Abstract
Lumen expansion driven by hydrostatic pressure occurs during many morphogenetic processes. Although it is well established that members of the Claudin family of transmembrane tight junction proteins determine paracellular tightness within epithelial/endothelial barrier systems, functional evidence for their role in the morphogenesis of lumenized organs has been scarce. Here, we identify Claudin5a as a core component of an early cerebral-ventricular barrier system that is required for ventricular lumen expansion in the zebrafish embryonic brain before the establishment of the embryonic blood–brain barrier. Loss of Claudin5a or expression of a tight junction-opening Claudin5a mutant reduces brain ventricular volume expansion without disrupting the polarized organization of the neuroepithelium. Perfusion experiments with the electron-dense small molecule lanthanum nitrate reveal that paracellular tightness of the cerebral-ventricular barrier decreases upon loss of Claudin5a. Genetic analyses show that the apical neuroepithelial localization of Claudin5a depends on epithelial cell polarity and provide evidence for concerted activities between Claudin5a and Na+,K+-ATPase during luminal expansion of brain ventricles. These data establish an essential role of a barrier-forming Claudin in ventricular lumen expansion, thereby contributing to brain morphogenesis.
Keywords: brain morphogenesis; lumen formation; Na+,K+-ATPase; tight junctions; cell polarity
Brain morphogenesis in the zebrafish embryo involves a well-studied ventricular lumen expansion process which occurs at early stages of development between 17 and 21 h after fertilization (hpf) (1). Genetic analyses showed that the osmoregulatory ion pump ATPase, Na+/K+ transporting, alpha 1 polypeptide (Atp1a1) is critically important for lumen expansion which suggests a role in the generation of hydrostatic pressure (1). However, the embryonic cerebral barrier expected to maintain luminal fluids and ions within the brain ventricles remains unknown. It is known that the blood–brain barrier forms 2 days after brain ventricle expansion (2). Also, the choroid plexus, which develops from the ependymal layer lining the ventral floor of the cerebral ventricle, is not functional at these stages and forms the blood-cerebrospinal fluid barrier only at later stages and in the adult fish (3, 4). To elucidate the nature of the cerebral-ventricular barrier system required for initial lumen expansion, we focused our attention on the neuroepithelium lining the cerebral cavities. We hypothesized that Claudins (Cldn) may contribute to the cerebral-ventricular barrier based on their established roles in the regulation of tight junction (TJ) barriers (5, 6). Claudins are characterized as either barrier- or pore-forming. In a recent study, Bagnat and colleagues demonstrated an involvement of the pore-forming Cldn15 in gut lumen expansion in the zebrafish embryo (7). Moreover, the C-terminal half of Clostridium perfringens enterotoxin (C-CPE), a polypeptide with inhibitory activity to several barrier-forming Claudins including Cldn3, Cldn4, and Cldn6, affected murine blastocoel cavity expansion, which is another lumen expansion process that requires hydrostatic pressure (8). Together, these studies implied an important function of Claudins in brain ventricular lumen expansion.
Here, we report that Cldn5a is essential for the establishment of an embryonic cerebral-ventricular barrier system within the neuroepithelium lining the brain ventricles. Tightening of this paracellular barrier requires Claudin-Claudin trans interactions via the second extracellular loop of Cldn5a. We propose that during ventricular lumen expansion, Cldn5a seals the neuroepithelial layer TJs and maintains the fluid pressure that depends on Atp1a1 activity. Therefore, concerted activities of these two proteins contribute to brain morphogenesis.
Results and Discussion
claudin5a Is Expressed Within the Brain During Ventricular Lumen Expansion.
There are at least 20 Claudin family members in teleosts (9, 10). Database searches (http://zfin.org/) combined with whole-mount in situ hybridization analyses revealed that two zebrafish clnd5 genes encoding the predicted orthologs of the human four-transmembrane pass TJ protein Cldn5 are expressed in the zebrafish embryo (cldn5a: gi|47086108|; cldn5b: gi|53733860|) (Fig. S1) and that zebrafish cldn5a has a strong neuroepithelial expression (Fig. 1A; http://zfin.org/) whereas cldn5b is endothelial-specific (zgc:103419; http://zfin.org/). At 14 hpf, which is just before brain ventricle formation and initial expansion, cldn5a is expressed within the developing central nervous system with a particularly strong expression in hindbrain and spinal cord regions and some weaker expression in some dorsal portions of the midbrain (Fig. 1A). By 30 hpf, expression of cldn5a is within the entire central nervous system and particularly strong within ventral neuroepithelial cells lining brain ventricles within the hindbrain and some regions of the midbrain and forebrain (Fig. 1A′). This expression pattern suggests a role of Cldn5a within developing ependymal cells lining the brain ventricles. An anti-mammalian Cldn5 antibody that recognizes both Cldn5a and Cldn5b isoforms in the zebrafish revealed strong expression of Cldn5 isoforms within the spinal cord and dorsal aorta (Fig. 1B). Overall, the zebrafish Cldn5a protein sequence is 56% identical and 73% similar to its human protein ortholog and identities are higher for the two extracellular loops (ECLs). These data, combined with the established role of murine Cldn5 in maintaining the integrity of the endothelial blood–brain barrier (11), suggested a potential involvement of Cldn5a in the formation of a cerebral-ventricular barrier in zebrafish.
Fig. 1.
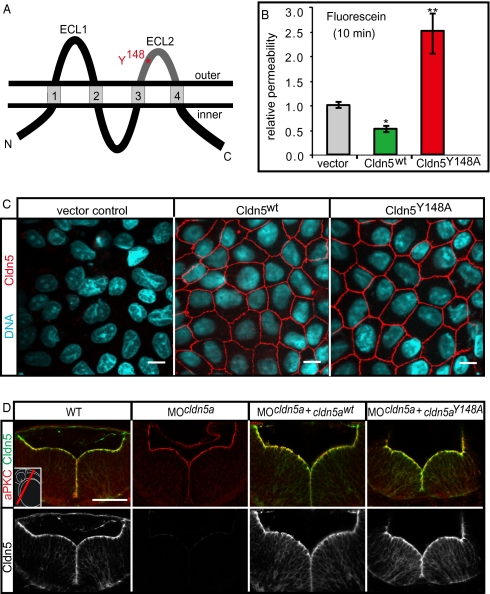
Loss of Claudin5a disrupts the neuroepithelial paracellular barrier function. (A and A′) Whole-mount in situ hybridization of claudin5a (cldn5a) expression at two developmental time points. Dorsal views onto the brain are shown magnified within the insets. (A) Before brain ventricle formation (14 hpf), cldn5a is strongly expressed within the developing central nervous system including the hindbrain (hb) and spinal cord (sc). Weaker expression of cldn5a is found within the dorsal midbrain region (mb). (A′) During ventricle expansion (30 hpf), cldn5a is strongly expressed within the spinal cord and the neuroepithelial ventricular zones of ventral hindbrain and midbrain. There is also strong expression in the forebrain (fb) ventricular zone. (B) Confocal microscopic images of cross-sections through the trunk region. Strong expression of Cldn5 proteins is present within the spinal cord (sc) and the dorsal aorta (da) but not the cardinal vein (cv) at 30 hpf. (C) Injection of MOcldn5a efficiently blocks expression of Cldn5a protein within the apical neuroepithelium lining the brain ventricles (V). Endothelial expression of Cldn5b is not affected in cldn5a morphants (arrows). (D–F) Electron micrographs of neuroepithelial cells covering the cerebral ventricles (V) after intraventricular injection of the electron-dense molecule lanthanum nitrate. In the WT, paracellular clefts are tight for the tracer, which accumulates in a dot-like pattern at the TJ (arrows). Knock-down of cldn5a results in diffusion of lanthanum nitrate into the paracellular space between cells (arrows). In rescue embryos, in which the cldn5a knock-down was rescued by concomitant cldn5a mRNA injection, electron dense material is confined to apico-lateral membranes of neuroepithelial cells similar to the distribution in WT embryos. (Scale bars: B and C, 50 μm; D, 2 μm.)
Loss of Claudin5a Disrupts the Neuroepithelial Paracellular Barrier Function.
To test whether zebrafish Cldn5a has a similar function for the integrity of the brain neuroepithelial layer lining the brain ventricles, we knocked down its expression by using antisense morpholino oligonucleotides (MOs) directed against the 5′UTR of the gene (12). The efficiency of the cldn5a knock-down was verified by using the anti-mammalian Cldn5 antibody (Fig. 1C). Tissue sections through the hindbrain ventricular zone revealed that in wild-type (WT) embryos Cldn5a colocalized together with the apical marker atypical protein kinase C (aPKC) (13) at apical surfaces of neuroepithelial cells lining the brain ventricles (Fig. 1C). Injection of MOcldn5a completely abrogated the neuroepithelial protein expression between 20 and 30 hpf without affecting the apical localization of aPKC (Fig. 1C and Fig. 2D). As expected, the endothelial Cldn5b expression was not affected in cldn5a morphants (Fig. 1C, arrows).
Fig. 2.
The TJ opening Claudin5aY148A mutant increases TJ permeability. (A) Schematic representation of the four-transmembrane pass TJ protein Claudin5a (Cldn5a). The two extracellular loops (ECL), facing the paracellular space, are involved in claudin-claudin trans interactions. The position of an aromatic residue within ECL2, which is conserved between all classic claudins (17) and which is essential for trans interactions, is highlighted in red. ECL2, which is conserved between all classic claudins (17) and which is essential for trans interactions. (B) Stable transfection of murine Cldn5wt reduces the paracellular permeability of fluorescein (filter culture) compared to the vector control, whereas transfection of the mutant Cldn5Y148A causes an increase of fluorescein permeability. Data represent mean ± SEM, n ≥ 10; *, P < 0.05, **, P < 0.01. (C) Stable transfection of murine Cldn5wt and Cldn5Y148A into MDCK-II cells. The Cldn5 variants are strongly enriched at cell-cell contacts as detected by immunocytochemistry against the N-terminal FLAG-tag. (Scale bars: 10 μm.) (D) Injection of mRNA encoding zebrafish Cldn5awt or Cldn5aY148A into cldn5a morphants results in the correct localization of the proteins at the ventricular surface of the 30 hpf neural tube as detected by confocal microscopy of sectioned immunohistochemical stainings. (Scale bar: 50 μm.)
Next, we assessed the possibility that the paracellular barrier of the neuroepithelial layer lining the brain ventricles could be affected by the loss of Cldn5a. To this end, we performed ventricular injections of lanthanum nitrate (molecular mass of 0.5 kDa) at 20 hpf and analyzed electron micrographs of hindbrain sections to detect the localization of this electron-dense material, which is routinely used to assess the integrity of the TJ (14, 15). Whereas in WT embryos, lanthanum nitrate was enriched within intercellular spaces apical from the TJ of neuroepithelial cells and was not detectable basally of the TJ (Fig. 1D), loss of Cldn5a resulted in the permeation of the marker throughout the intercellular space (Fig. 1E). The specificity of this effect could be verified by coinjection of MOcldn5a together with cldn5a mRNA lacking the MO target sequence (rescue), which completely restored the integrity of the TJ and prevented lateral diffusion of lanthanum nitrate (Fig. 1F). The finding that, upon loss of Cldn5a, lanthanum nitrate can diffuse through intercellular spaces is in fair agreement with earlier observations in Cldn5-deficient mice that revealed a leaky blood–brain barrier formed by endothelial cells for substances with a molecular mass of <0.8 kDa (11). Together, our analyses demonstrated that the early neuroepithelial layer lining brain ventricles represents a cerebral-ventricular barrier and that Cldn5a determines the permeability of hindbrain neuroepithelial cell TJs for small molecules.
A Tight Junction-Opening Mutant of Claudin5a Impairs Tight Junction Integrity and Affects Brain Ventricular Lumen Expansion.
To test the relevance of Cldn5a-mediated tightening of the TJ for ventricular lumen expansion and brain morphogenesis, we attempted to identify a specific Cldn5a mutant with a reduced tightening function. The ECL2 region has been shown to be involved in Claudin-Claudin trans interactions and formation of TJ strands between two opposing cells (16), and the essential residues are conserved between classic Claudins and between species as shown for Cldn5 and Cldn3 (Fig. 2A and Fig. S2) (17). All Cldn5 proteins contain the aromatic amino acid Tyr148, which has been shown to be essential for TJ strands formation of transfected human embryonic kidney (HEK) cells via an intercellular aromatic core (16). However, until now it has not been clarified whether the ECL2 contributes to TJ tightness. To test this possibility and, in particular, to assess the functional consequences of replacing Tyr148 for paracellular permeability, we used Madin-Darby canine kidney (MDCK-II) epithelial cells that stably express either murine Cldn5WT or mutant Cldn5Y148A (Fig. 2C). The tightness of the respective epithelial sheets was tested by measuring the transepithelial permeation of the tracer fluorescein (molecular mass of 376 Da). We found that the mutant protein was strongly expressed and correctly localized to apical junctions, just like the WT protein (Fig. 2C). As a result, the permeability of the MDCK-II cell layer to fluorescein significantly increased >5-fold for cells expressing Cldn5Y148A compared to cells expressing the WT form and was 2.5-fold higher for cells expressing Cldn5Y148A when compared to vector control transfected cells (Fig. 2B). Our data demonstrates that the ECL2 domain of a Claudin contributes to TJ tightness. Until now, evidence has only been provided for a tightening role of ECL1 (18). Together, these results suggest that the two ECLs contribute independently or cooperatively to the TJ. Moreover, these findings pointed at a dominant barrier-disrupting effect of murine Cldn5Y148A on TJ permeability and opened the possibility to use this mutant as a tool to specifically inhibit formation of a paracellular barrier.
Next, we investigated the effects of the zebrafish Cldn5aY148A mutant on the initial ventricular lumen expansion, which occurs ≈20 hpf (1). To assess the expression and localization of the mutant protein within the zebrafish embryo, we coinjected MOcldn5a, which is directed against the 5′ UTR, together with mRNAs that lack this target sequence and encode zebrafish Cldn5aWT or mutant Cldn5aY148A. We found that both Cldn5a proteins localized correctly to the apical side of neuroepithelial cells (Fig. 2D). The brain ventricular lumen was visualized and quantified by in vivo labeling of embryos using sodium green indicator in combination with recording of confocal microscopic Z-scan projection stacks (Fig. 3). Subsequently, 3D reconstructions were generated for a total of 4–5 embryos per sample group and volume measurements were generated by using Volocity software (Improvision). These measurements revealed a strong reduction of the mid- and hindbrain ventricular volume in cldn5a morphants (Fig. 3 B and F) and in embryos coinjected with MOcldn5a and cldn5aY148A mRNA (Fig. 3 E and F) when compared with WT (Fig. 3 A and F) or with embryos rescued by coinjection of MOcldn5a together with cldn5aWT mRNA (Fig. 3 D and F). We also coinjected MOcldn5a +p53 to prevent potential MO-induced p53-mediated apoptosis (19) and found that the ventricular lumen expansion was comparable to the knock-down of Cldn5a alone (Fig. 3 B, C, and F). Consistent with this finding, we also observed that loss of Cldn5a does not increase apoptotic cell death nor impair proliferation rates within the hindbrain region at 20 hpf. Whole-mount immunohistochemistry using antibodies against phosphorylated histone H3 to detect mitotic cells or against caspase 3 to detect apoptotic cells were comparable for both WT and cldn5a morphants [mitotic index determined for four embryos each (mean ± SD): WT (117/2,732) = 4.28 ± 0.15; MOcldn5a, (128/3,005) = 4.25 ± 0.28; MOcldn5a+p53 (123/2,787) = 4.41 ± 1.2; P = 0.886 (WT versus MOcldn5a), P = 0.888 (WT versus MOcldn5a+p53). Apoptotic events counted for five embryos each (mean ± SD): WT (5/1,386) = 0.36 ± 0.25; MOcldn5a (7/1,257) = 0.56 ± 0.11, P = 0.309]. Measurements taken at 30 hpf of live embryos injected with fluorescein isothiocyanate (FITC) 70-kDa dextran into the hindbrain ventricle revealed a sustained reduction of ventricle size in cldn5a morphants also at later stages of development (Fig. S3). Taken together, our data revealed that the early neuroepithelial layer lining the brain ventricles is part of an embryonic cerebral-ventricular barrier system at 20 hpf, which is required for the initial ventricular expansion process and that Claudin-Claudin trans interaction by Cldn5a provide an essential tightening role within this tissue.
Fig. 3.
Loss of Claudin5a affects expansion of the brain ventricular lumen at 20 hpf. (A–E) Shown are confocal microscopic Z-scans of in vivo sodium green indicator-labeled embryos. The ventricular lumen is indicated by false-coloring (orange). Indicated are ventricles of the midbrain (MBV) and hindbrain (HBV). (F) Quantifications of ventricular volume were generated from detailed 3D-reconstructions of confocal Z-scan sections for a total of 4–5 embryos per sample group by using Volocity software (unit size is [×106 μm3]). Data represent mean ± SEM, n ≥ 4, *, P = 0.016 in all three cases. (Scale bar: 100 μm.)
Claudin5a Localization Depends on Neuroepithelial Tissue Integrity.
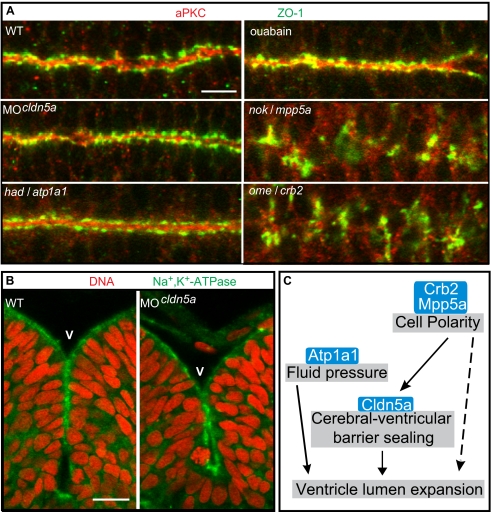
Loss of the cell polarity regulators Crumbs homolog 2 (Crb2) [in zebrafish known as Oko meduzy (Ome)] or Membrane protein, palmitoylated 5a (Mpp5a) [in zebrafish known as Nagie oko (Nok)] causes a severe disruption of the brain ventricular zone (20 –23) and a failure of brain ventricles to expand between 20 and 30 hpf (Fig. S4) (1). Similarly, heart and mindla1 (hadla1) embryos that are deficient for the osmoregulatory ion pump Atp1a1 display brain ventricle expansion defects by 30 hpf (Fig. S4) (1). The phenotypic similarities between these mutants and cldn5a morphants suggested a common molecular pathway involved in the establishment of the cerebral-ventricular barrier system. We therefore assessed whether loss of cell polarity or of Atp1a1, which is strongly expressed within the neuroepithelium (Fig. 4B), affected the apical localization of Cldn5a. Indeed, loss of Mpp5a or Crb2 affected the integrity of the neuroepithelium, disrupted the contiguous ventricular apical zone, and abolished the apical accumulation of Cldn5a, aPKC, and the focal localization of the junctional marker protein Zonula occludens-1 (ZO-1) (Fig. 4A and Fig. S4) (24). In contrast, loss of Atp1a1 did not disrupt the contiguous apical ventricular localization of Cldn5a, aPKC, or ZO-1 (Fig. 4A and Fig. S4). To further assess the role of the ion pump function for ventricular expansion, embryos were treated with ouabain, a potent inhibitor of Na+,K+-ATPase activity, between 18 and 30 hpf. By this treatment, ventricular lumen expansion was severely impaired (Fig. S5), although neuroepithelial tissue integrity was not affected (Fig. 4A). Together, these findings confirmed that neuroepithelial tissue polarity and the osmoregulatory function of Atp1a1 are a prerequisite for the expansion of brain ventricular lumen. Moreover, the correct localization of Cldn5a depends on neuroepithelial tissue polarity.
Fig. 4.
Loss of Claudin5a does not affect neuroepithelial tissue integrity. (A) Shown are apical views of immunohistochemical stainings onto the hindbrain ventricular zone of cell polarity mutants, Cldn5a- or Atp1a1-deficient embryos, or ouabain-treated embryos at 30 hpf. The neuroepithelial localization of Zonula occludens-1 (ZO-1) or aPKC is only affected in the cell polarity mutants nokm520/mpp5a and omem289/crb2. (Scale bar: 20 μm.) (B) Loss of Cldn5a does not affect the neuroepithelial localization of Na+,K+-ATPase which is labeled with the a6F antibody. Shown are confocal microscopic images of sections of immunohistochemical stainings of the hindbrain and ventricle. (Scale bar: 20 μm.) (C) Schematic diagram of developmental/cellular processes contributing to brain ventricle expansion. Our study suggests that the cell polarity regulators Crb2 and Mpp5a are essential for neuroepithelial integrity and maintenance of the TJ. Moreover, Crumbs complex proteins may be directly required for lumen formation (Results and Discussion). Tightness of the TJ is, at least in part, regulated by Cldn5a, which seals the neuroepithelial layer to maintain the fluid pressure, which may depend on the ion pump activity of Atp1a1. Ventricular fluid accumulation drives expansion of brain ventricles and tissue morphogenesis. crumbs2 (crb2); heart and mind (had); membrane protein, palmitoylated 5a (mpp5a); nagie oko (nok); oko meduzy (ome); Ventricle (V).
Embryos lacking Cldn5a or expressing the TJ-opening Cldn5aY148A mutant display significantly reduced brain ventricles, which is phenotypically reminiscent of cell polarity mutants, embryos lacking Atp1a1 or ouabain-treated embryos (Fig. S4 and S5). We therefore tested whether, conversely, loss of Cldn5a affects the polarized organization or integrity of the neuroepithelial ventricular zone. However, the focal localization of the junctional marker ZO-1 was not changed in cldn5a morphants compared with WT at 30 hpf (Fig. 4A). Moreover, loss of Cldn5a did not affect the neuroepithelial localization of Atp1a1 as detected by using the a6F antibody that recognizes different isoforms of the Na+,K+-ATPase (Fig. 4B) (25). These findings demonstrated that cell polarity and tissue integrity of the neuroepithelium are not disrupted in cldn5a morphants.
Our work sheds light on a poorly understood aspect of brain morphogenesis during early zebrafish development. We have shown that the embryonic neuroepithelial layer lining the developing brain ventricles is organized as a cerebral barrier and that Cldn5a-mediated paracellular tightness of this tissue is required for ventricular lumen expansion. Unlike in crb2 or mpp5a cell polarity mutants, tissue integrity of the neuroepithelial layer is not affected in cldn5a morphants. Therefore, Cldn5a tightens the cerebral-ventricular barrier by modifying the TJ physiology rather than by disrupting organization of the neuroepithelium. Based on our results, we favor a model according to which cell polarity regulators of the Crumbs complex are essential for ventricular lumen formation via regulation of neuroepithelial integrity, which is required for TJ formation and Cldn5a apical localization. In addition, Crumbs proteins have been shown to be required for the establishment of an apical membrane compartment, which is essential for lumen generation in cyst-formation assays in MDCK-II cells (26). A potential direct involvement of Crumbs complex proteins in brain ventricular lumen formation independent of apical Cldn5a localization remains to be tested. Cldn5a acts downstream of cell polarity regulation in sealing neuroepithelial TJ and maintaining ventricular fluid pressure. The generation of fluid pressure may depend on Atp1a1 activity during the brain ventricle expansion process between 17 and 30 hpf (Fig. 4C). This work on the barrier-forming Cldn5a and a published study (7) on the pore-forming Cldn15 highlight the diversity of mechanisms by which Claudin-mediated modulation of TJ physiology controls the maturation of different organ lumens during development.
It is a well established fact that the ventricular wall of the adult mammalian brain does not represent a cerebral barrier system and that neuroepithelial ependymal junctions are leaky (27). Therefore, the embryonic cerebral-ventricular barrier system described in our study should be considered transient and may only be essential for the initial ventricular expansion rather than for maintenance of ventricular shape. A similar maturational role has been described in the neonatal mouse kidney for Cldn6 and Cldn9, which are not expressed in the adult proximal tubuli (28). Both Cldn6 and Cldn9 are the closest relatives of Cldn5 in the phylogenetic tree of all claudins (17). Because Cldn5 is not obviously expressed in the ventricular zone of early mouse embryos between embryonic day (E) 9.5–E11.5 (Fig. S6; http://developingmouse.brain-map.org/), it is possible that other Claudins may serve a comparable function within the murine embryonic brain.
Based on our study, we envision that substances interfering with Cldn5-mediated paracellular barrier tightening could assist an improved drug delivery into the brain because Cldn5 is essential for blood–brain barrier integrity (11). Such substances could be identified using zebrafish brain ventricle expansion as an in vivo assay system.
Materials and Methods
Fish Stocks and Maintenance.
General zebrafish maintenance and embryo collection was carried out according to standard conditions. Embryos were staged at 28.5 °C. The following fish strains were used: AB, hadla1, nokm520, and omem289 (21, 23, 29).
DNA Constructs, Site-Directed Mutagenesis, and mRNA Synthesis.
Full-length cldn5a cDNA was produced by RT-PCR from 24 hpf embryonic cDNA by using the following primers and subcloned into pCS2+ expression vector:
cldn5a_fw: 5′-CCGCTCGAGATGGCCTCCGCGGCTTTG-3′,
cldn5a_re: 5′-GCTCTAGATCACACGTAATTCCTCTTGTC-3′.
Site-directed mutagenesis of cldn5aY148A was performed by using the QuikChange kit (Stratagene) using the following primers:
cldn5aY148A_fw: 5′-TATCATCTCCGACTTCGCTAACCCGCAGGTGCTGC-3′,
cldn5aY148A_re: 5′-GCAGCACCTGCGGGTTAGCGAAGTCGGAGATGATA-3′.
The resultant clones were cut with XhoI/XbaI and subcloned into the pCS2+ expression vector. For mRNA synthesis, the SP6 or T7 mMessage mMachine kits (Ambion) were used. Digoxigenin-UTP-labeled riboprobes for whole-mount in situ hybridizations were synthesized according to the manufacturer’s instructions (Roche) by using the pCS2+ vectors with full-length cldn5a.
Mammalian expression vectors for murine Cldn5 were based on pEYFP-N1 (Clontech). FLAG-cldn5-YFP was generated by amplification of murine cldn5 (accession number NM_013805) using pGTCl-5 as template and primers, one encoding the N-terminal FLAG epitope. To generate FLAG-cldn5, site-directed mutagenesis was performed as described in ref. 16 introducing a stop codon 3′ behind the cldn5 ORF. The amino acid substitution Y148A was again introduced by site-directed mutagenesis.
Zebrafish Microinjections and Morpholinos.
MOs (Gene Tools) were injected at a concentration of 150 μmol/L (for MOcldn5a) or 100 μmol/L (for MOp53). MO sequences were as follows:
MOp53: 5′-GCGCCATTGCTTTGCAAGAATTG-3′ (19),
MOcldn5a: 5′-AGGCCATCGCTTTCTTTTCCCACTC-3′.
For mRNA rescue experiments, 75–100 pg of mRNA and 150 μM of MOcldn5a were coinjected. All types of injections were performed by using the MPPI-2 pressure injector (Applied Scientific Instrumentation) and MM33 micromanipulator (Maerzhaeuser).
Whole-Mount in Situ Hybridization, Tissue Sectioning, and Immunohistochemistry.
Whole-mount in situ hybridizations were basically performed as described in ref. 30. For documentation, stained embryos were cleared in benzyl:benzoate (2:1) and embedded in Permount. Images were recorded on a Zeiss Axioplan microscope with ×10 objective by using a SPOT digital camera (Diagnostic Instruments) and Meta Morph software (Visitron). Images were processed with Photoshop software (Adobe). Immunohistochemistry was performed as described in ref. 13. For tissue sectioning of Cldn5a immunostainings, embryos were fixed in trichloroacetic acid for 1 h on ice and subsequently postfixed in 2% paraformaldehyde overnight for hardening of the tissue. Transverse sectioning was performed according to Trinh and Stainier (31). The following antibodies were used: mouse anti-mammalian Cldn5 (1:100; Invitrogen), rabbit anti-aPKCζ that recognizes both isoforms of aPKCs (1:100; Santa Cruz Biotechnology), rabbit anti-phospho-Histone H3 (1:800; Millipore), rabbit anti-Caspase 3 (1:200, BD Biosciences), mouse anti-ZO-1 (1:200; Zymed Laboratories), mouse anti-Na+,K+-ATPase (1:100; α6F, Hybridoma Bank), goat anti-rabbit Cy5 (1:100; Jackson ImmunoResearch Laboratories), goat anti-mouse FITC (1:100; Jackson ImmunoResearch Laboratories), rhodamine phalloidin (1:100, Invitrogen), 4′,6-Diamidino-2-phenylindole dihydrochloride (1:1000, Sigma). Cell proliferation and apoptosis quantifications were performed as described in ref. 1. To prevent bleaching during confocal microscopic imaging, samples were embedded in SlowFade Gold Antifade reagent (Invitrogen). Confocal images were obtained with the Zeiss LSM510 Meta confocal microscope by using ×40 and ×1 zoom. LSM 510 software was used to capture the images. Images were processed by using Photoshop software (Adobe).
Sequence Analysis and Phylogenetic Analysis.
We identified zebrafish cldn5a and cldn5b based on homology searches with Mus musculus cldn5 by using the NCBI BLAST search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The gene accession numbers of the claudin genes are as follows: cldn5a, GI47086108; cldn5b, GI53733860. Protein alignments and phylogenetic analysis were performed with MacVector software by using the ClustalW slow/accurate method. The accession numbers of protein sequences that were used for the phylogenetic analysis shown in Fig. S1 and the alignment shown in Fig. S2 are as follows: Cldn3/h (Danio rerio) NP_571842; Cldn3 (Mus musculus) NP_034032; Cldn3 (Homo sapiens) NP_001297; Cldn3b (Takifugu rubripes) AAT64048; Cldn5 (Mus musculus) NP_038833; Cldn5 (Homo sapiens) NP_003268; Cldn5a (Danio rerio) NP_998439; Cldn5b (Danio rerio) NP_001006044; Cldn5a (Takifugu rubripes) AAT64067; Cldn6 (Homo sapiens) NP_067018; Cldn6 (Mus musculus) NP_061247; Cldn9 (Homo sapiens) NP_066192; Cldn9 (Mus musculus) NP_064689.
Electron Microscopy.
Brain ventricles of zebrafish embryos were injected with ≈10 nL of 1% lanthanum nitrate together with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for immediate preservation of ventricular tissues. Subsequently, embryos were fixed with 2.5% glutaraldehyde (Paesel-Lorei) buffered in 0.1 M cacodylate buffer (pH 7.4). Thereafter, the whole fish was postfixed in 1% OsO4 in 0.1 M cacodylate buffer and then dehydrated in an ethanol series (50, 70, 96, and 100%). The 70% ethanol was saturated with uranyl acetate for contrast enhancement. Dehydration was completed in propylene oxide. The specimens were embedded in Araldite (Serva). Ultrathin sections were made on a FCR Reichert Ultracut ultramicrotome (Leica), mounted on pioloform-coated copper grids, contrasted with lead citrate, and analyzed and documented with an EM10A electron microscope (Zeiss).
Cell Culture.
MDCK-II cells were grown at 10% CO2 in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U penicillin, and 100 mg/mL streptomycin. Transfections of MDCK-II cells were performed with Lipofectamine 2000 (Invitrogen) or Cell Line Nucleofection Kit L (Amaxa). Stable lines were selected by adding 1 mg/mL G418 (Calbiochem). To exclude clonal variations, at least three independent cell clones were isolated of Cldn5wt, Cldn5Y148A, or vector control and were used for permeation studies. For immunocytochemistry, cells were cultured on glas coverslips for 5 days, fixed and stained as described in ref. 32 by using goat anti-rabbit Cy3 antibody (1:250) and DAPI (4′,6-diamidino-2-phenylindole dihydrochloride, 1:300, Molecular Probes). For confocal imaging, a Zeiss LSM 510 Meta confocal microscope and a ×100 objective was used.
Paracellular permeation studies were done with confluent cells on transwell filter inserts (Millipore). Cells were washed twice with PBS and incubated with Hanks’ balanced salt solution (Gibco); 100 μM Na-fluorescein was added to the apical side. One hundred-μL aliquots were removed from the basolateral reservoir 10 min after incubation. The fluorescein concentration was measured in a fluorescence microplate reader (Tecan). The permeability coefficient was calculated as 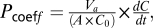 where V
a is volume in apical chamber, A is filter surface area, C
0 is initial apical fluorescein concentration, and dC/dt is initial slope of the concentration versus time curve (33).
where V
a is volume in apical chamber, A is filter surface area, C
0 is initial apical fluorescein concentration, and dC/dt is initial slope of the concentration versus time curve (33).
Brain Ventricle Volume Measurement and Pharmacological Treatment.
Sodium green indicator (Molecular Probes) was added directly to the fish medium at a final concentration of 5 μM. At 20 hpf, embryos were incubated in the medium for 1 h before mounting and imaging them under the microscope. Z-stack images of the whole ventricle zones were captured by using the Zeiss LSM Meta software. Three-dimensional projections and brain ventricle volume measurements were generated from the stack of images by using Volocity (Improvision) software, and the values of several experiments were analyzed and plotted in Microsoft Excel by using Mann–Whitney statistical testing. Injection of FITC 70-kDa dextran (Invitrogen) into 30 hpf brain ventricles was performed as described in ref. 1. Ouabain (Sigma) treatment was performed as described in ref. 29 from 18 to 30 hpf at 1 mM concentration. Differential interference contrast images were then taken on a Zeiss Axioplan microscope (Zeiss) by using a SPOT digital camera (Diagnostic Instruments) and Metamorph software (Zeiss). Images were processed with Adobe Photoshop 6.0. False-coloring of the ventricular lumen was performed by using Adobe Photoshop 6.0.
Whole-Mount In Situ Hybridization in Mice.
Embryos were dissected from pregnant mice at the designated time point and fixed overnight in 4% paraformaldehyde in PBS at 4°C. Whole-mount in situ hybridizations were carried out essentially as described in ref. 34. The localization of signals was studied in whole embryos or in plastic sections thereof. First-strand mouse cldn5 cDNA was used as in situ probe.
Supplementary Material
Acknowledgments
We are indebted to M. Furuse (Kyoto University, Japan), J. Herz, N.D. Lawson, J. Malicki, and B. Weinstein for sharing reagents and tools and to Robby Fechner for expert technical assistance with the fish facility. We thank Gabi Frommer-Kästle for expert help with electron microscopy. Nicole Cornitius and Jana Richter helped with the molecular laboratory work. We thank Alistair Garratt, Matthias Selbach, Michael Gotthardt, Erez Raz, and members of the S.A.-S. and I.E.B. laboratories for technical help, discussions, and comments on the manuscript, and in particular Justus Veerkamp for help with ventricular volume measurements. We thank M. Affolter and H. Belting for sharing unpublished results. This work was supported by the German Research Council Deutsche Forschungsgemeinschaft Grant BL308/7-3 (to I.E.B. and S.A.-S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911996107/DCSupplemental.
References
- 1.Lowery LA, Sive H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development. 2005;132:2057–2067. doi: 10.1242/dev.01791. [DOI] [PubMed] [Google Scholar]
- 2.Jeong JY, et al. Functional and developmental analysis of the blood-brain barrier in zebrafish. Brain Res Bull. 2008;75:619–628. doi: 10.1016/j.brainresbull.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 3.Bill BR, et al. Development and Notch signaling requirements of the zebrafish choroid plexus. PLoS One. 2008;3:e3114. doi: 10.1371/journal.pone.0003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Lecea M, Kondrychyn I, Fong SH, Ye ZR, Korzh V. In vivo analysis of choroid plexus morphogenesis in zebrafish. PLoS One. 2008;3:e3090. doi: 10.1371/journal.pone.0003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Furuse M, Moriwaki K. The role of claudin-based tight junctions in morphogenesis. Ann N Y Acad Sci. 2009;1165:58–61. doi: 10.1111/j.1749-6632.2009.04441.x. [DOI] [PubMed] [Google Scholar]
- 7.Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 8.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol. 2007;312:509–522. doi: 10.1016/j.ydbio.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 9.Kollmar R, Nakamura SK, Kappler JA, Hudspeth AJ. Expression and phylogeny of claudins in vertebrate primordia. Proc Natl Acad Sci USA. 2001;98:10196–10201. doi: 10.1073/pnas.171325898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res. 2004;14:1248–1257. doi: 10.1101/gr.2400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nitta T, et al. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 13.Horne-Badovinac S, et al. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 14.Whittembury G, Rawlins FA. Evidence of a paracellular pathway for ion flow in the kidney proximal tubule. Electromicroscopic demonstration of lanthanum precipitate in the tight junction. Pflugers Arch. 1971;330:302–309. doi: 10.1007/BF00588582. [DOI] [PubMed] [Google Scholar]
- 15.Wolburg H, et al. Epithelial and endothelial barriers in the olfactory region of the nasal cavity of the rat. Histochem Cell Biol. 2008;130:127–140. doi: 10.1007/s00418-008-0410-2. [DOI] [PubMed] [Google Scholar]
- 16.Piontek J, et al. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 17.Krause G, et al. Structure and function of claudins. Biochim Biophys Acta. 2008;1778:631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Wen H, Watry DD, Marcondes MC, Fox HS. Selective decrease in paracellular conductance of tight junctions: Role of the first extracellular domain of claudin-5. Mol Cell Biol. 2004;24:8408–8417. doi: 10.1128/MCB.24.19.8408-8417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bit-Avragim N, et al. Divergent polarization mechanisms during vertebrate epithelial development mediated by the Crumbs complex protein Nagie oko. J Cell Sci. 2008;121:2503–2510. doi: 10.1242/jcs.033167. [DOI] [PubMed] [Google Scholar]
- 21.Malicki J, Driever W. oko meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell-cell interactions of the retinal neuroepithelial sheet. Development. 1999;126:1235–1246. doi: 10.1242/dev.126.6.1235. [DOI] [PubMed] [Google Scholar]
- 22.Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16:945–957. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 23.Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31:150–157. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- 24.Itoh M, et al. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cells: cDNA cloning and immunoelectron microscopy. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond IA, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 26.Schlüter MA, et al. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Mol Biol Cell. 2009;20:4652–4663. doi: 10.1091/mbc.E09-02-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brightman MW, Reese TS. Junctions between intimately apposed cell membranes in the vertebrate brain. J Cell Biol. 1969;40:648–677. doi: 10.1083/jcb.40.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abuazza G, et al. Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol. 2006;291:F1132–F1141. doi: 10.1152/ajprenal.00063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu X, et al. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- 30.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 31.Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 32.Blasig IE, et al. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci. 2006;63:505–514. doi: 10.1007/s00018-005-5472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blasig IE, Mertsch K, Haseloff RF. Nitronyl nitroxides, a novel group of protective agents against oxidative stress in endothelial cells forming the blood-brain barrier. Neuropharmacology. 2002;43:1006–1014. doi: 10.1016/s0028-3908(02)00180-6. [DOI] [PubMed] [Google Scholar]
- 34.Hammes A, et al. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.