecture of the Anopheles gambiae species complex. PLoS One. 2007;2:e1249. doi: 10.1371/journal.pone.0001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Besansky NJ, et al. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: Evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 46.Nene V, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg MS, Kumar S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proc Natl Acad Sci USA. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 50.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 51.Morgenstern B. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 52.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;9:128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.