Abstract
RNA virus infection is recognized by retinoic acid-inducible gene (RIG)-I–like receptors (RLRs), RIG-I, and melanoma differentiation–associated gene 5 (MDA5) in the cytoplasm. RLRs are comprised of N-terminal caspase-recruitment domains (CARDs) and a DExD/H-box helicase domain. The third member of the RLR family, LGP2, lacks any CARDs and was originally identified as a negative regulator of RLR signaling. In the present study, we generated mice lacking LGP2 and found that LGP2 was required for RIG-I– and MDA5-mediated antiviral responses. In particular, LGP2 was essential for type I IFN production in response to picornaviridae infection. Overexpression of the CARDs from RIG-I and MDA5 in Lgp2 −/− fibroblasts activated the IFN-β promoter, suggesting that LGP2 acts upstream of RIG-I and MDA5. We further examined the role of the LGP2 helicase domain by generating mice harboring a point mutation of Lys-30 to Ala (Lgp2 K30A/K30A) that abrogated the LGP2 ATPase activity. Lgp2 K30A/K30A dendritic cells showed impaired IFN-β productions in response to various RNA viruses to extents similar to those of Lgp2 −/− cells. Lgp2 −/− and Lgp2 K30A/K30A mice were highly susceptible to encephalomyocarditis virus infection. Nevertheless, LGP2 and its ATPase activity were dispensable for the responses to synthetic RNA ligands for MDA5 and RIG-I. Taken together, the present data suggest that LGP2 facilitates viral RNA recognition by RIG-I and MDA5 through its ATPase domain.
Keywords: innate immunity, type I interferon, virus infection
RNA virus infection is initially recognized by host pattern recognition receptors, including retinoic acid-inducible gene (RIG)-I–like receptors (RLRs) and Toll-like receptors (TLRs), which induce antiviral responses such as the productions of type I IFNs and proinflammatory cytokines (1 –4). The RLR family comprises RIG-I, melanoma differentiation-associated gene 5 (MDA5) and LGP2. RLRs harbor a central DExD/H-box helicase domain and a C-terminal regulatory domain (RD). RIG-I and MDA5 also contain two N-terminal caspase recruitment domains (CARDs), whereas LGP2 does not. RIG-I recognizes relatively short double-stranded (ds) RNAs (up to 1 kb), and the presence of a 5′ triphosphate end greatly potentiates its type I IFN-inducing activity (5 –7). On the other hand, MDA5 detects long dsRNAs (more than 2 kb), such as polyinosinic polycytidylic acid (poly I:C). Analyses of Rig-I–deficient (Rig-I −/−) and Mda5 −/− mice have shown that RIG-I is essential for the production of type I IFNs in response to various RNA viruses, including vesicular stomatitis virus (VSV), Sendai virus (SeV), Japanese encephalitis virus (JEV), and influenza virus, whereas MDA5 is critical for the detection of picornaviridae such as encephalomyocarditis virus (EMCV) and mengovirus (8, 9). Some RNA viruses such as West Nile virus and reovirus are recognized by both RIG-I and MDA5 (10, 11). RIG-I is also reported to be involved in the recognition of foreign DNA in the cytoplasm through transcription of the DNA to dsRNA by polymerase III (12, 13).
The C-terminal RDs of RLRs are responsible for binding to dsRNAs (3). However, the functions of the helicase domains of the RLR family members have not yet been clarified. Although the RIG-I helicase domain has the ability to unwind dsRNA, this activity is not correlated with the level of IFN production (14). A recent report proposed that the RIG-I ATPase activity is required for translocation of RIG-I on dsRNA (15). The N-terminal CARDs of RIG-I and MDA5 trigger intracellular signaling pathways via IFN-β promoter stimulator (IPS)–1 (also known as MAVS, VISA, or CARDIF), an adaptor molecule possessing an N-terminal CARD (16). IPS-1 subsequently activates two IκB kinase (IKK)–related kinases, IKK-i, and TANK-binding kinase 1 (TBK1). These kinases phosphorylate IFN-regulatory factor (IRF) 3 and IRF7, which activate the transcription of genes encoding type I IFNs and IFN-inducible genes. The produced type I IFNs alert the surrounding cells by triggering signaling cascades that lead to phosphorylation and nuclear translocation of STAT1 (1, 2).
The third RLR family member LGP2, also known as Dhx58, harbors a DExD/H-box helicase domain and a C-terminal RD but lacks any CARDs (17). In vitro studies have suggested that LGP2 negatively regulates RIG-I–mediated dsRNA recognition (18). Several models have been proposed for the mechanisms of this inhibition. The first model is that LGP2 binds to viral dsRNA and prevents RIG-I– and MDA5-mediated recognition (18). The second model is that LGP2 inhibits multimerization of RIG-I and its interaction with IPS-1 via the RD of LGP2 (19). The third model is that LGP2 competes with IKK-i for recruitment to IPS-1, thereby suppressing RLR signaling (20). Structural analyses of the C-terminal domain of LGP2 have revealed that LGP2 can bind to the termini of dsRNAs more firmly than MDA5 (21 –23). A previous report showed that Lgp2 −/−mice exhibit enhanced production of type I IFNs in response to poly I:C stimulation and VSV infection, whereas the response to EMCV is suppressed (24). Therefore, the role of LGP2 in the negative or positive regulation of RLR signaling has not yet been fully clarified.
In the present study, we generated Lgp2 −/− mice and mice harboring a point mutation in the LGP2 helicase domain (K30A), and examined their responses to RNA viruses recognized by RIG-I and MDA5. Conventional dendritic cells (cDCs) and mouse embryonic fibroblasts (MEFs) obtained from Lgp2 −/− mice showed severely impaired IFN responses to infections with picornaviruses, which are recognized by MDA5. Furthermore, the responses to viruses recognized by RIG-I were also impaired in Lgp2 −/− cells. In contrast, the IFN productions in response to synthetic RNAs, poly I:C and RNA synthesized by T7 polymerase, were comparable between wild-type (WT) and Lgp2 −/− or Lgp2 K30A/K30A cells. Lgp2 −/− and Lgp2 K30A/K30A mice were highly susceptible to infection with EMCV. Taken together, the present results demonstrate that LGP2 acts as a positive, but not negative, regulator of RIG-I– and MDA5-mediated viral recognition.
Results
Generation of Mice Lacking Lgp2.
To investigate the physiological role of LGP2 in vivo, we established Lgp2 −/− mice (Fig. S1 A and B). As reported previously, the expression of Lgp2 mRNA was highly induced in response to IFN-β stimulation in MEFs (Fig. S1C) (17). Expression of Lgp2 mRNA was not detected in Lgp2 −/− MEFs, whereas the expression levels of Rig-I and Mda5 mRNAs were comparable between WT and Lgp2 −/− cells (Fig. S1C). The Lgp2 −/− progenies obtained from Lgp2 +/− intercrosses were lower than the expected Mendelian ratio (Fig. S2A), indicating that homozygous mutations of the Lgp2 gene cause embryonic lethality at a high frequency. In addition, adult female Lgp2 −/− mice showed an enlarged uterus filled with fluid resulting from vaginal atresia (Fig. S2 B and C).
Role of LGP2 in Type I IFN and Cytokine Productions in Response to RNA Viruses.
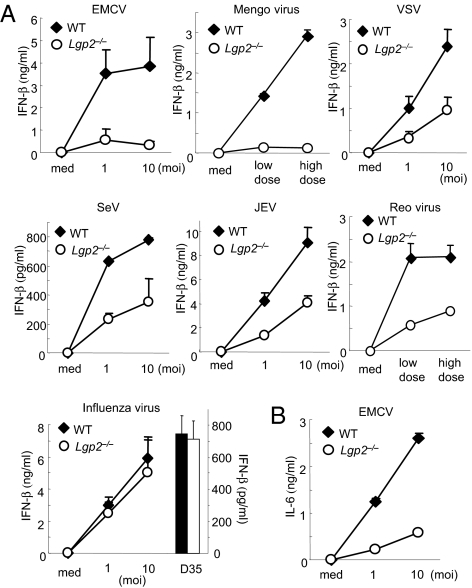
First, we examined the production of IFN-β in cDCs derived from bone marrow (BM) in the presence of GM-CSF after infection with a variety of RNA viruses (Fig. 1). The productions of IFN-β in response to picornaviridae, EMCV, and mengovirus were severely impaired in Lgp2 −/− cDCs compared with WT cells (Fig. 1A). IL-6 production induced by EMCV infection was also severely impaired in Lgp2 −/− cells (Fig. 1B). Furthermore, LGP2 was involved in the productions of IFN-β in response to several RNA viruses recognized by RIG-I, such as VSV, SeV, and JEV (Fig. 1A). IFN-β production in response to reovirus, a dsRNA virus, was also impaired in Lgp2 −/− cells (Fig. 1A). In contrast, the IFN-β productions in response to infection with influenza virus were comparable between WT and Lgp2 −/− cDCs (Fig. 1A). Stimulation with CpG-DNA, a TLR9 ligand, induced comparable amounts of IFN-β in WT and Lgp2 −/− cells (Fig. 1A).
Fig. 1.
Role of LGP2 in type I IFN production in response to various RNA viruses. (A and B) BM-derived cDCs from WT and Lgp2 −/− mice were exposed to the indicated viruses or treated with 1 μM CpG-DNA (D35) for 24 h. The concentrations of IFN-β (A) and IL-6 (B) in the culture supernatants were measured by ELISA. moi, multiplicity of infection; med, medium alone. Data are shown as means ± SD and are representative of at least three independent experiments.
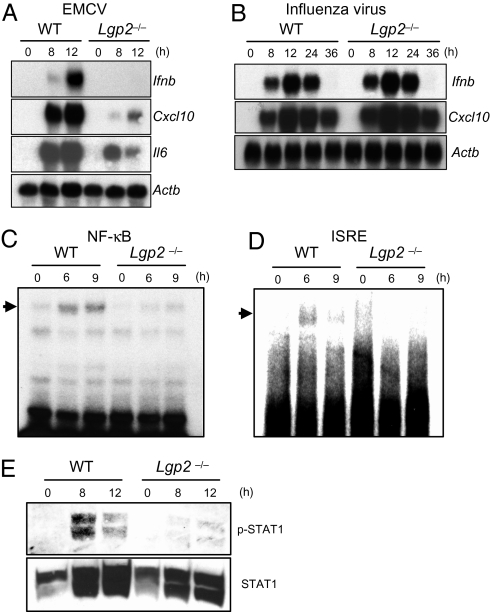
Next, we examined whether the defect in type I IFN production in response to EMCV was controlled at the mRNA level. The expressions of the genes encoding IFN-β, CXCL10 and IL-6 after infection with EMCV were severely impaired in Lgp2 −/− macrophages (Fig. 2A). However, the influenza virus-induced expressions of IFN-β and CXCL10 mRNAs were comparable between WT and Lgp2 −/− MEFs throughout the whole time course (Fig. 2B). Therefore, it is unlikely that LGP2 negatively regulates RIG-I–mediated responses, even during the later period of infection. These results indicate that LGP2 is involved in positive, but not negative, regulation of virus recognition by MDA5 and RIG-I, with the exception of influenza virus.
Fig. 2.
Role of LGP2 in the activation of signaling pathways leading to IFN-inducible gene expression. (A) Total RNAs extracted from WT and Lgp2 −/− macrophages infected with EMCV were subjected to Northern blot analyses for the expressions of Ifnb, Cxcl10, Il6, and Actb mRNAs. (B) WT and Lgp2 −/− MEFs were infected with influenza virus followed by isolation of the total RNA. The expressions of Ifnb, Cxcl10, and Actb mRNAs were determined by Northern blot analyses. (C and D) Nuclear extracts were prepared from WT and Lgp2 −/− macrophages infected with EMCV for the indicated periods. The binding activities of DNA to NF-κB (C) and ISREs (D) were determined by EMSAs. (E) Cell lysates were prepared from WT and Lgp2 −/− macrophages infected with EMCV and probed with anti–phospho-STAT-1 and anti-STAT1 antibodies. The data are representative of at least three independent experiments.
Cell Type-Specific Involvement of LGP2 in EMCV Recognition.
We previously showed that RIG-I– and MDA5-dependent RNA virus recognition occurs in cDCs but not in plasmacytoid dendritic cells (pDCs) (8). To determine whether LGP2 functions in a cell type–specific fashion, B220−CD11c+ cDCs and CD11c+B220+ pDCs were purified from WT and Lgp2 −/− splenocytes. EMCV-induced IFN-β production was severely impaired in Lgp2 −/− splenic cDCs compared with WT cDCs, whereas splenic pDCs from WT and Lgp2 −/− mice produced comparable amounts of IFN-β (Fig. S3). These data indicate that LGP2 functions in cDCs but not in pDCs.
LGP2 Is Essential for Triggering RLR Signaling Pathways.
To investigate whether LGP2 regulates the primary responses to RNA virus infections, we examined the activation of intracellular signaling pathways. Electrophoretic mobility shift assays (EMSAs) revealed that the activations of NF-κB and IFN-stimulated regulatory elements (ISREs) in response to EMCV infection were severely impaired in Lgp2 −/− cells (Fig. 2 C and D). Furthermore, the phosphorylation of STAT1 was abrogated in Lgp2 −/− cells (Fig. 2E). Nevertheless, the expressions of Lgp2 in response to IFN-β treatment were not altered in Rig-I −/− and Mda5 −/− cells (Fig. S4). These results suggest that LGP2 is required for the initial recognition of EMCV, leading to activation of transcription factors involved in the expression of type I IFNs.
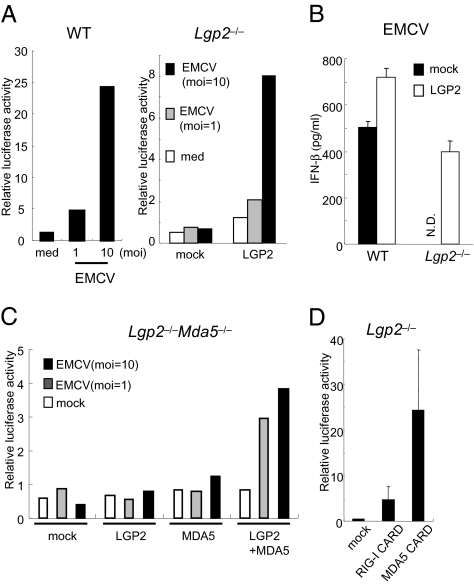
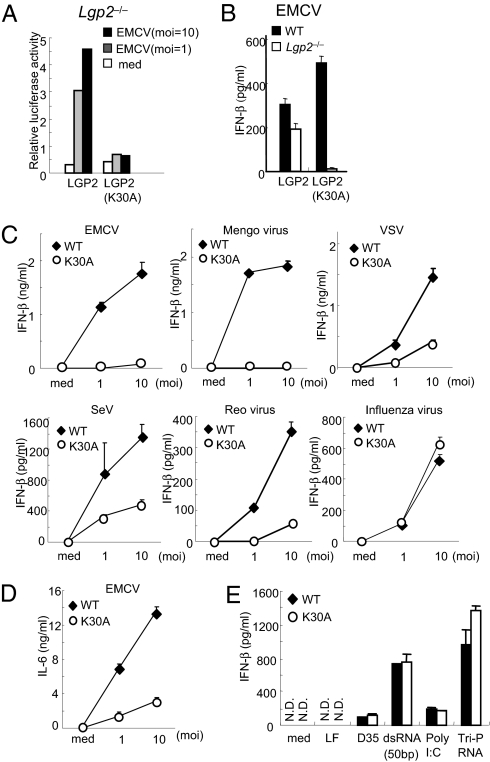
Next, we examined whether the expression of Lgp2 could rescue the virus-mediated IFN responses. IFN-β–dependent reporter gene expression was induced in response to EMCV infection in WT, but not in Lgp2 −/−, MEFs (Fig. 3A). Expression of exogenous LGP2 in Lgp2 −/− cells restored the EMCV-induced IFN-β promoter activity as well as IFN-β production (Fig. 3 A and B). Although overexpression of either LGP2 or MDA5 alone in Lgp2 −/− Mda5 −/− MEFs failed to confer EMCV-induced IFN-β promoter activity, coexpression of both LGP2 and MDA5 restored EMCV responsiveness (Fig. 3C). Overexpression of the CARDs from RIG-I or MDA5 in Lgp2 −/− MEFs activated the IFN-β promoter (Fig. 3D), suggesting that LGP2 functions upstream of RIG-I and MDA5.
Fig. 3.
LGP2 acts in the upstream of RIG-I and MDA5. (A) WT and Lgp2 −/− MEFs were transiently transfected with the IFN-β promoter construct together with expression plasmids encoding LGP2. The cells were infected with EMCV for 8 h and then lysed. The cell lysates were analyzed by a luciferase assay. (B) WT and Lgp2 −/− MEFs were infected with a retrovirus expressing Lgp2. At 2 days after infection, the cells were exposed to EMCV for 24 h. The IFN-β concentrations in the culture supernatants were measured by ELISA. N.D., not detected. (C) Lgp2 −/− Mda5 −/− MEFs were transiently transfected with the IFN-β promoter reporter construct together with the indicated expression plasmids. After 24 h, the cells were infected with EMCV for 8 h and then lysed. The cell lysates were analyzed by a luciferase assay. (D) Lgp2 −/− MEFs were transiently transfected with the IFN-β promoter construct together with expression plasmids encoding the CARDs of RIG-I or MDA5 and then lysed at 48 h after transfection. The cell lysates were analyzed by a luciferase assay.
Normal IFN Responses of LGP2−/− cells to Exogenously Transfected RNAs.
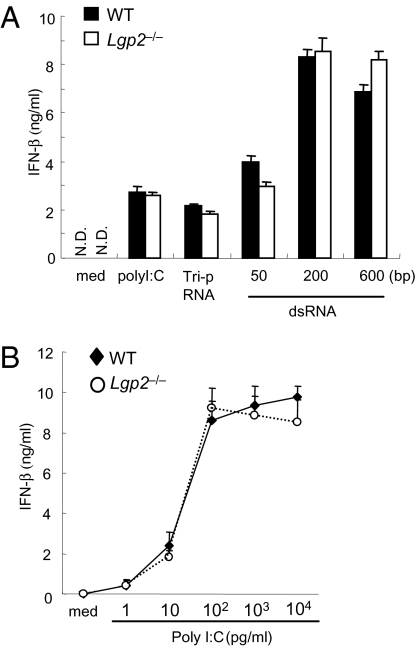
We examined the responses of Lgp2 −/− cells to synthetic RNAs recognized by RIG-I and MDA5. Unexpectedly, WT and Lgp2 −/− cDCs produced comparable amounts of IFN-β in response to poly I:C, in vitro–transcribed dsRNA and RNA with a 5′ triphosphate end (Tri-P) (Fig. 4A). Similarly, Lgp2-deficiency did not affect the IFN-β productions in response to the synthesized RNAs in fibroblasts (Fig. 4A). In addition, no differences were observed in the responses to the various concentrations of poly I:C examined (Fig. 4B). These data suggest that LGP2 is dispensable for the recognition of synthesized dsRNA and 5′ triphosphate RNA.
Fig. 4.
Role of LGP2 in the recognition of exogenously transfected RNAs. (A) WT and Lgp2 −/− MEFs were stimulated with triphosphate RNA, in vitro–transcribed dsRNA (1 μg/mL) or poly I:C complexed with Lipofectamine 2000 for 24 h. The IFN-β concentrations in the culture supernatants were measured by ELISA. med, medium; N.D., not detected. Data are shown as the means ± SD of triplicate samples. Similar results were obtained in three independent experiments. (B) WT and Lgp2 −/− MEFs were transfected with the indicated amounts of poly I:C complexed with Lipofectamine 2000. The IFN-β concentrations in the culture supernatants were measured by ELISA.
Function of LGP2 ATPase Domain in Type I IFN Responses to Virus Infections.
The recognition of dsRNA and RNA viruses by RIG-I or MDA5 requires ATPase activity (17, 25). To examine the role of the LGP2 ATPase activity in LGP2-mediated virus recognition, we reconstituted Lgp2 −/− MEFs with WT LGP2 or LGP2-K30A harboring a Lys-to-Ala point mutation in the Walker ATP-binding motif using a retrovirus system. Expression of WT LGP2 in Lgp2 −/− MEFs conferred IFN-β promoter activity as well as IFN-β production in response to EMCV, whereas expression of LGP2-K30A failed to confer these responses to EMCV infection (Fig. 5 A and B).
Fig. 5.
Essential role of the LGP2 ATPase activity in the recognition of RNA viruses. (A) Lgp2 −/− MEFs were transiently transfected with the IFN-β promoter construct together with expression plasmids encoding LGP2 or LGP2 (K30A). The cells were infected with EMCV for 8 h and then lysed. The cell lysates were analyzed by a luciferase assay. (B) WT and Lgp2 −/− MEFs were infected with retroviruses expressing LGP2 or LGP2 (K30A). At 2 days after infection, the cells were exposed to EMCV for 24 h and the IFN-β concentrations in the culture supernatants were measured by ELISA. (C and D) WT and Lgp2 K30A/K30A (K30A) mice were exposed to the indicated RNA viruses for 24 h. The concentrations of IFN-β (C) and IL-6 (D) in the culture supernatants were measured by ELISA. (E) WT and Lgp2 K30A/K30A cDCs were transfected with the indicated RNAs for 24 h. The concentrations of IFN-β in the culture supernatants were measured by ELISA. moi, multiplicity of infection; med, medium alone; LF, lipofectamine alone. Data are shown as means ± SD and are representative of at least three independent experiments.
To further examine the role of the LGP2 ATPase activity in vivo, we generated mice harboring the LGP2 K30A point mutation (Fig. S5 A and B). Expression of Lgp2 mRNA was comparably induced in response to IFN-β stimulation in WT and Lgp2 K30A/K30A MEFs (Fig. S5C). We confirmed the insertion of the point mutation by sequencing analysis (Fig. S5D). The Lgp2 K30A/K30A mice were born at the expected Mendelian ratio, and did not show any developmental defects. We examined the IFN-β productions in cDCs in response to infections with RNA viruses. The IFN-β productions in response to infections with EMCV, mengovirus, VSV, SeV, and reovirus, but not with influenza virus, were severely impaired in Lgp2 K30A/K30A cDCs compared with WT cells (Fig. 5C). The IL-6 production in response to EMCV infection was also impaired in Lgp2 K30A/K30A cDCs (Fig. 5D). The defects observed in Lgp2 K30A/K30A cDCs were as severe as those observed in Lgp2 −/− cDCs, suggesting that the ATPase activity of LGP2 is essential for the recognition of viruses. The productions of IFN-β in response to transfections of synthetic RNAs and poly I:C were comparable between WT and Lgp2 K30A/K30A cells (Fig. 5E), further confirming that LGP2 is not involved in the responses to the transfection of synthetic RNAs. Taken together, these results indicate that the ATPase activity of LGP2 is essential for LGP2 to function as a positive regulator in MEFs. This finding is in marked contrast to in vitro experiments in which overexpression of WT LGP2 and LGP2-K30A in HEK293 cells suppressed RIG-I–mediated IFN-β promoter activity (Fig. S6), suggesting that overexpression of LGP2 in HEK293 cells nonspecifically inhibits the RIG-I–mediated pathway.
Role of LGP2 and Its ATPase Activity in Antiviral Host Defenses In Vivo.
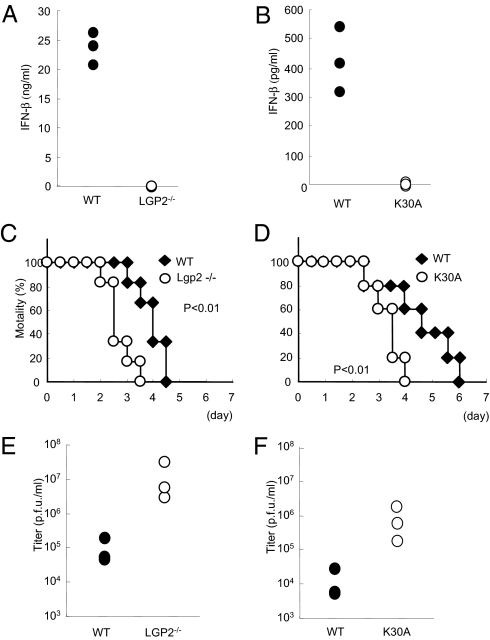
Finally, we assessed the role of LGP2 and its ATPase activity in antiviral responses in vivo. When Lgp2 −/− mice were challenged with EMCV, IFN-β production was not detected in their sera (Fig. 6A). Furthermore, Lgp2 −/− mice were highly susceptible to EMCV infection compared with their littermate controls (Fig. 6C). Consistent with the increased susceptibility to EMCV, the viral titer in the heart was remarkably higher in Lgp2 −/− mice than in control mice (Fig. 6E). Similar to the results for Lgp2 −/− mice, Lgp2 K30A/K30A mice showed severe defects in IFN-β production in response to EMCV infection (Fig. 6B). Lgp2 K30A/K30A mice were highly susceptible to infection with EMCV, with highly increased viral titers in their hearts (Fig. 6 D and F). These data indicate that LGP2 also plays a key role in the host defenses against RNA viruses recognized by MDA5 in vivo.
Fig. 6.
Role of LGP2 in host defense against EMCV infection in vivo. (A and B) WT and littermate Lgp2 −/− mice (A) or WT and littermate Lgp2 K30A/K30A (K30A) mice (B) were i.v. inoculated with 1 × 107 pfu EMCV. Serum samples were obtained at 4 h after injection, and the IFN-β concentrations were determined by ELISA. (C and D) Survival rates of WT and Lgp2 −/− mice (C) or WT and littermate Lgp2 K30A/K30A mice (D) intraperitoneally infected with 1 × 102 pfu EMCV were monitored every 12 h for 5 days. (E and F) WT and littermate Lgp2 −/−mice (E) or WT and littermate Lgp2 K30A/K30A mice (F) were infected i.p. with 1 × 102 pfu EMCV. After 48 h, the mice were killed and the virus titers in their hearts were determined by a plaque assay.
Discussion
The present data clearly demonstrate that LGP2 acts as a positive regulator of MDA5- and RIG-I–mediated viral recognition, except for influenza virus. These findings are in contrast to the conclusions deduced from previous in vitro studies and a report on Lgp2 knockout mice generated by another group (17, 18, 20, 24). LGP2 is particularly important for the recognition of picornaviruses, including EMCV and mengovirus, among RNA viruses. Analyses of the activation status of signaling molecules revealed that LGP2 was involved in the primary recognition of EMCV upstream of MDA5. LGP2 was also involved in the recognition of RNA viruses recognized by RIG-I, such as VSV and SeV, although the defects in the responses to these viruses observed in Lgp2 −/− cells were not as severe as the defects in the responses to picornaviruses. Surprisingly, Lgp2 −/− cells responded efficiently to synthetic RNA compounds, including poly I:C, dsRNA transcribed in vitro using T7 polymerase and 5′ triphosphate RNA.
Cells from Lgp2 K30A/K30A mice showed severe defects in the IFN responses to RNA viruses to extents similar to those of Lgp2 −/− cells. Furthermore, expression of the LGP2-K30A mutant protein in Lgp2 −/− cells failed to restore the EMCV responsiveness. These results clearly demonstrate that the ATPase domain of LGP2 is a prerequisite for its function in recognizing RNA virus infection. Recent advances in studies on DExD/H-box proteins have revealed that these proteins are involved in all aspects of RNA metabolism including translation initiation, mRNA splicing, and nuclear transport (26). Although DExD/H-box proteins, including RIG-I, are known to exhibit ATP-dependent RNA helicase activity in vitro, many DExD/H-box proteins have a more general RNA conformational change activity, rather than just a duplex-unwinding activity. In this regard, it is tempting to speculate that LGP2 functions to modify viral RNA by removing proteins from viral ribonucleoprotein (RNP) complexes or unwinding complex RNA structures to facilitate MDA5- and RIG-I–mediated recognition of dsRNA. Picornaviruses replicate in association with the cytoplasmic membranes of infected cells (27). It is therefore possible that LGP2 makes viral RNP complexes more accessible to MDA5 and RIG-I by changing their intracellular localization.
RLRs contain a C-terminal regulatory domain that is responsible for the binding to dsRNAs. The recent solution of the RLR C-terminal regulatory domain structures showed that the LGP2 and RIG-I C-terminal domains have a large basic surface, formed by the RNA-binding loop, and that the LGP2 C-terminal domain binds to the termini of dsRNAs (14, 21 –23, 28). Although the MDA5 C-terminal domain also has a large basic surface, it is extensively flat because of the open conformation of the RNA-binding loop (21). Consequently, the RNA-binding activity of MDA5 is much weaker than those of RIG-I and LGP2. The present study has demonstrated that LGP2 is more profoundly required for the recognition of RNA viruses detected by MDA5 than for those detected by RIG-I. MDA5 may require LGP2 for efficient recruitment of viral dsRNAs to facilitate the initiation of signaling, and LGP2 appears to be more important for MDA5 than for RIG-I, possibly because of differences in their affinities for dsRNAs.
Although LGP2 is involved in the responses to various RNA viruses, influenza virus infection induced normal type I IFN production in Lgp2 −/− cells. Type I IFN production in response to influenza virus was dependent on RIG-I but not on MDA5. We (5) and Pichlmair et al. (29) previously showed that phosphatase treatment of genomic RNA derived from influenza virus completely abolishes its type I IFN–inducing activity via RIG-I, indicating that a phosphate group at the 5′ end of the influenza virus genome is responsible for RIG-I–mediated recognition. Therefore, the 5′ triphosphate RNA present on viral genomes may be readily accessible to RIG-I without modification by LGP2.
Venkataraman et al. (24) reported that LGP2 acts as a negative regulator for the recognition of VSV and poly I:C, and a positive regulator for EMCV-induced IFN responses in macrophages by generating Lgp2 −/− mice. Their results are contradictory to our present findings in terms of the responses to poly I:C and viruses recognized by RIG-I. Although we do not have a clear explanation for these discrepancies, expression of LGP2 in Lgp2 −/− cells restored the responses to VSV and EMCV. Furthermore, we found that both Lgp2 −/− and Lgp2 K30A/K30A cells showed defects in IFN production in response to certain viruses recognized by RIG-I and showed normal responses to dsRNA specimens. Therefore, we believe that LGP2 acts as a positive, but not negative, regulator of RIG-I– and MDA5-dependent recognition of RNA virus infection and plays a pivotal role in antiviral responses in vivo.
Although some of the female Lgp2 −/− mice showed a defect in the development of the vagina, Lgp2 K30A/K30A mice did not exhibit any developmental abnormalities. Although the ATPase domain was essential for antiviral responses, the vaginal atresia was regulated by LGP2 independently of its ATPase activity. Given that Rig-I −/− mice showed fetal liver apoptosis at day 13, it will be interesting to analyze the role of the RIG-I ATPase activity in the control of development. Further studies are required to determine the roles of the RLR family members in controlling mammalian development.
Given that many RLR signaling molecules are inhibited by viral components, LGP2 may also be a target of the escape mechanisms exerted by various RNA viruses. Future studies aimed at identifying the mechanisms by which LGP2 modifies viral RNP complexes will help us to understand the roles of the innate immune system in intracellular virus recognition, and will lead to the development of new strategies to manipulate antiviral responses.
Materials and Methods
Generation of Lgp2 −/− and Lgp2 K30A/K30A Mice.
The Lgp2 gene was isolated from genomic DNA extracted from embryonic stem (ES) cells (GSI-I) by PCR. The targeting vector was constructed by replacing a 4-kb fragment encoding the Lgp2 ORF (including the DExH/H box) with a neomycin-resistance gene cassette (neo), and inserting herpes simplex virus thymidine kinase (HSV-TK) driven by the PGK promoter into the genomic fragment for negative selection. After the targeting vector was transfected into ES cells, G418 and gancyclovir double-resistant colonies were selected and screened by PCR, and recombination was confirmed by Southern blotting. Homologous recombinants were microinjected into C57BL/6 female mice, and heterozygous F1 progenies were intercrossed to obtain Lgp2 −/− mice. Lgp2 −/− and littermate control mice were used for subsequent experiments.
A point mutation was inserted into a genomic fragment harboring the exon encoding Lys-30 of murine Lgp2 by site-directed mutagenesis (Clontech) to replace this residue with Ala. A targeting vector was constructed with this genomic fragment and electroporated into ES cells. Homologous recombinants were selected and microinjected into C57BL/6 female mice, and heterozygous F1 progenies were crossed with CAG-Cre transgenic mice to excise the neo cassette. Next, the CAG-Cre transgene was removed from Lgp2 +/K30A mice by crossing the mice with C57BL/6 mice. Lgp2 K30A/K30A and littermate control mice were used for subsequent experiments.
All animal experiments were carried out with the approval of the Animal Research Committee of the Research Institute for Microbial Diseases (Osaka University).
Mice, Cells, and Reagents.
Rig-I −/− and Lgp2 −/− Mda5 −/− MEFs were prepared from embryos on 129Sv and C57BL/6 backgrounds derived at 11.5 days post coitum. BM-derived DCs were generated in RPMI medium 1640 containing 10% FCS, 50 mM 2-mercaptoethanol, and 10 ng/mL GM-CSF (PeproTech). pDCs and cDCs were isolated from the spleen by MACS using anti-B220 and anti-CD11c microbeads (Miltenyi Biotech). Poly I:C was purchased from Amersham Biosciences. RNAs were complexed with the cationic lipid Lipofectamine 2000 (Invitrogen) and added to cells. A/D-type CpG-oligodeoxynucleotides (D35) were synthesized by Hokkaido System Science. In vitro–transcribed dsRNA and triphosphate RNA were described previously (5, 9). Antibodies against phospho-STAT1 and STAT1 (Cell Signaling) were used for Western blotting as described previously (8).
Viruses.
VSV, VSV lacking an M protein variant (NCP), influenza virus ΔNS1, JEV, EMCV, and mengovirus were described previously (9). SeV lacking V protein (V–) was kindly provided by Dr. A. Kato. Reovirus was kindly provided by Dr. T. Dermody.
Northern Blotting.
Total RNA was extracted from peritoneal macrophages infected with EMCV or MEFs infected with influenza virus using TRIzol reagent (Invitrogen). The obtained RNA was electrophoresed, transferred to nylon membranes, and hybridized with various cDNA probes. To detect the expression of Lgp2 mRNA, a 326-bp fragment (772–1098) was used as a probe.
EMSA.
Peritoneal macrophages (3 × 106) were infected with EMCV for various periods. Nuclear extracts were purified from the cells using lysis buffer (10 mM Hepes-KOH pH 7.8, 10 mM KCl, and 10 mM EDTA, pH 8.0), incubated with specific probes for NF-κB or ISRE DNA-binding sites, electrophoresed, and visualized by autoradiography.
Luciferase Assay.
MEFs were transiently transfected with a reporter construct containing the IFN-β promoter together with an empty vector (Mock) or expression constructs for several genes using Lipofectamine 2000. As an internal control, the cells were transfected with a Renilla luciferase construct. The transfected cells were left untreated (medium alone) or infected with EMCV for 8 h. The cells were then lysed and subjected to a luciferase assay using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Retroviral Expression.
Murine LGP2 and LGP2K30A cDNAs were individually cloned into the pLZR-IRES/GFP retroviral vector (30). Retroviruses were produced by transient transfection of the constructs into PlatE cells. MEFs were separately infected with the retroviruses expressing LGP2 and LGP2 (K30A). At 2 days after infection, the cells were exposed to EMCV for 24 h, and the IFN-β concentrations in the culture supernatants were measured by ELISA.
Plaque Assay.
At 48 h after EMCV infection, hearts were prepared and homogenized in PBS. The virus titers in the hearts were determined by a standard plaque assay as described previously (9). After centrifugation, the supernatants were serially diluted and added to plates containing HeLa cells. Cells were overlaid with DMEM containing 1% low-melting point agarose and incubated for 48 h. The numbers of plaques were counted.
Measurement of Cytokine Production.
Culture supernatants were collected, and the cytokine concentrations were measured using ELISA kits for IFN-β (PBL Biomedical Laboratories) and IL-6 (R&D Systems) according to the manufacturers’ instructions.
Statistical Analysis.
The statistical significance of differences between groups was determined by Student's t test, and survival curves were analyzed by the log-rank test. Values of P < 0.05 were considered to indicate statistical significance.
Supplementary Material
Acknowledgments
We thank all of the colleagues in our laboratory, E. Kamada for secretarial assistance, and Y. Fujiwara, M. Kumagai, and R. Abe for technical assistance. We thank Drs. A. Kato and T. Dermody for providing viruses. This work was supported by the Special Coordination Funds of the Japanese Ministry of Education, Culture, Sports, Science and Technology, and grants from the Ministry of Health, Labour and Welfare in Japan, the Global Center of Excellence Program of Japan, and the National Institutes of Health (P01 AI070167).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1261.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912986107/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 10.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 15.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 17.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 18.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahasi K, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: Identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pippig DA, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 26.Linder P. Dead-box proteins: A family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salonen A, Ahola T, Kääriäinen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 29.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Vela A, et al. Transplanted long-term cultured pre-BI cells expressing calpastatin are resistant to B cell receptor-induced apoptosis. J Exp Med. 2001;194:247–254. doi: 10.1084/jem.194.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.