Abstract
Developing T cells face a series of cell fate choices in the thymus and in the periphery. The role of the individual T cell receptor (TCR) in determining decisions of cell fate remains unresolved. The stochastic/selection model postulates that the initial fate of the cell is independent of TCR specificity, with survival dependent on additional TCR/coreceptor “rescue” signals. The “instructive” model holds that cell fate is initiated by the interaction of the TCR with a cognate peptide-MHC complex. T cells are then segregated on the basis of TCR specificity with the aid of critical coreceptors and signal modulators [Chan S, Correia-Neves M, Benoist C, Mathis (1998) Immunol Rev 165: 195–207]. The former would predict a random representation of individual TCR across divergent T cell lineages whereas the latter would predict minimal overlap between divergent T cell subsets. To address this issue, we have used high-throughput sequencing to evaluate the TCR distribution among key T cell developmental and effector subsets from a single donor. We found numerous examples of individual subsets sharing identical TCR sequence, supporting a model of a stochastic process of cell fate determination coupled with dynamic patterns of clonal expansion of T cells bearing the same TCR sequence among both CD4+ and CD8+ populations.
Keywords: CDR3, clonal expansion, immune repertoire, T cell receptor
Following production of their T cell receptors (TCRs), T cells experience several developing stages. An encounter with a cognate peptide-MHC complex can induce naïve T (Tn) cells expressing the CD45RA isomer to begin to express CD45RO. Cells expressing both isomers are considered transitional in nature (Tt), thus cells identified on the basis of CD45RA expression alone include Tn and Tt and can thus be referred to as Tn+t. Cells expressing only CD45RO have passed into the memory (Tm) compartment, where they can lay quiescent awaiting repeat stimulation by the same or similar peptide-MHC complexes. Activated T cells (Ta) driven to effector function lose expression of both CD45RA and RO and express CD69. During different developing stages, T cells also face a series of cell fate choices: CD4+CD8+ cells commit to either the CD4+ helper (Th) or CD8+ cytotoxic (Tc) lineages, a choice closely associated with binding to MHC class II or class I peptide complexes, respectively. Subsequently, CD4+ T cells can develop into regulatory (Tr) CD25+ cells, or into CD25−CD294− Th1 (IFN-γ producing) or CD25−CD294+ Th2 (IL-4 producing) effector subsets. Other choices are also available (1, 2).
Although it is generally accepted that the TCR expressed by the developing T lineage cell will determine the response to a specific peptide-MHC complex, the role of the individual TCR in determining decisions of cell fate remains unresolved. To address these issues, we have coupled high-throughput sequencing techniques (3, 4) to high volume antibody covered superparamagnetic polystyrene bead isolation of defined T cell subsets with semiquantitative PCR amplification of the complementarity determining region 3 regions (CDR3) from mRNA molecules. CDR3 sequences, composed by the V(D)J combination, form the center of the antigen binding site where they often play a critical role in defining the affinity and specificity of the receptor for individual peptide-MHC complexes (5) of both the TCRα and TCRβ chains. Our goal was to produce comprehensive, unrestricted profiles of TCR diversity for key subsets of T cells isolated from the blood of a healthy individual at sequence-level resolution.
Results
In total, approximately 1.67 million effective sequence reads, which correspond to sequenced cDNA molecules, were generated for eight distinct T cell populations isolated from peripheral blood from a healthy, east Asian male, age 48, who had no known illnesses at the time of blood donation and reported feeling normal and well during the month before the sampling of his blood (Table 1). The first amplification sampled CD3+ T cells in general (pan T) (Figs. S1 and S2). Four additional amplifications (Tc, Tr, Th1, and Th2) sampled T cell subsets with divergent effector functions; the final three amplifications (Tn+t, Ta, and Tm) sampled T cells at different stages of T cell development (SI Text, Figs. S1 and S3, and Tables S1–S3). From these sequence reads, about 1.48 million CDR3 intervals were identified, totaling 169,977 and 113,290 unique CDR3 intervals for TCRα and TCRβ chains, respectively. With a few exceptions, a highly random pattern of germline VJ gene segment combinations was observed in the pan T sample (Fig. S2). Altogether, we identified 2,505 VJ combinations among 70,005 unique CDR3 nucleotide sequences (Fig. S2), which accounted for about 87% of the 2,874 potential Vα and Jα and Vβ and Jβ combinations predicted to yield functional rearrangements as cataloged in the ImMunoGeneTics (IMGT) database (6).
Table 1.
Sequence reads and CDR3 for different subsets of T cells
| Subset | Cell count | Effective read* | Total CDR3 | Unique CDR3† | |||
| TCRα | TCRβ | ||||||
| aa | na | aa | na | ||||
| Tr | 6.30 × 107 | 206,087 | 179,354 | 34,804 | 38,773 | 22,906 | 23,654 |
| Th1 | 1.84 × 108 | 174,046 | 150,122 | 29,471 | 32,518 | 19,644 | 20,061 |
| Th2 | 1.94 × 107 | 105,567 | 91,369 | 14,038 | 15,301 | 6,250 | 6,447 |
| Tc | 1.69 × 108 | 221,832 | 200,412 | 16,654 | 18,214 | 9,310 | 9,735 |
| Tn+t | 9.52 × 107 | 213,054 | 191,121 | 22,728 | 24,652 | 13,947 | 14,373 |
| Ta | 8.89 × 106 | 187,494 | 167,727 | 9,052 | 10,084 | 3,873 | 4,129 |
| Tm | 1.45 × 107 | 168,301 | 146,762 | 16,302 | 18,049 | 15,081 | 15,536 |
| pan T | 3.77 × 107 | 283,241 | 251,665 | 37,857 | 42,045 | 26,981 | 27,960 |
| pan T‡ | — | 80,246 | 71,765 | 15,638 | 16,622 | 10,308 | 10,483 |
| pan T§ | — | 30,579 | 27,263 | 7,794 | 8,130 | 5,334 | 5,416 |
| Total | — | 1,670,447 | 1,477,560 | 137,751 | 169,977 | 106,903 | 113,290 |
| Public¶ | — | 1,311 | 1,222 | 203 | 210 | 916 | 938 |
Tr, T regulatory cell (CD4+CD25+); Th1, T helper cell 1 (CD4+CD25−CD294−); Th2, T helper cell 2 (CD4+CD25−CD294+); Tc, T cytotoxic cell (CD8+); Tn+t, naïve and transitional T cell (CD45RA+); Ta, activated T cell (CD45-RO-CD69+); Tm, memory T cell (CD45RA-RO+); aa, amino acids; na, nucleic acids.
*An effective read is a read that can be mapped with both V and J germline segments.
†A unique CDR3 sequence is a nonredundant fragment of amino acids (aa) or nucleic acids (na), which is in a stop-codon-free reading frame containing both translated conserved motifs (SI Text).
‡,§pan T samples were processed along with pan B cells and T cell counts for these two samples were not recorded.
¶Public sequence data set was compiled by combining relevant cDNA sequences in both the GenBank and the IMGT database. Reported here are those passed through the analysis pipeline.
To avoid the distortion by dominant clones as results of immune responses, each unique CDR3 sequence was counted as one regardless of how many copies were observed when we examined the pattern of V, D, and J domain usage; CDR3 length; addition of nontemplated nucleotides; trimming of nucleotide at the V, D, and J coding ends; or amino acids usage in CDR3 intervals. Among the seven T cell developmental or effector subsets examined, we found no statistically significant difference in TCR Vα, Jα, Vβ, Dβ, or Jβ utilization (SI Text). For the CDR3s, we found no statistically significant difference in the distribution of CDR3 lengths, addition of nontemplated (N) nucleotides, or trimming of nucleotides at the V(D)J coding ends among different T cell subsets (Table S4). The frequency of use of individual amino acids within CDR3 intervals was indistinguishable between the subsets of cells (Fig. S4). This similarity in V(D)J recombination products irrespective of subset is consistent with the view that the differentiation of T cells into the subsets that we examined is neutral to V(D)J recombination.
Different T cells forming identical TCR CDR3 nucleotide sequences during development are so remote (SI Text) that individual TCRβ CDR3 nucleotide intervals can be used as clonal markers. Although our approach to evaluating TCR diversity has allowed us to sample the repertoire with several orders of magnitude of increased resolution, the complexity of a T cell population that has been estimated to approach 1014 cells per individual (SI Text) still precludes the possibility of measuring its diversity directly. Previous attempts used either extrapolation from less than 10,000 sequences or indirect molecular measurements (7 –10). The compound Poisson process model has been used to estimate human gene number by evaluating the results of large-scale EST sequencing (11). Using this same approach, we estimated the diversity of the TCR α and β repertoires expressed by our study subject to include 0.47 × 106 and 0.35 × 106 unique TCRα and TCRβ CDR3 nucleotide sequences, respectively (Table 2). This represents about one third of the extent of TCR diversity previously estimated by a combination of spectratyping and sequencing analysis (7).
Table 2.
Numbers of TCRα and TCRβ CDR3 sequences of T cell subsets in peripheral blood based on sequencing data
| Subset | Predicted TCRα | Predicted TCRβ |
| Tc | 50,873 | 30,376 |
| Tr | 89,920 | 58,325 |
| Th1 | 69,298 | 57,072 |
| Th2 | 27,674 | 12,715 |
| Tn+t | 74,851 | 47,507 |
| Tm | 36,595 | 35,254 |
| Ta | 19,494 | 7,563 |
| Overall T* | 466,757 | 348,519 |
Also listed in Table 2 is the estimated diversity of the CDR3 repertoire by T cell subset. Among the three developmental subsets (Tn+t, Ta, and Tm) evaluated, the Ta subset exhibited the least diversity, which was defined as the estimated number of unique CDR3 intervals, for both the TCRα and TCRβ repertoires. Among the various effector populations, the Th2 subset appeared least diverse and the Tr population exhibited the greatest diversity of all.
When evaluated by the extent of clonal expansion as manifest by the frequency of clones (a clone is defined as a unique CDR3 fragment) having >100 reads, a striking divergence was observed between the four effector T cell subpopulations (Fig. 1). More than 60% of the Tc sequences belonged to clones having >100 reads, whereas the Tr and Th2 subsets exhibited few to none such clones. The Th1 population proved intermediate between these two groups. Divergence in the frequency of clones having >100 reads was also observed when T cells were evaluated by developmental stage. The Tn+t and Ta subsets exhibited patterns of clones having >100 reads similar to those observed in the Tc population, whereas the Tm subset matched Th2 and Tr subsets for minimal numbers of such clones. The TCRα and TCRβ CDR3 sequences that dominated Tc subset also dominated the Tn+t, Ta, and pan T subsets (Table S5).
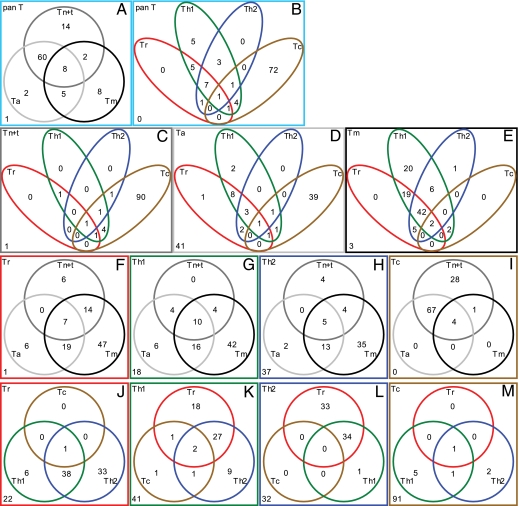
Fig. 1.
Abundance of CDR3 sequences versus cumulative frequency for TCRα (Left) and TCRβ (Right). Red: Tr; pink: Th1; green: Th2; cyan: Tc; gray: Tn+t; black: Ta; blue: Tm; and orange: pan T. As the total number of CDR3 sequences for different subsets of T cells are different, uniform sampling procedure was applied to each subset of T cells to bring the same number of TCRα or TCRβ CDR3, respectively.
We identified the 10 most common TCRα and TCRβ sequences in the pan T cell population and compared them to the seven T cell subsets whose repertoire we had amplified (Table S5). Many of those dominant CDR3 intervals were seen in several subsets. For example, the TCRα CDR3 interval “APEAMGGSEKLV” was the second most common sequence in pan T, Tn+t, Ta, and Tc. This sequence was also present in the Th1 population. In the Tm population, clones common to the Th1 subset were overrepresented; furthermore, of those clones present in the Tn+t, Ta, and panT that were not predominant in the Tc population, several were predominant in the Th1 population. Conversely, clones common to the Th2 population were over-represented in the Tr subset.
When clone sequences are found in common across amplified products from varied subpopulations, the possibility of technique artifact or random accident must be addressed. We ruled out these possibilities in part due to cell isolation protocol and in part through statistical analysis. First, cell subsets within the same group (effector group: Th1, Th2, Tr, and Tc or developmental group: Tn+t, Ta, and Tm) were exclusive according to the cell isolation protocol (see Materials and Methods). Second, the possibility that the detection of identical CDR3s in these two T cell subsets as the result of cell contamination was largely ruled out by the vigorous cross-contamination detection procedure (SI Text). Third, the shared CDR3 sequences are long and contain several N-nucleotide additions, thus it is unlikely that shared CDR3 sequences could be generated by independent events. For instance, the CDR3 sequence “APEAMGGSEKLV” was generated with 11 N-nucleotides addition and 7 nucleotides trimmed at the 5′ end of TRAJ57 and the random chance generating CDR3 sequences akin to this one is estimated as 1.1e-11.
To further assess how commonly TCR sequences were shared among clonally expanded cells, we extended our analysis to the 100 most abundant TCRβ (Fig. 2) and TCRα (Fig. S5) CDR3 sequences. Both analyses yielded highly similar results and supported the data obtained with the top ten sequences as well as provided additional insights.
Fig. 2.
(A–M) The 100 most abundant TCRβ CDR3 sequences of a particular subset (labeled at the top left corner of each box) common to those in subsets of T cells of either different developing stages (gray: Tn+t; dark grey: Ta; and black: Tm) or different fates (red: Tr; green: Th1; blue: Th2; and brown: Tc). The numbers of CDR3 sequences that are unique to each subset are shown in the nonoverlapping sections. The number of CDR3 sequences that are common to any two, three, and four of these subsets are indicated in the relevant overlapping areas. The number of CDR3 sequences that are not found in those examined subsets is labeled at the bottom left corner of each box.
Fig. 2A shows the composition of the 100 most abundant TCRβ CDR3 sequences in the pan T samples at three different developing stages (Tn+t, Ta, and Tm). Of the top 100 most dominant TCRβ CDR3 sequences in the pan T cell amplification, 84 were found in Tn+t subset, indicating the dominance of Tn+t in the overall T cell population. In addition, 68 of 100 dominant TCRβ CDR3 sequences in the pan T samples are common to both Tn+t and Ta, suggesting that the expanded clones were primarily the product of recent antigenic stimulation. Approximately one-quarter of the pan T TCRβ CDR3 sequences were common to those in memory T cells, suggesting that this population was a mixture of both recently generated and longstanding clones. When compared by effector T cell subset (Fig. 2B). Approximately four-fifths of the common pan T clones were found in the Tc subset. Overlap was observed between the Tc, Th1, Th2, and Tr subsets.
Among the Tn+t population (Fig. 2C), more than 90% of the clones were found in the Tc population. However, only 43 of the 100 most common Ta CDR3s could be identified among Tc clones, with the Th1, Th2, and Tr subsets increasing their representation with mixed sharing among the four subsets (Fig. 2D).
Among the Tm population (Fig. 2E), only 4 clones could be found in the Tc subset, whereas the Th1 subset encompassed 91 clones. An even more dramatic overlap of TCR sequences among the Th1, Th2, and Tr subsets became apparent, with 44 sequences present in all three.
When effector cells were examined by developmental stage (Fig. 2 F–I), Tr cells dominated in the Tm population comprising 87 clones. Th1 and Th2 cells were also over-represented in the Tm population, whereas Tc cells were over-represented in the Tn+t and Ta subsets.
Effector cells were also examined by other effector cells (Fig. 2 J–M). There are more overlaps between Tr and Th2. There are substantial number of overlaps between Th1, Th2, and Tr, but a limited number of overlap between CD8+ (Tc) and CD4+ (Tr, Th1, and Th2) T cells.
Discussion
Repertoire diversity is a fundamental determinant of the competence of the immune system. The loss of diversity of an immune repertoire has been linked to aging (12) and implicated in various disease states (13 –15). Previous methods extrapolate the full diversity of the human immune repertoire from only a small fraction of VJ combinations, which in turn was chosen at random, thus making it difficult to determine whether the actual diversity or the extent of clonal amplification of the repertoire could have been quantified. Here the 454 technology platform has enabled the rapid sequencing of millions of DNA fragments at low cost and without cloning bias (3). By combining this method with the ARM-PCR (16), which allows millions of highly similar sequences to be amplified in a semiquantified matter from a complex mixture, our approach has provided us with the necessary arsenal to characterize the majority of antigen receptor sequences at the level of sequence resolution.
Our analysis has yielded literally hundreds of thousands of sequence reads with tens of thousands of unique sequences. The various T cell subsets examined exhibit many common features in terms of germline gene segment usage, CDR3 length, number of N-nucleotide additions, nibbling at ends of germline gene segments, and amino acids usage at the CDR3 intervals, which is consistent with the fact that the fate of different subset T cells are determined after the expression of TCRs.
This first view of the expanded repertoire presents us with a dramatic view of T cell subset population dynamics. We identified a number of clones with very high frequency sequence reads among the cells from the CD8+ population. The donor had no known illnesses and reported feeling normal and well during the month before the time of sampling. Although it is possible that he had activated a subpopulation of CD8+ T cells due to an acute subclinical infectious challenge (17, 18), the possibility that a set of CD8+ T cells had undergone an independent expansion unrelated to a recent exogenous antigenic challenge (19 –23) cannot be excluded.
CD8+ T cells dominate in the Tn+t cell population, and a substantial number of both CD4+ and CD8+ T cells become activated after an antigenic challenge. From these, many CD4+ T cells and only a small portion of CD8+ T cells differentiate into memory T cells. Thus it is possible that a recent or ongoing antigenic challenge precipitated the expansion of those CD4+ and CD8+ T cells that shared the identical TCR sequences, perhaps due to a dominant effect of specific antigen epitopes. However, we cannot rule out the possibility that these outgrowths also reflect the early appearance of age-related CD4+ and CD8+ clonal expansions sharing a common origin (24). These possibilities are not mutually exclusive because clonally expanded cells might have a greater likelihood of begin activated by the same or different antigens.
Among the clonally expanded T cells of different fates, the majority of Tr, Th1, and Th2 cells are related to memory T cells, whereas the majority of Tc cells are related to naïve T cells. The more moderate expansion of Th1 cells may reflect the effect of Tc expansion, as Tc cells secret IFN-γ (25), which promotes the proliferation of Th1 subsets while inhibiting the proliferation of Th2 subsets (26). The Th1 and Th2 populations contribute the majority of the memory T cell compartment with some sharing with the naïve/transitional and activated subsets.
Our results call attention to the substantial number of TCR α and β CDR3 sequences that are clonal expanded and common in T cell subsets of different fates. There is a significant amount of sharing of TCR sequence among the Th1, Th2, and Tr populations. These findings would suggest semicoordinated expression of Th1, Th2, and Tr cells among clonally expanded populations within these subsets, suggesting that the choices of Th1, Th2, and Tr outcomes are stochastic and can be driven by the same or highly similar antigenic stimulus. An ancestral T cell thus appears to have the potential to develop into different cells with the same specificity but opposite effects, such as Th1, Th2 versus Tr cells, which have potent inhibitory effects on immune responses to foreign antigens and the development of autoimmunity (27, 28). Shared specificity through identical CDR3 sequences between effector T cells might provide an important communication avenue to maintain the homeostasis of immune system response, providing paired signals at the cellular level.
Our studies present a cross-sectional view of the TCR repertoire among one pan T and seven distinct subpopulations of T cells identified on the basis of characteristic surface markers. Of these, one amplification (pan T) sampled CD3+ T cells in general, and three amplifications (Tn+t, Ta, and Tm) sampled T cells at different stages of development. The remaining four amplifications (Tc, Tr, Th1, and Th2) sampled T cell subsets with divergent effector functions which, until recently, were thought to be relatively fixed. However, recent studies have indicated that fate decisions in T helper cells, and especially in the Tr population, may be more plastic than previously appreciated (29 –31). It is thus additionally possible that some of the shared TCR sequences among effector T cell subsets reflect alterations in cell fate that occurred after antigen exposure created a clonal expansion in cells bearing a different effector phenotype.
Harder to explain are the rare, but dominant, clones, where sequences commonly found in the CD8+ T cell population are also found in the Th1 and Th2 subsets. It is unclear whether identical TCRβ CDR3 sequences can recognize the same or different antigens presented by different MHC molecules; however, the sharing of the same clonal specificity between CD4+ and CD8+ cells has been noticed previously (32). Our cell sorting process leaves open the possibility of a CD4+CD8+ population, but it is difficult to conceive of this population, which is typically viewed as the product of recent thymic activity, entering into the CD25+ compartment as well as Th1 and Th2. Conversely, it is possible that the population of cells bearing shared TCR sequence regardless of effector function or MHC segregation may represent benign clonal outgrowths of cells that had undergone a transforming mutation in the thymus.
We also identified many expanded CDR3 reads that could not be assigned to any of the functional subsets studies. For example, there were 41 expanded TCRβ CDR3s in the activated T cell pool that could not be found in the Tr, Th1, Th2, or Tc subsets, suggesting that they might belong to other subsets not identified by the sorting process that we used.
Among 70,005 unique CDR3 nucleotide sequences, we identified 2,505 VJ combinations among the TCRα and TCRβ VJ repertoire that were expressed in our donor subject. This accounted for about 87% of the 2,874 potential Vα and Jα, and Vβ and Jβ combinations. The expressed TCRα and TCRβ repertoires are heavily influenced by rearrangement frequency and by both positive and negative selection events in the thymus and the periphery (33 –40). At present we cannot distinguish between differences in rearrangement frequency (33) or the effects of either positive or negative selection events in the thymus or in the periphery (34 –40) as the proximate cause of the absence of these particular combinations. In either case, it is clear that our donor subject does not have full access to the potential TCR diversity encoded by the germline repertoire. Whether this absence creates “holes” in the repertoire that can influence the development of disease and whether the same or different “holes” can be found in other members of the population remains to be determined.
In conclusion, we have demonstrated a successful approach for determining the entire diversity of immune repertoire with sequence level resolution. The enormous data generated by this approach has corroborated previous estimates of the diversity of T cell repertoire at a direct level. This systematic approach provides a useful tool for assessing immune competence, tracking T cell expansion kinetics, and identifying antigen-specific T cell clones in patients with infection or cancer. Understanding those is likely to facilitate the development of immunotherapy for the treatment of viral infections and tumors.
Materials and Methods
Isolation of T Cell Subsets.
Informed consent was obtained from the blood donor. T cell isolations were performed using superparamagnetic polystyrene beads (Miltenyi) coated with monoclonal antibodies specific for the particular T cell subset (Fig. S1).
From whole blood, mononuclear cells were obtained by Ficoll Prep, followed by anti-CD14 microbeads to remove monocytes. This monocytes-depleted, mononuclear cell fraction was then used as a source for specific T cell subset fractions.
Cytotoxic CD8+ T cells were isolated by negative selection with anti-CD4 multisort beads (Miltenyi Biotec), followed by positive selection with anti-CD8 beads. CD4+ T cells were isolated by positive selection with anti-CD4 beads. Anti-CD25 beads (Miltenyi Biotec) were used to select CD4+CD25+ regulatory T cells. From the CD4+CD25− flow through, anti-CD56 beads was used to remove CD4+CD56+ NKT cells. From the CD4+CD56-flow through, anti-CD294 beads were added to select CD4+CD25−CD294+ Th2 cells and the flow through CD4+CD25−CD294− were collected as Th1 cells.
Isolated CD4+ T cells and CD8+ T cells according to the protocols listed above were pooled together, and anti-CD45RA microbeads were added to isolate a combined population of CD45RA+ naive and transitional T cells (Tn+t). Anti-CD45RO beads were added to the CD45RA− pass-through to select for CD45RA-RO+ memory T cells. Anti-CD69 microbeads were added to the flow-through CD45RA-RO− cells to isolate CD45RA-RO−CD69+ activated T cells.
All isolated cell populations were immediately resuspended in RNAprotect reagent (Qiagen).
ARM-PCR Procedure.
For each target, a set of nested sequence specific primer was designed (Forward-out, Fo; Forward-in, Fi; Reverse-out, Ro; and Reverse-in, Ri). A pair of common sequence tags was linked to all internal primers (Fi and Ri). Once these tag sequences were incorporated into PCR products in the first few amplification cycles, an exponential phase of the amplification could be carried out with a pair of communal primers, called superprimers, which can pair with the tag sequences (16). In the first round of amplification, only sequence-specific nested primers were used. The nested primers were then removed by exonuclease and the first-round PCR products were used as templates for a second round of amplification by adding communal primers and a mixture of fresh enzyme and dNTP.
Aligning Immune Repertoire Sequences.
To assign rearranged mRNA sequences to their germline V, D, and J counterparts, we developed a tool called IRmap which is similar to the Germline Query program (41). The IRmap program uses the pyromap program (42), a modification of the Smith-Waterman algorithm, adapted to the 454 sequencing error pattern. The 454 sequencing platform outputs a Phred-equivalent quality score for every position in a read. Quality scoring in the 454 sequencing platform was originally designed to measure the confidence that the homopolymer length at that position is correct (3). The quality score of a position is also a good measurement of confidence that the correct base is called at any position, as with a traditional Phred score. By incorporating the quality score into the Smith-Waterman algorithm, the program can improve the mapping accuracy between individual reference sequences and the 454 read (42). This improved mapping algorithm allowed us precisely determine alignments between 454 reads and germline V, D, and J segment alignments in the IMGT/GENE-DB (6) reference directory of human T cell receptor α and β gene products (available on the IMGT server http://www.imgt.org). The IRmap program systematically searches the directory of germline V and J gene segments for the best matches and masks the region of the query sequence that aligns to V and J segments and searches for Dβ segments in the intervening sequences subsequently.
Germline Dβ segments are very short and the aligned fragments are even shorter because of nibbling and somatic mutation. Thus, although V and J identity were easily obtained, Dβ assignment proved difficult because the scores of the alignments between germline Dβ and the sequencing reads proved too small to be distinguishable from random noise. To assign Dβ segments onto sequencing reads reliably, we first calculate cutoff scores to assign Dβ segments to an mRNA through a simulation experiment. A set of 10,000 sequences was randomly generated with the equal frequency for A, C, G, T for a particular length. The simulated sequences were aligned to each germline Dβ segment, and the 99th percentile score is set as the cutoff score for that particular Dβ segment. For each length ranging from 10 to 100 bases, the cutoff scores were calculated. We filtered out alignments with Dβ segments of the score less than the cutoff value.
Define CDR3 Interval.
Both TCRα and TCRβ transcripts have the conserved amino acid sequence Y[YFLI]C at the 3′ end of the V gene segment and [FW]GXGT (X stands for 1 of 20 amino acids) within the J segments. The CDR3 interval was identified as comprising all of the amino acids between these two conserved motifs.
Supplementary Material
Acknowledgments
We thank Dr. Chris Gunter and Dr. Michael Mindrinos for critical reading of the manuscript and Mr. Lonnie McMillian for inspiring conversations. This work was supported by funding from the HudsonAlpha Institute for Biotechnology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited into NCBI sequence read archive (accession no. SRA010149).
This article contains supporting information online at www.pnas.org/cgi/content/full/0913939107/DCSupplemental.
References
- 1.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 2.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul WE. Fundamental Immunology. 6th Ed. Lippincott Williams & Wilkins, Philadelphia, PA; 2008. p. 1632. [Google Scholar]
- 6.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33(Database issue):D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arstila TP, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 8.Casrouge A. Size estimates of the alpha beta TCR repertoire of naive mouse splenocytes. J Immunol. 2001;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 9.Baum PD, McCune JM. Direct measurement of T-cell receptor repertoire diversity with AmpliCot. Nat Methods. 2006;3:895–901. doi: 10.1038/NMETH949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogle BM, et al. Direct measurement of lymphocyte receptor diversity. Nucleic Acids Res. 2003;31:e139. doi: 10.1093/nar/gng139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JP, et al. Gene capture prediction and overlap estimation in EST sequencing from one or multiple libraries. BMC Bioinformatics. 2005;6:300. doi: 10.1186/1471-2105-6-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naylor K, et al. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174:7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 13.Wagner UG, Koetz K, Weyand CM, Goronzy JJ. Perturbation of the T cell repertoire in rheumatoid arthritis. Proc Natl Acad Sci USA. 1998;95:14447–14452. doi: 10.1073/pnas.95.24.14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peggs KS, Verfuerth S, D’Sa S, Yong K, Mackinnon S. Assessing diversity: Immune reconstitution and T-cell receptor BV spectratype analysis following stem cell transplantation. Br J Haematol. 2003;120:154–165. doi: 10.1046/j.1365-2141.2003.04036.x. [DOI] [PubMed] [Google Scholar]
- 15.Manca F, et al. Rational reconstitution of the immune repertoire in AIDS with autologous, antigen-specific, in vitro-expanded CD4 lymphocytes. Immunol Lett. 1999;66:117–120. doi: 10.1016/s0165-2478(98)00168-0. [DOI] [PubMed] [Google Scholar]
- 16.Han J, et al. Simultaneous amplification and identification of 25 human papillomavirus types with Templex technology. J Clin Microbiol. 2006;44:4157–4162. doi: 10.1128/JCM.01762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantaleo G, et al. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 18.Callan MF, et al. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus In vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hingorani R, et al. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J Immunol. 1993;151:5762–5769. [PubMed] [Google Scholar]
- 20.Monteiro J, et al. Oligoclonality in the human CD8+ T cell repertoire in normal subjects and monozygotic twins: Implications for studies of infectious and autoimmune diseases. Mol Med. 1995;1:614–624. [PMC free article] [PubMed] [Google Scholar]
- 21.Morley JK, Batliwalla FM, Hingorani R, Gregersen PK. Oligoclonal CD8+ T cells are preferentially expanded in the CD57+ subset. J Immunol. 1995;154:6182–6190. [PubMed] [Google Scholar]
- 22.Eiraku N, et al. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotropic virus type I-infected individuals. J Immunol. 1998;161:6674–6680. [PubMed] [Google Scholar]
- 23.Batliwalla F, Monteiro J, Serrano D, Gregersen PK. Oligoclonality of CD8+ T cells in health and disease: Aging, infection, or immune regulation? Hum Immunol. 1996;48:68–76. doi: 10.1016/0198-8859(96)00077-8. [DOI] [PubMed] [Google Scholar]
- 24.Wack A, et al. Age-related modifications of the human alphabeta T cell repertoire due to different clonal expansions in the CD4+ and CD8+ subsets. Int Immunol. 1998;10:1281–1288. doi: 10.1093/intimm/10.9.1281. [DOI] [PubMed] [Google Scholar]
- 25.Slifka MK, Whitton JL. Antigen-specific regulation of T cell-mediated cytokine production. Immunity. 2000;12:451–457. doi: 10.1016/s1074-7613(00)80197-1. [DOI] [PubMed] [Google Scholar]
- 26.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 27.Taams LS, et al. Antigen-specific T cell suppression by human CD4+CD25+ regulatory T cells. Eur J Immunol. 2002;32:1621–1630. doi: 10.1002/1521-4141(200206)32:6<1621::AID-IMMU1621>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 28.McHugh RS, Shevach EM, Thornton AM. Control of organ-specific autoimmunity by immunoregulatory CD4(+)CD25(+) T cells. Microbes Infect. 2001;3:919–927. doi: 10.1016/s1286-4579(01)01453-8. [DOI] [PubMed] [Google Scholar]
- 29.Sundrud MS, et al. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J Immunol. 2003;171:3542–3549. doi: 10.4049/jimmunol.171.7.3542. [DOI] [PubMed] [Google Scholar]
- 30.Peck A, Mellins ED. Plasticity of T-cell phenotype and function: The T helper type 17 example. Immunology. 2009 doi: 10.1111/j.1365-2567.2009.03189.x. 2009 Nov 17. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Imberti L, Sottini A, Signorini S, Gorla R, Primi D. Oligoclonal CD4+ CD57+ T-cell expansions contribute to the imbalanced T-cell receptor repertoire of rheumatoid arthritis patients. Blood. 1997;89:2822–2832. [PubMed] [Google Scholar]
- 33.Wilson A, Maréchal C, MacDonald HR. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J Immunol. 2001;166:51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- 34.Aude-Garcia C, et al. Pairing of Vbeta6 with certain Valpha2 family members prevents T cell deletion by Mtv-7 superantigen. Mol Immunol. 2000;37:1005–1012. doi: 10.1016/s0161-5890(00)00106-1. [DOI] [PubMed] [Google Scholar]
- 35.Blackman MA, Marrack P, Kappler J. Influence of the major histocompatibility complex on positive thymic selection of V beta 17a+ T cells. Science. 1989;244:214–217. doi: 10.1126/science.2784868. [DOI] [PubMed] [Google Scholar]
- 36.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 37.Kappler JW, Staerz U, White J, Marrack PC. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 38.MacDonald HR, Lees RK, Schneider R, Zinkernagel RM, Hengartner H. Positive selection of CD4+ thymocytes controlled by MHC class II gene products. Nature. 1988;336:471–473. doi: 10.1038/336471a0. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald HR, et al. T-cell receptor V beta use predicts reactivity and tolerance to Mlsa-encoded antigens. Nature. 1988;332:40–45. doi: 10.1038/332040a0. [DOI] [PubMed] [Google Scholar]
- 40.Gulwani-Akolkar B, et al. Do HLA genes play a prominent role in determining T cell receptor V alpha segment usage in humans? J Immunol. 1995;154:3843–3851. [PubMed] [Google Scholar]
- 41.Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: A systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J Mol Biol. 1997;270:587–597. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.