Abstract
Control of organ size by cell proliferation and survival is a fundamental developmental process, and its deregulation leads to cancer. However, the molecular mechanism underlying organ size control remains elusive in vertebrates. In Drosophila, the Hippo (Hpo) signaling pathway controls organ size by both restricting cell growth and proliferation and promoting cell death. Here we investigated whether mammals also require the Hpo pathway to control organ size and adult tissue homeostasis. We found that Mst1 and Mst2, the two mouse homologs of the Drosophila Hpo, control the sizes of some, but not all organs, in mice, and Mst1 and Mst2 act as tumor suppressors by restricting cell proliferation and survival. We show that Mst1 and Mst2 play redundant roles, and removal of both resulted in early lethality in mouse embryos. Importantly, tumors developed in the liver with a substantial increase of the stem/progenitor cells by 6 months after removing Mst1 and Mst2 postnatally. We show that Mst1 and Mst2 were required in vivo to control Yap phosphorylation and activity. Interestingly, apoptosis induced by TNFα was blocked in the Mst1 and Mst2 double-mutant cells both in vivo and in vitro. As TNFα is a pleiotropic inflammatory cytokine affecting most organs by regulating cell proliferation and cell death, resistance to TNFα-induced cell death may also contribute significantly to tumor formation in the absence of Mst1 and Mst2.
Keywords: Hippo signaling, tumor suppressor, Yap phosphorylation
During development and regeneration, the determinants of vertebrate organ size are poorly understood, but include extrinsic and intrinsic factors that coordinate cell proliferation and cell death (1 –4). Recent studies in Drosophila suggest that the Hippo (Hpo) signaling pathway plays an important role in organ size control (5, 6). Central to the Hpo pathway is a kinase cascade consisting of Hpo, Salvador (Sav), Warts (Wts), and Mats (6, 7). Mutations in any of these genes resulted in overgrowth of adult appendages, including eyes and wings (8 –16). Hpo, also called dMst (9), is the Drosophila homolog of mammalian Ste20 family kinases Mst1 and Mst2, and forms a complex with the WW-repeat scaffolding protein Sav to phosphorylate and activate the downstream kinase Wts/Lats. Wts/Lats acts in association with a small regulatory protein Mats to restrict cell growth and proliferation and promote cell death primarily by regulating Yorkie (Yki), the Drosophila homolog of mammalian transcriptional coactivator YAP and TAZ (17 –19). Hpo/Wts-mediated phosphorylation of Yki restricts its nuclear localization (20 –22), and Yki interacts with the TEAD/TEF family transcription factor Scalloped (Sd) to regulate Hpo pathway target genes (21, 23, 24).
The Hpo pathway is evolutionarily conserved, and the vertebrate Hpo pathway has been implicated in regulating cell contact inhibition, organ size, and tumorigenesis (6, 7). Yap is the primary effector of the mammalian Hpo pathway (20, 25–27). YAP in human is a candidate oncogene that is amplified in several types of tumor (28, 29). YAP protein levels and/or nuclear localization are elevated in many human cancers (20, 30). Yap nuclear localization can be regulated by cell-cell contact through Lats-mediated phosphorylation (20, 25). Overexpression of an activated and nuclear localized form of Yap in mouse liver increased liver size (20, 26), which ultimately resulted in hepatocellular carcinoma (20). As in Drosophila, the TEAD family of transcription factors mediates the function of Yap and Taz in mammalian cells (31 –34).
Here we have addressed the physiological role of the mammalian Hpo pathway mediated by Mst1 and Mst2 during embryonic development and adult tissue homeostasis by genetically removing Mst1 and Mst2 in mouse embryos and adult tissues. We found that Mst1 and Mst2 play redundant roles and are required for mouse embryonic development, size control of some organs, and tumor suppression. We also provide evidence that resistance to TNFα-induced cell death may contribute significantly to tumor formation in the absence of Mst1 and Mst2.
Results
Mst1 and Mst2 Play Redundant Roles in Mammalian Embryonic Development.
To investigate the function of the mammalian Hpo signaling pathway in vivo (6, 7), we generated conditional alleles of Mst1 (Mst1c) and Mst2 (Mst2c). The mutant Mst1Δ and Mst2Δ alleles were derived from the conditional alleles using the Sox2-Cre line (35) (Fig. S1A). Because the sequences encoding the kinase domains were deleted in the Mst1Δ and Mst2Δ alleles, and Mst1 and Mst2 proteins were not detected in the Mst1Δ/Δ and Mst2Δ/Δ embryos (Fig. S1B), the Mst1Δ and Mst2Δ are null alleles. The Mst1Δ/Δ and Mst2Δ/Δ mice were viable, morphologically normal, and fertile. This is due to the functional redundancy of Mst1 and Mst2 in embryonic development as the Mst1Δ/Δ;Mst2Δ/Δ mice were embryonically lethal (Table S1 and Fig. S2). The double-mutant embryos were reduced in size, developmentally delayed, and died between E9.5 and 11.5 (Table S2 and Fig. S2A), possibly due to hematopoietic defects, because the yolk sac and the embryo were very pale compared with the control (Fig. S2B). The Mst1Δ/Δ;Mst2Δ/+ and Mst1Δ/+;Mst2Δ/Δ mice were also viable, morphologically normal, and fertile. These findings suggest that Mst1 and Mst2 play redundant and essential roles in early embryonic development, and that one copy of either Mst1 or Mst2 is sufficient to support normal embryonic development.
We then examined Yap phosphorylation in primary mouse embryonic fibroblast (MEF) cells and hepatocytes. Yap phosphorylation was significantly reduced when Mst1 and Mst2 were removed in hepatocytes, but not in MEF cells (Fig. S1C). Using a TEAD/Sd luciferase reporter (21), however, we found that the transcriptional activity of Yap was increased in the Mst1/Mst2 MEF cells (Fig. S1D), suggesting that Mst1 and Mst2 may be required differentially for Yap phosphorylation and TEAD activity inhibition in vivo.
Mst1 and Mst2 Regulate the Size of Certain Organs.
Because early lethality of the Mst1Δ/Δ;Mst2Δ/Δ mutant embryos precludes the assessment of the role of Mst1 and Mst2 at later stages of development, particularly in regulating mammalian organ size and tissue homeostasis, we employed tissue-specific Cre lines to remove Mst1 and Mst2 in specific organs. Surprisingly, when Mst1 and Mst2 were removed from the developing limb mesoderm by the Prx1-Cre (36) (Fig. S3 A and B), the limb was actually shorter with shorter skeletal elements (Fig. S3C). However, there were no obvious defects in skeletal development (Fig. S3D) except that the cell size appeared to be smaller and cell density was increased in the cartilage of the Mst1Δ/Δ;Mst2c/Δ;Prx1-Cre embryo (Fig. S3 D and E). Chondrocyte proliferation, as examined by Ki67 expression, was significantly increased in the mutant embryo (Fig. S3F). Interestingly, in the mutant, Ki67 staining was also detected in the enlarged hypertrophic chondrocytes, which had exited cell cycle and undergone terminal differentiation in the control embryo. Thus, the mammalian Hpo signaling pathway is not required for hypertrophic differentiation of chondrocytes, but is required for these cells to exit cell cycle during hypertrophy. Cell death examined by the TdT-mediated dUTP nick end labeling (TUNEL) assay was detected in the hypertrophic chondrocytes at the chondro-osseous junction in the control embryo, whereas in the mutant, it was reduced (Fig. S3G). Despite increased cell proliferation and reduced cell death, the cartilage size of the mutant was not increased, indicating that cell proliferation and death are not the only determining factors in vertebrate organ size control.
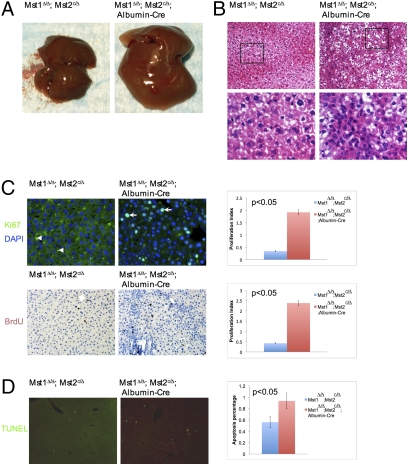
To test whether loss of Mst1 and Mst2 can lead to increased sizes in other organs, we removed Mst1 and Mst2 in all organs in newborn mouse pups with the tamoxifen (TM)-inducible CAGGCre-ER line (37). Injecting TM into the mouse pups induced substantial gene deletion in the liver, stomach, small intestine, skin, lung, heart, spleen, and kidney when assessed by molecular analyses and the R26R mouse line (38) (Fig. S3 H–J). No obvious change was observed in the size of total body, limb, and skeleton in the Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice. However, the liver and stomach were always enlarged 1–7 months after TM-induced gene inactivation. The heart and spleen were also enlarged, albeit less frequently. We never observed enlarged lung, kidney, and intestines (n = 8). Furthermore, removing Mst1 and Mst2 specifically in the liver by the Albumin-Cre (39) resulted in significant liver enlargement and dysplasia at 1 month of age (Fig. 1A). Hepatocytes in the mutant were smaller and cell densities were higher. Some cells showed vacuolated cytoplasm (Fig. 1B). Cell proliferation was increased (Fig. 1C), but cell death was only slightly increased in the mutant liver (Fig. 1D). These findings suggest that Mst1 and Mst2 are required to restrict cell proliferation, but their control of organ size is tissue specific in mammals.
Fig. 1.
Removal of Mst1 and Mst2 in the liver. (A) Livers from 1-month-old mice. (B) H&E staining of liver sections. The boxed region is shown in higher magnification in the lower panel. The cells in the Mst1Δ/Δ;Mst2c/Δ;Albumin-Cre liver were smaller and the density was higher. (C) Cell proliferation shown by Ki67 (green, arrow) and BrdU staining (brown, Lower). More Ki67- or BrdU-positive cells were found in the Mst1Δ/Δ;Mst2c/Δ;Albumin-Cre liver. Autofluorescence blood cells were indicated by arrowhead. (Right) Statistical analysis. (D) TUNEL assay of liver sections. A slight increase in cell death (arrow) was detected in the mutant liver as quantified in the right panel.
Loss of Mst1 and Mst2 Leads to Tumor Formation in the Liver.
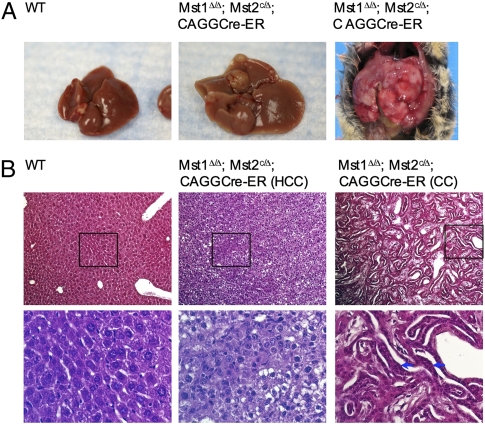
To test whether increased cell proliferation in the absence of Mst1 and Mst2 eventually led to tumor formation, we let the TM-injected Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice and their wild-type littermate mice age. By 6 months, liver tumors developed in all Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice (6/6) (Fig. 2A). No tumor was found in the heart, lung, kidney, small intestine, or skin. In addition, all TM-injected wild-type mice were normal and tumor free. The liver tumor nodules displayed characteristics of hepatocellular carcinoma (HCC). The cells in the nodule exhibited abnormal hematoxylin and eosin (H&E) staining, cellular pleiomorphism (some enlarged cells were mixed with small cells), and/or loss of cytoplasmic staining (Fig. 2B). In some liver tumor samples, cholangiocarcinoma (CC), composed of mainly multilayered biliary epithelial cells, was also found (Fig. 2B). Significantly increased cell proliferation was found in the liver of the Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice (Fig. 3A). However, cell death was also increased in the liver tumors (Fig. 3B), possibly secondary to cell overproliferation. These findings suggest that Mst1 and Mst2 suppress tumors by restricting cell proliferation. To test whether tumors form spontaneously as a result of loss of heterozygosity of Mst1 or Mst2, we let the Mst1Δ/Δ;Mst2c/Δ, Mst1Δ/Δ;Mst2c/c mice and their Mst1Δ/+;Mst2Δ /+ control littermates age (n > 30). By 12 months, only liver tumors were found in the Mst1Δ/Δ;Mst2c/Δ mice (2/31) (Fig. S4A). Because Mst2 could not be detected in the tumor nodules in the Mst1Δ/Δ;Mst2c/Δ liver, the tumor likely resulted from loss of Mst2 heterozygosity in the absence of Mst1 (Fig. S5B). These findings show that Mst1 and Mst2 are potent tumor suppressors, in the absence of which liver tumors form preferentially.
Fig. 2.
Liver tumors form in the absence of Mst1 and Mst2. (A) Livers from 6-month-old TM-injected wild-type and mutant mice. Two of the mutant livers with tumor nodules are shown. (B) H&E staining of liver sections from the liver samples in A. Boxed regions are shown in the lower panel in higher magnification. Both hepatocyte carcinoma (HCC) and chonlangiocyte carcinoma (CC) were observed. Multilayered epithelial cells are indicated by arrows.
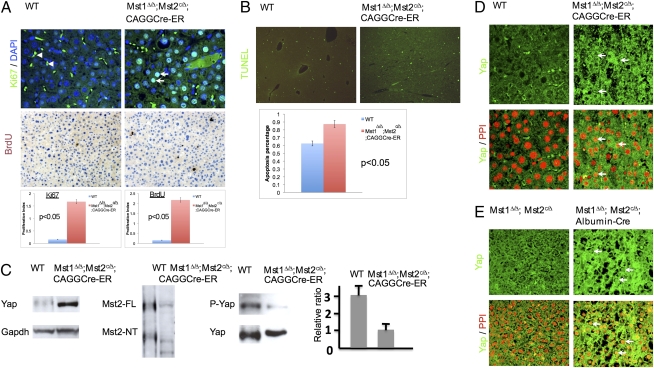
Fig. 3.
Increased cell proliferation and reduced Yap phosphorylation in the liver tumor. (A) Ki67 (green) and BrdU staining (brown) of liver sections from 6-month-old TM-injected wild-type and Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice. In the HCC regions, both Ki67- and BrdU-positive cell numbers were increased as shown by quantification. DAPI (blue) stains the nucleus. Autofluorescence blood cells were indicated by arrowhead. (B) TUNEL assay of liver sections as in A. Cell death (green) was increased in the HCC region as shown by quantification. (C) Western blot of samples from normal (wild-type) and tumor (Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER)-bearing livers. GAPDH was control for total protein, and Yap phosphorylation levels were normalized by the total Yap protein. The phosphorylated YAP to total YAP protein ratio was significantly reduced in the mutant as quantified by densitometry. (D and E) Immunohistochemistry of Yap (green) on the liver sections. Yap protein levels and nuclear localization (arrow) were increased in the mutants. (Lower) Nucleus stained red by propidium iodide (PPI, red).
Because expression of an active form of Yap (YapS127A) leads to increased liver size and ultimately liver tumor formation (20), we tested whether increased cell proliferation and tumor formation in the liver of the Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice are associated with reduced Yap phosphorylation by analyzing liver samples with immunohistochemistry and Western blots. YAP protein levels were increased and Yap phosphorylation was significantly reduced in the primary Mst1Δ/Δ;Mst2Δ/Δ hepatocytes and liver tumor nodules (Fig. S1C and Fig. 3C). In addition, Yap nuclear localization was increased in pretumor hepatocytes (Fig. 3E) and liver tumor nodules (Fig. 3D and Fig. S4B).
Cellular and Molecular Alterations in the Mst1 and Mst2 Mutant Liver.
To understand the molecular mechanism underlying tumor formation in the absence of Mst1 and Mst2, we examined expression of genes implicated in Yap-regulated proliferation and cell death in the liver (21). In both the liver tumor nodules and pretumor mutant liver samples (Fig. S4 C and D), Survivin mRNA expression was significantly up-regulated. We did not observe consistent up-regulation of cMyc, cIAP, and Sox4 expression. Interestingly, we also found that in the liver tumor nodules and pretumor mutant liver tissue, β-catenin levels were increased and nuclear localization of β-catenin was observed (Fig. S4 E and F). In addition, loss of Mst1 and Mst2 led to increased β-catenin transcription activity in the MEF cells (Fig. S4G). These findings suggest that the Wnt/β-catenin signaling activity, which is oncogenic by itself (40 –42), was also increased in the absence of Mst1 and Mst2.
The presence of both HCC and CC in the liver of Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice suggests that these tumors may have originated from liver progenitor cells that give rise to both hepatocytes and cholangiocytes (biliary epithelial cells). When a stem/progenitor cell on its way to differentiation develops into cancer, this will give rise to tumors with a full range of phenotypes with varying hepatocellular and cholangiacellular differentiation characteristics (43). Liver stem cell/progenitor cells are hard to find in the wild-type adult liver, and they express cytokeratin 19 (CK-19) and alpha-fetoprotein (AFP) (43 –47). However, in the liver of the Mst1Δ/Δ;Mst2c/Δ;CAGGCre-ER mice, the number of CK-19- and AFP-expressing cells was dramatically increased (Fig. S4H), suggesting that the HCCs and CCs might result from overproliferation of the stem/progenitor cells.
Loss of Mst1 and Mst2 Renders Liver Resistant to TNFα-Induced Apoptosis.
As cytokines like TNFα play very important roles in regulating hepatocyte proliferation and survival (48, 49), and loss of Mst1 and Mst2 led to liver tumor formation preferentially, we examined TNFα response in the Mst1 and Mst2 mutant cells. Both Mst1Δ/Δ;Mst2Δ/Δ MEF cells and hepatocytes were resistant to TNFα-induced cell death (Fig. S5A). In support of this, we found that in both MEF cells and hepatocytes, TNFα-induced JNK activation was weakened and AKT activation was prolonged, suggesting that JNK and AKT activation by TNFα in regulating cell death might, to a certain extent, requires Mst1 and Mst2 (Fig. S5 B and C). Other immediate readouts of TNFα signaling, including IκB degradation, were normal (Fig. S5D).
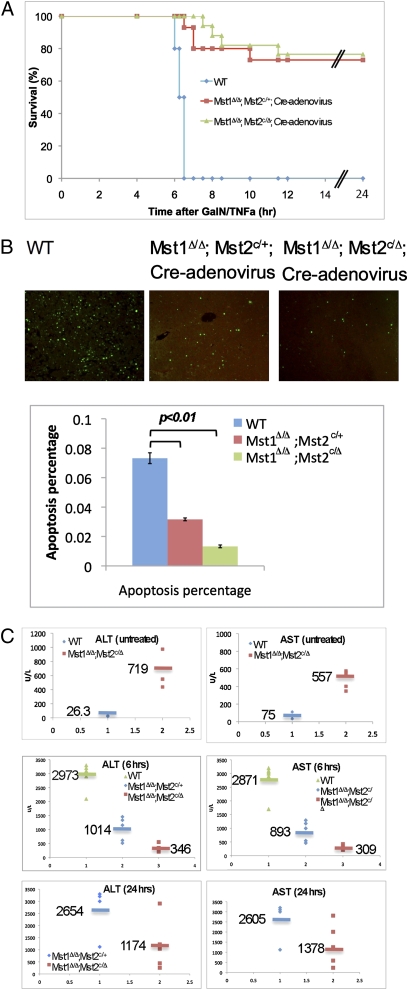
To test whether loss of Mst1 and Mst2 renders liver resistant to TNFα-induced cell death, we first removed the Mst1 and Mst2 in the liver by injecting the Cre-adenovirus into the Mst1Δ/Δ;Mst2c/Δ, Mst1Δ/Δ;Mst2c/+ and wild-type mice. The adenovirus preferentially infects the liver as shown by the injected GFP-adenovirus (Fig. S6A). Acute liver cell death was induced in vivo by injecting TNFα to mice (50). Though all wild-type mice died by 6.5 h postinjection (PI), all Mst1Δ/Δ;Mst2c/Δ littermates were completely protected from TNFα-induced liver injury and appeared healthy until 24 h or even 2 months PI (Fig. 4A). Interestingly, most of the Mst1Δ/Δ;Mst2c/+ mice also survived at 24 h PI and appeared to be healthy (Fig. 4A). Apoptosis was strongly increased in the wild-type livers, but significantly and progressively reduced in the Mst1Δ/Δ;Mst2c/+ and Mst1Δ/Δ;Mst2c/Δ livers (Fig. 4B). Higher circulating concentrations of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in wild-type mice at 6 h PI confirmed the development of liver injury (Fig. 4C). The Mst1Δ/Δ;Mst2c/Δ mice had ALT and AST levels similar to those of untreated ones, whereas the AST and ALT levels in the Mst1Δ/Δ;Mst2c/+ mice were only slightly increased (Fig. 4C). By 24 h PI, the AST and ALT levels were much higher in the Mst1Δ/Δ;Mst2c/+ mice (Fig. 4C). Therefore, the deletion of Mst1 and Mst2 rendered mice resistant to TNFα cytotoxicity in a gene-dose dependent manner. We also notice that the AST and ALT levels in the untreated Mst1Δ/Δ;Mst2c/Δ mice were higher than those in the wild-type control mice (Fig. 4C), suggesting that there was liver inflammation in the Mst1Δ/Δ;Mst2c/Δ mice.
Fig. 4.
TNFα-induced liver toxicity was blocked by loss of Mst1 and Mst2. (A) Survival of Cre-adenovirus-injected wild-type mice (n = 10), Mst1Δ/Δ;Mst2c/+ mice (n = 10), and Mst1Δ/Δ;Mst2c/Δ mice (n = 10) after treatment with GalN and TNFα. (B) TUNEL analysis of liver tissue isolated 6 h after administration of GalN and TNFα. Results represent the staining of four mice from each group. (C) Serum concentration of liver enzymes in nontreated mice or at 6 h and 24 h after treatment with GalN and TNFα (n = 5 mice per group). ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The requirement of Mst1 and Mst2 in promoting cell death was further confirmed by activating the death receptor Fas, a potent inducer of liver apoptosis, through injecting the Jo-2 antibody, a Fas agonist (51 –53). All wild-type control mice died by 4.5 h PI, whereas all Mst1Δ/Δ;Mst2c/Δ and Mst1Δ/Δ;Mst2c/+ mutant mice survived beyond 16 h PI (Fig. S6B). Again, cell death was massive in the wild-type liver, but much reduced in the Mst1Δ/Δ;Mst2c/Δ and Mst1Δ/Δ;Mst2c/+ mutant liver (Fig. S6C). Taken together, these findings suggest that Mst1 and Mst2 are required to promote cell death triggered by external signals such as TNFα in vivo.
Discussion
Here we show that the mammalian Hpo homologs Mst1 and Mst2 are required to control cell proliferation and cell death in both embryonic development and adult tissue homeostasis. Loss of Mst1 and Mst2 leads to increased organ size and tumor formation preferentially in the liver. Mst1 and Mst2 are potent tumor suppressor genes; a single copy of either Mst1 or Mst2 can significantly inhibit tumor formation in the liver.
How appropriate organ sizes are achieved and maintained in development and tissue repair or regeneration is a question that has fascinated many biologists. Organ sizes are controlled by both intrinsic and extrinsic factors, and it is conceivable that extrinsic factors need to act through intrinsic pathways. Mst1 and Mst2 are key players in a critical intracellular signaling pathway controlling cell proliferation and cell death during organ size control and tumorigenesis. Whether Mst1 and Mst2 activities are controlled by extracellular cues or whether they are constitutively active intrinsic regulators remains elusive. In Drosophila, the atypical cadherin Fat (Ft) has been identified as a potential receptor regulating the Hpo pathway (54 –56). However, in mammalian cells, the role of Fats in regulating tissue growth has not been demonstrated. Here we have identified cytokines like TNFα as critical extracellular cues that may act through the mammalian Hpo pathway to regulate cell death. These findings also suggest that Mst1 and Mst2 may play important roles regulating inflammation.
It is important to note that when Mst1 and Mst2 were removed by ubiquitously expressed inducible Cre recombinase, not all organs increased their sizes. Organ sizes are determined by at least the following factors: cell number, size, and density. It has been shown that different vertebrate organs employ distinct mechanisms to control organ sizes (57). For instance, the pancreas organ size is determined by the number of progenitor cells that are set aside early in development, whereas the liver size is not constrained by the early progenitor cell numbers. In both liver and cartilage, we observed reduced cell size and increased cell density when cell proliferation was increased. Thus, it appears that organs can offset the impact of cell number changes on final organ sizes by altering cell size and density, although the underlying feedback mechanism is not clear. In the cartilage, such feedback mechanism is so robust such that the final cartilage is even smaller in the Mst1 and Mst2 mutant limb. In addition, vertebrate organs are more complex in that each organ is composed of a distinct set of different cell types. Interactions among these different cell types also contribute significantly to final organ size. For instance, the limb size is controlled by epithelial-mesenchymal interactions mediated by factors such as the fibroblast growth factors (Fgfs) (58, 59). Such interactions may also be subject to feedback regulations when cell proliferation or cell death is intrinsically altered, for instance, by loss of Mst1 and Mst2.
The rapid development of liver tumor caused by loss of Mst1 and Mst2 or Yap overactivation suggests that Yap is a primary effector of the mammalian Hpo signaling pathway in vivo. Intriguingly, Yap phosphorylation was much less dependent on Mst1 and Mst2 in MEF cells than in hepatocytes, suggesting that Yap phosphorylation does not entirely depend on Mst1 and Mst2. In mice, other Ste20 kinase family members that have lower sequence homologies to Hpo outside of the kinase domain may also phosphorylate YAP, albeit less efficiently. Tissue-specific regulation of Yap phosphorylation and Yap activity may explain why tumors form more quickly in the liver than in other organs, such as the intestine (Fig. S7).
HCC is the fifth most frequent cancer worldwide and the third leading cause of cancer death due to the lack of effective treatment options (60). Surgical removal or liver transplantation is the only curative treatments for HCC, but neither is applicable to most patients with advanced disease. Liver cancer may respond differently from other cancers to standard treatments because of the intrinsic regenerative ability of the liver. The development of HCC and CC is invariably associated with chronic liver damages that can target liver progenitor cells, leading to their expansion and transformation (43, 61). HCCs expressing progenitor cell markers like CK-19 have a more aggressive clinical course. Some studies show that even 5% CK-19 positive cells already affect the outcome of the patient (43). Thus, determining the precise cellular origin of liver tumors is critical in developing the most effective therapeutic strategies. Importantly, we have identified Hpo signaling pathway as a significant target in treating aggressive liver tumors. First, the liver tumors formed in the Mst1 and Mst2 mutants are likely to be caused by increased proliferation of liver stem/progenitor cells that can give rise to both hepatocytes and cholangiocytes. Second, Mst1 and Mst2 mutant cells are resistant to TNFα-induced cell death. TNFα is a major cytokine/growth factor in the liver that promotes liver progenitor cell growth, and it is produced during inflammation and partial hepatectomy (PH) (47). It is conceivable that loss of cell death response to TNFα will significantly augment its proproliferation effect in liver tumorigenesis.
Materials and Methods
Mouse Strains.
The Sox2-cre (35), CAGGCre-ER (37), and R26R (38) mouse lines have been described previously. Genotyping was done by PCR using genomic DNA prepared from the mouse yolk sac or tail.
Tamoxifen (TM) Induction in Mice.
TM (Sigma) was dissolved in corn oil (Sigma) at a concentration of 30 mg/mL. A single dose of tamoxifen (0.2 mg/g body weight) was injected intraperitoneally at postnatal day 10 (P10). Organ samples were fixed in 4% paraformaldehyde overnight at 4 °C and embedded in paraffin and sectioned at 6 μm thickness.
Histological Analysis and Western Blot.
Primary antibodies used in immunohistochemistry include anti-Ki67 mouse monoclonal IgG (Dako) at 1:50, anti-Yap rabbit polyclonal IgG (Santa Cruz) at 1:50, anti-phospho-Yap rabbit polyclonal IgG (Cell Signaling) at 1:50, anti-β-catenin mouse monoclonal IgG (Transduction Lab), anti-Mst2 rabbit polyclonal IgG (Cell Signaling) at 1:50, anti-CK-19 mouse IgG (Dako) at 1:50, and anti-AFP goat IgG (Dako) at 1:25. The signal was detected using FITC-conjugated secondary antibodies (Molecular Probes). Apoptosis was detected as described (62). Additional antibodies used in Western blot: anti-JNK (BD Pharmingen); anti-phospho-JNK (Cell Signaling); anti-Akt and anti-phospho-Akt (Cell Signaling); and anti-GAPDH (Sigma).
Quantitative RT-PCR.
Quantitative PCR was performed to measure the relative mRNA levels using Platinum SYBR Green kit (Invitrogen). Samples were normalized with β-tubulin and actin expression.
Cre-adenovirus and GalN/ TNFα Injection.
Cre-adenovirus was injected at 2 × 109 pfu/mL in 0.15 mL sterile saline into adult mice via retro-orbital sinus using a 27- to 29-gauge needle. The mice were housed for a week before TNFα/GalN injection. The TNFα-induced liver toxicity experiments were performed as described (51). TNFα was administered via retro-orbital sinus. Blood was collected at 6 h and 24 h respectively. Serum was separated and analyzed for aminotransferase activities.
Isolation and Culture of Primary Mouse Hepatocytes and MEF Cells.
Primary hepatocytes were isolated by a two-step collagenase perfusion of the liver (63) from 1- to 2-month-old mice. MEF cells were isolated from E12.5–E14.5 embryos and cultured in DMEM containing 10% FBS.
Apoptosis and Proliferation Assay.
Apoptosis assays of MEF cells were performed by flow cytometry after staining the cells with annexin-V-FITC (Pharmingen) and propidium iodide (Roche) following treatment with 10 ng/mL TNFα and 10 μg/mL cycloheximide for 16 h. Hepatocytes were treated with 30 ng/mL TNFα and 10 μg/mL cycloheximide for 8 h, and fragmented DNA were determined with the Cell Death Detection ELISA Kit (Roche). Proliferation was assessed by the percentage of Ki67- or BrdU-positive cells.
Adenoviral Infection and Luciferase Assay.
Luciferase activity was measured 24–36 h after transfection according to the Dual-Luciferase Reporter Assay System (Promega). The histograms are presented as the average ± SD from three independent transfections. For adenoviral infection, primary hepatocytes or MEF cells were incubated with Cre- adenovirus at 1 × 109 pfu/mL for 2 h in suspension, and analyzed 48–72 h later.
Supplementary Material
Acknowledgments
We thank the Yang Laboratory members for stimulating discussions during the course of the work; Drs. Randy Johnson and Dae-Sik Lim for sharing unpublished results; Drs. Zheng-Gang Liu and Xinwei Wang for insights into TNF signaling and liver tumors; and Julia Fekecs for help in preparing the figures. This work is supported by the Intramural Research Program of the National Human Genome Research Institute. (Y. Y.) and the Welch Foundation grant I-1603 (J. J. ).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911409107/DCSupplemental.
References
- 1.Twitty VC, Schwind JL. The growth of eyes and limbs transplanted heteroplastically between two species of Amybystoma. J Exp Zool. 1931;59:61–86. [Google Scholar]
- 2.Wolpert L. Cellular basis of skeletal growth during development. Br Med Bull. 1981;37:215–219. doi: 10.1093/oxfordjournals.bmb.a071705. [DOI] [PubMed] [Google Scholar]
- 3.Müller-Sieburg CE, Cho RH, Sieburg HB, Kupriyanov S, Riblet R. Genetic control of hematopoietic stem cell frequency in mice is mostly cell autonomous. Blood. 2000;95:2446–2448. [PubMed] [Google Scholar]
- 4.Robin C, et al. An unexpected role for IL-3 in the embryonic development of hematopoietic stem cells. Dev Cell. 2006;11:171–180. doi: 10.1016/j.devcel.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Yue T, Jiang J. Hippo signaling pathway and organ size control. Fly (Austin) 2009;3:68–73. doi: 10.4161/fly.3.1.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 9.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pantalacci S, Tapon N, Léopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 11.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 13.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 14.Tapon N, et al. Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 16.Lai ZC, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Goulev Y, et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 27.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 28.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinhardt AA, et al. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi S, Tenzen T, McMahon AP. Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis. 2003;37:51–53. doi: 10.1002/gene.10225. [DOI] [PubMed] [Google Scholar]
- 36.Logan M, et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 38.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 39.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 40.Takigawa Y, Brown AM. Wnt signaling in liver cancer. Curr Drug Targets. 2008;9:1013–1024. doi: 10.2174/138945008786786127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra L, et al. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roskams T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- 44.Thorgeirsson SS. Hepatic stem cells in liver regeneration. FASEB J. 1996;10:1249–1256. [PubMed] [Google Scholar]
- 45.Fausto N. Hepatocyte differentiation and liver progenitor cells. Curr Opin Cell Biol. 1990;2:1036–1042. doi: 10.1016/0955-0674(90)90153-6. [DOI] [PubMed] [Google Scholar]
- 46.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 47.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 48.Fausto N, Laird AD, Webber EM. Liver regeneration. 2. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 1995;9:1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- 49.Wullaert A, van Loo G, Heyninck K, Beyaert R. Hepatic tumor necrosis factor signaling and nuclear factor-kappaB: Effects on liver homeostasis and beyond. Endocr Rev. 2007;28:365–386. doi: 10.1210/er.2006-0031. [DOI] [PubMed] [Google Scholar]
- 50.Pobezinskaya YL, et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9:1047–1054. doi: 10.1038/ni.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogasawara J, et al. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 52.Leu JI, Crissey MA, Taub R. Massive hepatic apoptosis associated with TGF-beta1 activation after Fas ligand treatment of IGF binding protein-1-deficient mice. J Clin Invest. 2003;111:129–139. doi: 10.1172/JCI16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang YA, Zhang GM, Feigenbaum L, Zhang YE. Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell. 2006;9:445–457. doi: 10.1016/j.ccr.2006.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 55.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Willecke M, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 58.Verheyden JM, Sun X. An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature. 2008;454:638–641. doi: 10.1038/nature07085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scherz PJ, Harfe BD, McMahon AP, Tabin CJ. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- 60.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 61.Alison MR, Lovell MJ. Liver cancer: The role of stem cells. Cell Prolif. 2005;38:407–421. doi: 10.1111/j.1365-2184.2005.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrow JR, et al. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev. 2003;17:394–409. doi: 10.1101/gad.1044903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun R, Jaruga B, Kulkarni S, Sun H, Gao B. IL-6 modulates hepatocyte proliferation via induction of HGF/p21cip1: Regulation by SOCS3. Biochem Biophys Res Commun. 2005;338:1943–1949. doi: 10.1016/j.bbrc.2005.10.171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.