Abstract
In response to infection, invertebrates process replicating viral RNA genomes into siRNAs of discrete sizes to guide virus clearance by RNA interference. Here, we show that viral siRNAs sequenced from fruit fly, mosquito, and nematode cells were all overlapping in sequence, suggesting a possibility of using siRNAs for viral genome assembly and virus discovery. To test this idea, we examined contigs assembled from published small RNA libraries and discovered five previously undescribed viruses from cultured Drosophila cells and adult mosquitoes, including three with a positive-strand RNA genome and two with a dsRNA genome. Notably, four of the identified viruses exhibited only low sequence similarities to known viruses, such that none could be assigned into an existing virus genus. We also report detection of virus-derived PIWI-interacting RNAs (piRNAs) in Drosophila melanogaster that have not been previously described in any other host species and demonstrate viral genome assembly from viral piRNAs in the absence of viral siRNAs. Thus, this study provides a powerful culture-independent approach for virus discovery in invertebrates by assembling viral genomes directly from host immune response products without prior virus enrichment or amplification. We propose that invertebrate viruses discovered by this approach may include previously undescribed human and vertebrate viral pathogens that are transmitted by arthropod vectors.
Keywords: arboviruses, piRNAs, siRNAs, viral immunity, massively parallel sequencing
The Dicer family of host immune receptors mediates antiviral immunity in fungi, plants, and invertebrate animals by RNA interference (RNAi) or RNA silencing (1 –3). In this immunity, a viral dsRNA is recognized by Dicer and diced into siRNAs. These virus-derived siRNAs are then loaded into an RNA silencing complex to act as specificity determinants and to guide slicing of the target viral RNAs by an Argonaute protein (AGO) present in the complex. Dicer proteins typically contain an RNA helicase domain, a PAZ domain shared with AGOs, and two tandem type III endoribonuclease (RNase III) domains. Dicer cleaves dsRNA with a simple preference toward a terminus of dsRNA, producing duplex small RNA fragments of discrete sizes progressively from the terminus (4).
In addition to siRNAs, micro-RNAs (miRNAs) and PIWI-interacting RNAs (piRNAs) guide RNA silencing in similar complexes but with distinct AGOs (4 –6). In Drosophila melanogaster, miRNAs and siRNAs are predominantly 22 and 21 nucleotides in length, dependent on Dicer-1 (DCR1) and DCR2 for their biogenesis, and act in silencing complexes containing AGO1 and AGO2 in the AGO subfamily, respectively (4 –6). In contrast, ∼24–30-nt piRNAs are Dicer-independent and require AGO3, Aubergine (AUB), and PIWI in the PIWI subfamily for their biogenesis (4 –6). Genetic analyses (7 –10) have clearly demonstrated a role for D. melanogaster DCR2 in the immunity and biogenesis of viral siRNAs targeting diverse positive-strand (+) RNA viruses, including Flock house virus (FHV), cricket paralysis virus, Drosophila C virus (DCV), and Sindbis virus (SINV). Cloning and sequencing of small RNAs from FHV-infected Drosophila cells further indicate that the viral dsRNA replicative intermediates (vRI-dsRNAs) are the substrate of DCR2 and the precursor of viral siRNAs (11, 12). Drosophila susceptibility to Drosophila X virus (DXV), which contains a dsRNA genome, is influenced by components from both the siRNA (e.g., AGO2, R2D2) and piRNA (e.g., AUB, PIWI) pathways (13). However, detection of small RNAs derived from any dsRNA virus has not been reported yet (1, 13).
Virus-derived small RNAs were first detected in plants infected with a +RNA virus (14). The Dicer proteins involved in the production of siRNAs targeting both +RNA viruses and DNA viruses have been identified in Arabidopsis thaliana (2, 3), and plants encode AGOs in the AGO subfamily but none from the PIWI subfamily (15). Cloning and sequencing of plant viral siRNAs suggest that they may be processed either from vRI-dsRNAs or hairpin regions of single-stranded RNA precursors (16 –20). Production of viral siRNAs has also been demonstrated in fungi, silkworms, mosquitoes, and nematodes in response to infection with +RNA viruses, and viral small silencing RNAs produced in fungi and mosquitoes have recently been cloned and sequenced (21 –25).
The available data thus illustrate that accumulation of virus-derived small silencing RNAs is a common feature of an active immune response to viral infection in diverse eukaryotic host species. In this study, we found that viral small silencing RNAs produced by invertebrate animals are overlapping in sequence and can assemble into long contiguous fragments of the invading viral genome from small RNA libraries sequenced by next-generation platforms. Based on this finding, we developed an approach of virus discovery in invertebrates by deep sequencing and assembly of total small RNAs (vdSAR) isolated from a host organism of interest. Use of this approach revealed mix infection of Drosophila cell lines and adult mosquitoes by multiple RNA viruses, five of which were previously undescribed. Analysis of small RNAs from mix-infected Drosophila cells showed that infection of all three distinct dsRNA viruses triggered production of viral siRNAs with features similar to siRNAs derived from +RNA viruses. Our study also revealed production and assembly of virus-derived piRNAs in Drosophila cells, suggesting a previously undescribed function of piRNAs in viral immunity. Unique features of vdSAR and its potential in discovering invertebrate and arthropod-borne animal and human viral pathogens are discussed.
Results
Virus Genome Sequencing by Assembly of Viral siRNAs Produced in Invertebrate Hosts.
The type III dsRNA-specific ribonuclease Dicer preferentially cleaves long dsRNA substrates from a terminus, such that dsRNA precursors with a defined terminus are processed into siRNAs in 21-nt phases (26, 27). However, sequencing of small RNAs by the 454 platform from Drosophila cells acutely infected with FHV showed recently that the DCR2-dependent 21-nt viral siRNAs are not produced in phase (11). Thus, we tested the idea that viral siRNAs produced by the host immune system might be overlapping in sequence by determining if the sequenced FHV siRNA fragments could be assembled back into the RNA genome of FHV.
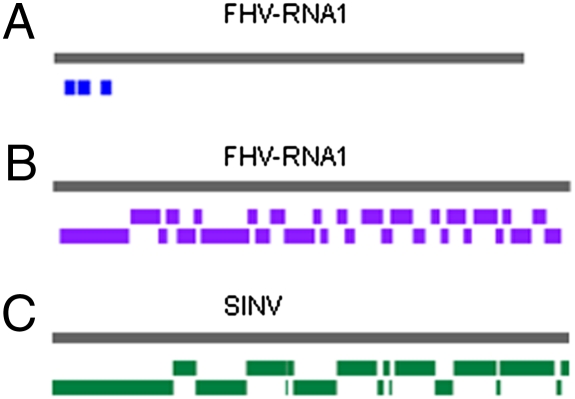
We chose the Velvet program (28) developed for genome assembly from short reads and set 17 nucleotides as the minimal overlapping length (k-mer) required to join two small RNAs into a contig. Assembly of the sequenced 1,177 FHV siRNAs (11) by the Velvet program yielded three contigs of 54, 73, and 52 nucleotides in length, which contained 27, 47, and 35 siRNAs, respectively (Fig. 1A). This indicated that viral siRNAs produced in infected fruit fly cells were indeed overlapping in sequence. All three assembled contigs were clustered in the 5′-terminal region of the genomic RNA1 of FHV (Fig. 1A), to which more than 60% of the RNA1-specific siRNAs were previously mapped (11).
Fig. 1.
Position and distribution of FHV and SINV siRNA contigs assembled from small RNAs sequenced from Drosophila S2 cells infected with the B2-deletion mutant of FHV (11) (A), a transgenic C. elegans strain in the RNAi-defective 1 (rde-1) mutant background carrying an FHV RNA1 replicon in which the coding sequence of B2 was replaced by that of GFP (29) (B), and adult mosquitoes infected with SINV (22) (C). Note that the length of RNA genomes was not drawn to scale.
The nematode Caenorhabditis elegans carrying a self-replicating genomic RNA1 of FHV produces viral siRNAs detectable by Northern blot hybridization (29). A total of 1,236,800 small RNAs 19–25 nucleotides in length were sequenced from the nematodes by the Illumina platform (Illumina 2G Analyzer at the campus Genomics Institute Core Facility for Genomics). In addition to 321,568 (26%) known C. elegans miRNAs, the library contained 5,957 (0.48%) and 1,455 (0.12%) reads that were 100% identical/complementary to and differed by one nucleotide from the replicating FHV genome, respectively. The viral siRNAs were divided equally into (+) and (−) polarities, and the most abundant viral siRNAs of both polarities were 23 nucleotides in length (Fig. S1), consistent with the size determined by Northern blot hybridization (29). Notably, assembly of the viral siRNAs cloned from C. elegans animals yielded 29 contigs that covered 93% of the FHV RNA1 by 36 times (Fig. 1B). There was also overlap with the neighboring contig for 21 of the 29 FHV contigs, and lack of further assembly by the Velvet program was because the overlap length was shorter than the defined k-mer value (17 nucleotides).
Cloning and sequencing of viral siRNAs produced in mosquitoes (Aedes aegypti) infected with the arthropod-borne SINV were reported recently (22). The library contained 525,457 perfectly matched SINV small RNAs and 68,669 SINV small RNAs with one mismatch. We found that assembly of the SINV small RNAs contained in the library generated 19 siRNA contigs with only five true gaps and that 99% of the 10-kb genome of SINV was covered by 1,029 times (Fig. 1C).
These findings illustrate that high-volume sequencing of total small RNAs from infected hosts yielded viral siRNA assemblies to cover almost the entire viral genomes by multiple times. Therefore, we conclude that viral siRNAs produced by the three invertebrate hosts are overlapping in sequence and could be used for viral genome assembly. Because production of viral siRNAs is an inevitable immune response of many eukaryotic hosts to virus infection (1 –3), we thought that it might be possible to discover viruses by deep sequencing and assembly of total small RNAs accumulated in an organism of interest.
Discovery of Four RNA Viruses from a Drosophila Schneider 2 Cell Line.
To test if vdSAR worked, we analyzed two duplicate small RNA libraries constructed from a Drosophila Schneider 2 (S2) cell line previously maintained in Gerald Rubin’s laboratory (University of California, Berkeley, CA) termed S2-GMR (12). These libraries contain 6,454,759 small RNAs 18–28 nucleotides in length in total, of which 1,092,833 molecules are unique. We first ran the small RNA assembly program Velvet using a k-mer of 17 and obtained 1,639 contigs in total. The National Center for Biotechnology Information (BLASTN) identified three groups of contigs that were identical or highly homologous to the nucleotide sequence entries of the nonredundant databases of the NCBI. The first group of 1,032 contigs was mapped to the genome of D. melanogaster, and 62% of those contigs overlapped transposon loci, suggesting that fruit fly endogenous siRNAs are also overlapping in sequence.
The second group of 49 contigs (33–401 nucleotides in length) was found to correspond to the bipartite dsRNA genome of DXV. Seventy-eight percent of DXV genome segment A (23 contigs) and 91% of DXV segment B (26 contigs) were obtained from the small RNA assemblies, yielding a total of 5.55 kb of consensus RNA sequence with 73 times of coverage. Because the viral genome assembled from the sequenced small RNAs showed 98% identity to DXV (NC_004177, NC_004169) at both the nucleotide and protein sequence levels, we conclude that the S2-GMR cell line was persistently infected with DXV.
The third group included 57 contigs that exhibited strong homology to the bipartite +RNA genome of Tn5 cell line virus (TCLV). TCLV is a recently described member of the genus Alphanodavirus and shares 89.3% and 84% nucleotide sequence identity with RNA1 and RNA2 of FHV in the same genus, respectively (30). Assembly of siRNAs yielded 2,196 and 1,048 nucleotides in length for RNA1 and RNA2, respectively. The remaining parts of the bipartite genome were obtained by RT-PCR and RACE-PCR from S2-GMR cells originally obtained from Gerald Rubin, producing complete RNA1 and RNA2 molecules of 3,107 and 1,416 nucleotides in length. The identified virus, designated American nodavirus (ANV), was most closely related to TCLV, with 94% and 92% identities to RNA1 and RNA2 of TCLV, respectively. ANV also shared 89% and 82% identities to RNA1 and RNA2 of FHV, which explained why it was thought previously that the cells were persistently infected by FHV (12). In addition to the RNA-dependent RNA polymerase (RdRP) and coat protein (CP), both TCLV and ANV encode the RNAi suppressor (protein B2) of 106-aa residues. However, the three viral proteins exhibited similar levels of sequence variation among ANV, TCLV, and FHV, suggesting that ANV represents a unique species of Alphanodavirus.
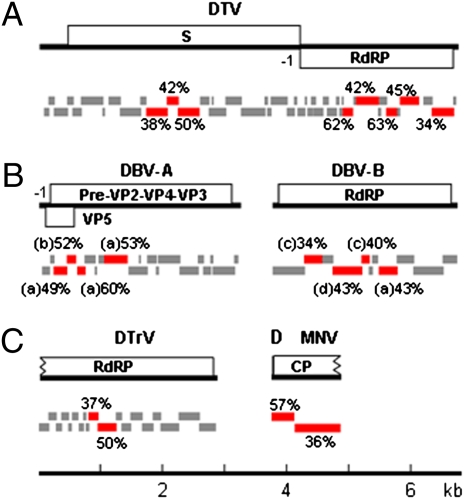
Three additional clusters of virus-specific contigs were identified among the remaining 501 assembled contigs by the National Center for Biotechnology Information (BLASTX) comparison with the known viral proteins in the NCBI (cutoff: 1e-3). Eight contigs in the first cluster (Fig. 2A, red bars) encoded proteins with 34–62% identities to either the RdRP (five contigs of 1,410 nucleotides in length) or the structural protein (three contigs of 899 nucleotides in length) of Penaeid shrimp infectious myonecrosis virus (PsIMNV) (31). PsIMNV is an unassigned member in the Totiviridae, which includes three established genera of viruses with a linear dsRNA genome (32). The complete genome of the identified virus, designated Drosophila totivirus (DTV), was obtained from S2-GMR cells by RT-PCR using primers designed according to the sequences of the eight contigs and their relative positions mapped in the genome of PsIMNV (Fig. 2A) and by RACE-PCR. The genome of DTV was 6,780 nucleotides long and encoded CP and RdRP ORFs that overlapped by 205 nucleotides. Although the RdRPs of DTV and PsIMNV shared only 37.6% identities, phylogenetic analysis of the viral RdRPs in the Totiviridae showed that DTV and PsIMNV formed a distinct cluster outside of the known three genera (Fig. S2A). We thus suggest a unique genus in the Totiviridae to include DTV and PsIMNV.
Fig. 2.
Discovery of dsRNA viruses DTV (A) and DBV (B) and +RNA viruses DTrV (C) and MNV (D) from S2-GMR cells by virus discovery by deep sequencing and assembly of total small RNAs (vsSAR). Red bars refer to the virus-specific contigs initially identified by % sequence similarities of their encoded proteins to a viral protein in the databases. The contigs of DTV, DTrV, and MNV showed the highest similarities to Penaeid shrimp infectious myonecrosis virus (PsIMV), EEV, and WNV, respectively. However, four different members in the Birnaviridae were identified as the closest to DBV contigs: (a) Infectious bursal disease virus (IBDV), (b) DXV, (c) Marine birnavirus (MAV), and (d) Blotched snakehead virus (BSV). Gray bars refer to the contigs that were assembled from small RNAs of S2-GMR cells and subsequently mapped to specific viruses after the complete genomes were obtained. Note that the length of RNA genomes was drawn to scale and the ORFs encoded by the partial genome of DTrV (3,005 nucleotides in length) and MNV (1,130 nucleotides in length) were incomplete.
The second siRNA cluster also contained eight contigs (Fig. 2B) encoding proteins with homology to various members of the Birnaviridae, which contain a bipartite dsRNA genome (33). Four of those contigs with a combined length of 1,224 nucleotides in total were mapped to the RdRP (VP1) coding region, whereas the remaining contigs with a combined length of 888 nucleotides mapped to the second segment of the birnaviral genome that encodes for the structural proteins (Fig. 2B). The complete bipartite genome of the identified birnavirus, designated Drosophila birnavirus (DBV), was recovered from S2-GMR cells by PCR, as described previously, and cloned. Segment A of DBV was 3,258 nucleotides long and encoded a polyprotein homologous to the known birnaviral structural proteins and an N-terminal overlapping protein, which, however, exhibited no similarities to the N-terminal overlapping proteins encoded by several birnaviruses (33). Segment B was 3,014 nucleotides long and encoded the viral RdRP (Fig. 2B). Sequence and phylogenetic analyses indicate that DBV is clearly distinct from all the known birnaviruses, including DXV, the only reported birnavirus isolated from an insect host (33). For example, neither of the predicted RdRP and structural proteins of DBV shares greater than 31% identities with any member of the three known birnaviral genera (Fig. S2B). Thus, we suggest that DBV represents a unique species and genus in the Birnaviridae.
The last siRNA cluster contained two contigs (Fig. 2C) encoding proteins homologous to the RdRP of Euprosterna elaeasa virus (EEV) from the Tetraviridae, members of which contain a +RNA genome. The combined length of the two contigs was 892 nucleotides. Repeated attempts to recover the viral genome from the S2-GMR cell line established at the University of California, Riverside, by RT-PCR were unsuccessful. However, inclusion of an additional small RNA library [NCBI–Gene Expression Omnibus (GEO): accession no. GSM 272653] constructed from Drosophila Kc cell line originally obtained from Gerald Rubin (12) in the assembly yielded a long contiguous contig 3,005 nucleotides in length. This long contig contained the 2 initially identified contigs and 17 additional contigs in the S2-GMR libraries that did not exhibit detectable homology to known viral proteins (Fig. 2C, red and gray bars). The assembled siRNA consensus sequence encoded a protein of 984 residues, which shared ≈29% identities with the RdRP of both EEV and Thosea asigna virus (TAV) in the Tetraviridae. Further phylogenetic analysis of the RdRPs in the Tetraviridae (Fig. S2C) suggests that the identified virus, designated Drosophila tetravirus (DTrV), represents a unique species in the Tetraviridae.
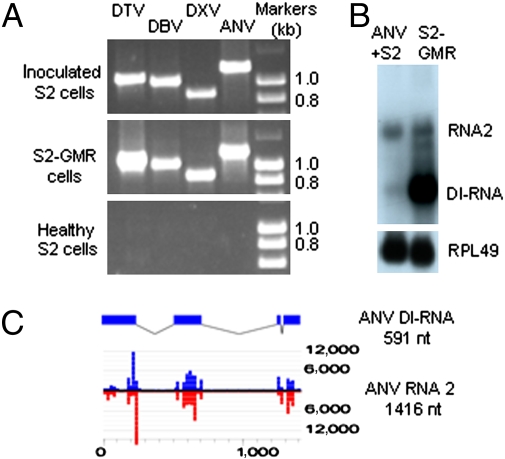
These results indicate that the S2-GMR cells used for library construction were persistently infected with five RNA viruses belonging to four different virus families. As expected, we showed that the S2-GMR cell line established subsequently at the University of California, Riverside, indeed contained infectious DXV, ANV, DTV, and DBV, but not DTrV, by inoculation of healthy S2 cells followed by RT-PCR analysis (Fig. 3A). These results explained that although we were successful in obtaining the full-length genome sequences of ANV, DTV, and DBV, our repeated attempts to recover DTrV failed. We found that 388,289 (6%) of the total 6,454,759 reads from the S2-GMR cells and 220 of the 1,639 assembled contigs were mapped to the five viruses. The most predominant species of small RNAs derived from either the three dsRNA viruses (DTV, DBV, and DXV) or the two +RNA viruses (ANV and DTrV) was 21 nucleotides in length (Fig. S3A), and the ratios of (+) and (−) 21-nt viral siRNAs were approximately equal (Fig. S3B). These features of virus-derived small RNAs were similar to those of FHV-derived siRNAs produced by DCR2 (11), suggesting a shared biogenesis pathway for viral siRNAs targeting +RNA and dsRNA viruses in D. melanogaster.
Fig. 3.
S2-GMR cells contained four infectious RNA viruses. (A) DTV, DBV, DXV, and ANV were all detected by RT-PCR in noncontaminated S2 cells 4 days after inoculation with the supernatant of the S2-GMR cells. Healthy S2 cells and the S2-GMR cells were used as controls. Primers used for RT-PCR were expected to yield specific products of 1,087, 1,030, 865, and 1,212 base pairs in length from DTV, DBV, DXV, and ANV, respectively. (B) Detection of an abundant DI-RNA derived from ANV RNA2 in S2-GMR cells (Right) that were much less abundant in S2 cells after inoculation with the supernatant of S2-GMR cells (Left) by Northern blot hybridizations using a probe recognizing the 3′-terminal 120 nucleotides of RNA2. (C) Structure of the cloned DI-RNA of ANV (Upper) and mapping of the perfectly matched 21-nt siRNAs sequenced from S2-GMR cells to the positive (blue) and negative (red) strands of ANV RNA2 (20-nt windows) (Lower).
There were major differences in the relative abundance of siRNAs derived from each of the five viruses, with 56%, 18.1%, 17.1%, 5.7%, and 3.4% of the total viral siRNA assigned to ANV, DTV, DBV, DXV, and DTrV, respectively. Further analysis indicated that the highest siRNA density targeting ANV was most likely attributable to the presence of a 591-nt defective-interfering RNA (DI-RNA) derived from ANV. We cloned the DI-RNA and found that 51% of the total viral siRNAs of the S2-GMR cells was mapped to the three siRNA peaks of ANV RNA2, which corresponded precisely to the regions of RNA2 (nucleotides 1–245, 515–712, 1,250–1,277, and 1,297–1,416) present in the DI-RNA (Fig. 3 B and C). Northern blot hybridization (Fig. 3B) revealed that the DI-RNA replicated to high levels in S2-GMR cells (Fig. 3B, Right) but to a much lower level in fresh healthy S2 cells inoculated with the supernatant of the S2-GMR cells (Fig. 3B, Left), indicating that high replication levels of DI-RNA may be a key feature of the mix viral infection in the S2-GMR cells. In addition to the DI-RNA-derived siRNA peaks in ANV RNA2, the distribution of viral siRNAs was not uniform along the remaining viral genomic RNAs (Fig. S4), as noted previously (16 –20). However, our analyses indicated that the high siRNA density regions of the viral RNA genomes were associated with neither unusual adenine+uridine (AU) content nor strong secondary structures.
Assembly of siRNAs and Virus Discovery in Mosquitoes and C. elegans
We next determined if vdSAR also worked in other invertebrates. The mosquito small RNA library reported by Myles et al. (22) contained 3,771,297 reads of 18–26 nucleotides in length, representing 756,219 unique sequences. Except for the 19 contigs mapped to the Sinbis viral genome, no additional virus-specific contigs were identified by BLASTN. However, BLASTX searches of the remaining 435 contigs of small RNAs identified two contigs (Fig. 2D) encoding proteins exhibiting 54% and 72% similarities to the CP precursor of Wuhan nodavirus (WNV). WNV, an insect virus identified recently, is an unassigned member of the Nodaviridae (34, 35). The combined length of the two contigs was 1,103 nucleotides, and the encoded protein covered 83% of, and shared 41.6% identity with, the WNV CP precursor. Thus, the identified virus may represent a unique virus, designated as mosquito nodavirus (MNV). Phylogenetic analysis of the nodaviral CPs indicates that MNV does not belong to either of the established genera in the Nodaviridae (Fig. S2D).
The total small RNAs we sequenced from C. elegans strain N2 were assembled into 117 contigs in total. However, except for the 29 FHV-specific contigs, no additional virus-specific contigs were identified by either BLASTN or BLASTX. Similarly, none of the 172 contigs assembled from a large library of 10,964,021 small RNAs constructed from mix stages of C. elegans (36) exhibited detectable similarities to known viruses. This suggests that the common laboratory strain of C. elegans might not be persistently infected with an RNA virus of sufficient homology detectable by vdSAR.
Detection and Assembly of Virus-Derived piRNAs in Drosophila Ovary Somatic Sheet Cells.
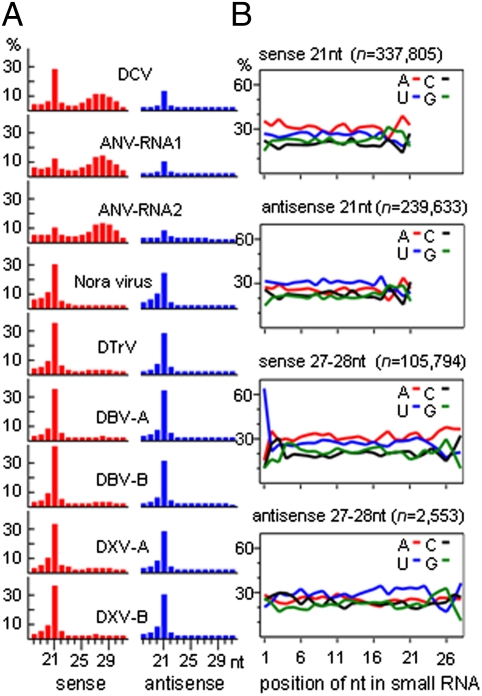
We further carried out assembly of the small RNAs sequenced recently from a Drosophila ovary somatic sheet (OSS) cell line (37). Unlike S2 cells isolated originally from late embryonic stages, which do not express any of the three PIWI subfamily members, OSS cells produce abundant primary piRNAs of 24–30 nucleotides in addition to siRNAs and miRNAs because of the expression of the PIWI protein (37 –39). BLASTN searches of the assembled contigs readily identified six RNA viruses in the OSS cells. These include DXV, ANV, DBV, and DTrV, all of which were also detected in S2-GMR cells, as well as DCV and Nora virus. DCV and Nora virus belong to different +RNA virus families and both share similarities with picornaviruses. A common source of virus contamination for the two cell lines might be extracts from infected flies used in cell culture (37). BLASTX searches of the remaining assembled contigs did not identify additional viruses. Of the total 36,389,371 reads from the OSS cells, 3.3% were mapped to the six viruses. Among the 1,184,811 total viral siRNAs, 31.4%, 26.9%, 17%, 13.5%, 7.1%, and 4% came from DCV, Nora, DXV, DBV, ANV, and DTrV, respectively. Thus, ANV was not the predominant target for dicing in the OSS cells, and, consistently, mapping of the siRNAs to individual genomic RNAs did not identify the three siRNA peaks corresponding to the regions specific to the DI-RNA of RNA2 detected in the S2-GMR cells.
Notably, we found a previously underscribed population of virus-derived small RNAs in the OSS cells (Fig. 4A) that was not detected in S2-GMR cells. We suggest that they represent virus-derived piRNAs because of the following three shared features with the endogenous primary piRNAs detected in OSS cells (37 –39). First, these viral piRNAs were 24–30 nucleotides in length with two peaks at 27 and 28 nucleotides (Fig. 4A). Second, viral piRNAs exhibited strong 5′-uridine bias (∼63%) but no preference for adenine at the 10th position (Fig. 4B); thus, they were distinct from Drosophila ovary secondary piRNAs loaded in AGO3, 73% of which have adenine at the 10th position (38). Third, viral piRNAs were almost exclusively (95%) of one polarity (Fig. 4A). By comparison, viral siRNAs were shorter than viral piRNAs and exhibited no strand bias or preference for a particular nucleotide at any position. In addition, the relative abundance of viral piRNAs was highly variable among the six RNA viruses persistently infecting the OSS cells. Viral piRNAs targeting ANV and DCV were much more abundant than those targeting the remaining four viruses. Strikingly, ANV-specific piRNAs were more than twice as abundant as viral siRNAs in the OSS cells. Nevertheless, piRNAs of all six viruses were highly biased for sense reads, corresponding to either the genomic RNA of +RNA viruses (ANV, DCV, DTrV, and Nora virus) or the mRNA-sense strand of the dsRNA viruses (DXV and DBV), and these viral piRNAs exhibited 5′-uridine bias in only sense but not antisense reads (Fig. 4B).
Fig. 4.
Size distribution (A) and aggregate nucleotide composition (B) of virus-derived small RNAs in Drosophila OSS cells. For each viral genome or genome segment, the percent of either sense (red bar) or antisense (blue bar) viral small RNAs of distinct sizes over total reads with a length of 18–31 nucleotides with a perfect match is shown in A. Percent aggregate nucleotide compositions for all viral reads of 21-nt siRNA or 27-nt + 28-nt piRNAs were calculated, with the total numbers of reads in each size shown in parentheses.
We next determined if these six viruses could be identified by assembly of viral piRNAs in the absence of viral siRNAs. To this end, 19,334,507 reads (3,298,838 unique sequences) from 25–30 nucleotides were sorted out of the OSS cell libraries. We found that all six viruses were identified by assembly of these siRNA-free piRNA reads followed by BLASTN, regardless of their relative piRNA abundance. Twenty-eight contigs were mapped to ANV, covering 94% and 99% of RNA1 and RNA2, respectively. Ninety-two percent of the DCV genome was represented by 68 assembled contigs. For the remaining four viruses that yielded fewer piRNAs, a total of 205 virus-specific piRNA contigs were identified, covering 83–95% of either the complete genomes of DBV, DXV, and Nora virus or the partial DTrV genome (Fig. S5). However, BLASTX searches of the remaining assembled contigs did not identify additional viruses, including DTV, consistent with the results from the assembly of viral siRNAs.
Discussion
In this study, we describe vdSAR, an approach for virus discovery in invertebrates by deep sequencing and assembly of viral small silencing RNAs produced by host immune machinery in response to infection. vdSAR was based on the observation that viral small silencing RNAs produced by fruit fly, mosquito, and nematode cells were all overlapping in sequence. In this approach, total small RNAs were isolated from a host, sequenced in a single Illumina lane, and assembled into contigs by the Velvet program. Virus-specific contigs were identified by searching the nonredundant nucleotide sequence entries of the National Center for Biotechnology Information (NCBI) both before (BLASTN) and after (BLASTX) in silico translation, and the complete genomes of the viruses identified could subsequently be recovered by PCR and cloned. Use of vdSAR revealed persistent mix infection of Drosophila S2-GMR and OSS cell lines by five and six RNA viruses, respectively. Viral siRNA contigs were also assembled and identified from acutely infected Drosophila S2 cells and adult mosquitoes (Fig. 1 A and C). However, no virus was identified by vdSAR from the N2 laboratory strain of C. elegans. Thus, it may be necessary to examine field isolates (40) for virus discovery in C. elegans, because laboratory maintenance of worm strains often involves multiple rounds of bleaching to start worm cultures from eggs by removing larvae and adult animals and associated microbial contamination.
Five of the viruses assembled from fly and mosquito small RNAs were unique and include three +RNA viruses and two dsRNA viruses. Except for ANV, DBV, DTV, DTrV, and MNV, all exhibited low sequence identities (25–42%) to known viruses that were detectable only in short regions of the encoded viral proteins. As a result, none of the four viruses could be assigned into an existing virus genus. This suggests that vdSAR is capable of discovering viruses that are only distantly related to known viruses. It should be pointed out that viruses discovered by vdSAR from invertebrates may include those human and vertebrate viral pathogens that are transmitted by arthropod vectors. An article published during preparation of this paper reported the identification of two unique DNA viruses in sweet potato plants by an approach similar to vdSAR (41), indicating that vdSAR works in both plants and invertebrates.
Analysis of the recently reported small RNA libraries made in Drosophila OSS cells identified virus-derived piRNAs that have not been previously described in any other host species. This finding suggests that piRNAs may have an antiviral role, in addition to their role in genome defense against transposons (4 –6). However, it is interesting to note that these viral piRNAs are also overlapping in sequence and can be used for viral genome assembly in the absence of viral siRNAs. This suggests that vdSAR is likely to be effective for hosts or host tissues that may produce viral piRNAs only.
Discovery of animal viruses is often hindered by difficulties in their amplification in cell culture and/or lack of their cross-reactivity in serological and nucleic acid hybridization assays to known viruses. Many viruses have been recently discovered in environmental and clinical samples using metagenomic approaches, in which viral particles are first partially purified and viral nucleic acid sequences are randomly amplified before subcloning and sequencing (42 –44). Both the metagenomic approaches and vdSAR are culture-independent and can identify viruses that share only low sequence similarities with the known viruses. By comparison, vdSAR requires neither viral particle purification nor viral nucleic acid sequence amplification. Moreover, vdSAR involves sequencing of the fraction of host small RNAs and data mining of only those small RNAs that can assemble into contigs, such that both the amount of sequencing and data complexity are greatly reduced. Importantly, vdSAR assembles viral genomes from the products of an active host immune response to infection. Thus, only the replicating and infectious viruses that induce the immune response are identifiable by vdSAR.
Given the genetic and structural diversity of the characterized viruses, it is possible that there are viral and subviral pathogens yet to be discovered that exhibit no similarity to any of the known viruses detectable by the available bioinformatic tools. These pathogens would readily escape detection by the current homology-dependent metagenomic approaches and vdSAR. Indeed, a number of the assembled contigs from the Drosophila cells exhibit no detectable similarity to entries in the NCBI database. In this regard, the unique features of vdSAR may facilitate development of bioinformatic tools for selecting particular contigs for virus discovery. For example, small RNA densities, small RNA size distribution patterns, and positive/negative strand ratios of small RNAs in the assembled contigs that are consistent with viral small silencing RNAs may all be considered as indicators of contigs with a viral origin.
Materials and Methods
Cell Culture.
Culture, virus infection of S2 cells, and Northern blot hybridization were as described (11). The S2-GMR cell line was kindly provided by one of the authors (E.C.L.). The supernatant of S2-GMR cells established at the University of California, Riverside, was used for infection of fresh healthy S2 cells.
Sequencing, Assembly, and Analysis of Small RNA Libraries.
The small RNA library of C. elegans was constructed as described (45) and sequenced by the Illumina 2G Analyzer. Other small RNA libraries were retrieved from the GEO database. The genome sequence of D. melanogaster and a repeat annotation file were downloaded from the University of California, Santa Cruz Genome Browser website. The nonredundant protein (nr) and nucleotide (nt) sequence databases were downloaded from the NCBI (updated on January 12, 2009). The Velvet program (28) was downloaded from the European Bioinformatics Institute (EBI). Mapping of small RNAs and assembled contigs to fly and viral genomes was done with the BLASTN program using the standard parameters in genome assembly (contigs or viral contig with ≥90% similarity and ≥90% coverage of contigs). Assembled contigs were also examined for similarity of their encoded proteins to databases using the BLASTX program. Additional data analyses were carried out with in-house Perl scripts. The computation analyses were carried out using the campus Genomics Institute Core Facility for Bioinformatics.
RT-PCR, RACE-PCR, and Sequencing.
RT and PCR were used to fill the gaps between siRNA contigs using primers designed according to the consensus sequences of the specific contigs involved and their relative positions mapped in the closely related viral genome. RT-PCR products were sequenced directly by conventional Sanger dideoxyl sequencing. In accordance with the instructions of the manufacturer (Invitrogen), 5′ RACE was carried out. For 3′-RACE, the total RNA was isolated from fly cells by the TRIzol protocol; denatured at 65°C for 5 min; and ligated to a preadynated 3′ adaptor, ppACACTCGGGCACCAAGGA (linker2; IDT Company) with T4 RNA ligase-truncated fragment (New England Biolabs) (46). Following ethanol precipitation, the ligation products were reverse-transcribed by SuperScript III (Invitrogen), amplified by PCR. Before Sanger dideoxyl sequencing, 5′ RACE and 3′ RACE products were cloned in pGEM-T easy vector (Promega). The Phred-Phrap-consed package (47-49) was used for virus genome assembly.
Phylogenetic Analysis.
The Mega 4 package (50) was used to build the phylogenetic trees. Alignment of proteins was performed with the Clustal W method, and the phylogenetic tree was calculated by using the neighbor-joining method. The reliability of each branch was evaluated with bootstrap (1,000 times repeat). We have recently become aware that Drosophila tetravirus (DTrV) described in this study is identical in sequence to Drosophila A virus reported by Karyn Johnson and colleagues (51).
Supplementary Material
Acknowledgments
We thank Kevin Myles for providing the published small RNA libraries made from mosquitoes. This work was supported by National Institutes of Health grant AI052447 (to S.D.) and National Research Initiative of the US Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2007-35319-18325 (to S.D.).
Footnotes
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. GQ342961 (DTV), GQ342962 (DBV-A), GQ342963 (DBV-B), GQ342964 (DTrV), GQ342965 (ANV-RNA1), GQ342966 (ANV-RNA2), and GU144510 (MNV)].
The authors are declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911353107/DCSupplemental.
References
- 1.Aliyari R, Ding SW. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol Rev. 2009;227:176–188. doi: 10.1111/j.1600-065X.2008.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlotshwa S, Pruss GJ, Vance V. Small RNAs in viral infection and host defense. Trends Plant Sci. 2008;13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siomi MC, Saito K, Siomi H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 2008;582:2473–2478. doi: 10.1016/j.febslet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 6.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 8.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li HW, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 11.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynt A, Liu N, Martin R, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci USA. 2009;106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 15.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Molnár A, et al. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho T, Pallett D, Rusholme R, Dalmay T, Wang H. A simplified method for cloning of short interfering RNAs from Brassica juncea infected with Turnip mosaic potyvirus and Turnip crinkle carmovirus. J Virol Methods. 2006;136:217–223. doi: 10.1016/j.jviromet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Qi X, Bao FS, Xie Z. Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS One. 2009;4:e4971. doi: 10.1371/journal.pone.0004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donaire L, et al. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 2009;392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Wang XB, et al. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0904086107. 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105:19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez-Vargas I, et al. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol. 2008;82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans . PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li TC, Scotti PD, Miyamura T, Takeda N. Latent infection of a new alphanodavirus in an insect cell line. J Virol. 2007;81:10890–10896. doi: 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulos BT, Tang KF, Pantoja CR, Bonami JR, Lightner DV. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J Gen Virol. 2006;87:987–996. doi: 10.1099/vir.0.81127-0. [DOI] [PubMed] [Google Scholar]
- 32.Hanizlik TN, et al. Totiviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy—Eighth Report of the International Committee on Taxonomy of Viruses. San Diego: Academic; 2005. pp. 873–883. [Google Scholar]
- 33.Delmas B, et al. Birnaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy—Eighth Report of the International Committee on Taxonomy of Viruses. San Diego: Academic; 2005. pp. 561–569. [Google Scholar]
- 34.Liu C, et al. Isolation and RNA1 nucleotide sequence determination of a new insect nodavirus from Pieris rapae larvae in Wuhan city, China. Virus Res. 2006;120:28–35. doi: 10.1016/j.virusres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, et al. Sequence analysis of coat protein gene of Wuhan nodavirus isolated from insect. Virus Res. 2006;121:17–22. doi: 10.1016/j.virusres.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau NC, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 39.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 40.Troemel ER, Félix MA, Whiteman NK, Barrière A, Ausubel FM. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 2008;6:2736–2752. doi: 10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreuze JF, et al. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology. 2009;388:1–7. doi: 10.1016/j.virol.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- 43.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 2008;4:e1000163. doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 45.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Q, et al. Poly A-transcripts expressed in HeLa cells. PLoS One. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 48.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 49.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 51.Ambrose RL, et al. Drosophila A virus is an unusual RNA virus with a T = 3 icosahedral core and permuted RNA-dependent RNA polymerase. J Gen Virol. 2009;90:2191–2200. doi: 10.1099/vir.0.012104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.