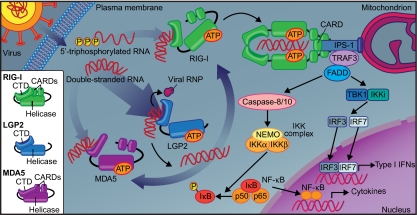
We live under constant attack by viruses, and understanding both how viruses invade and how we fight them may help us to take action against them. In the host, the battle against viral invasion begins with recognition, and a report on LGP2, a member of the RIG-I-like receptor family, by Satoh et al. in this issue of PNAS (1) sheds new light on the mechanisms of cytoplasmic virus recognition. This report provides evidence, in contrast to previous findings, that LGP2 functions as a positive regulator of virus recognition and subsequent antiviral responses (Fig. 1).
Fig. 1.
Model for LGP2 function in potentiating RIG-I and MDA5 antiviral signaling. Upon infection with an RNA virus, viral dsRNA or 5′-triphosphorylated RNA can activate RIG-I or MDA5 by binding to their CTDs. LGP2, as demonstrated by Satoh et al., facilitates recognition of viral RNA by RIG-I and MDA5, possibly by directly altering RNA conformation or dislodging viral ribonucleoproteins (RNPs) in an ATP-dependent manner to expose RNA. Once activated, RIG-I undergoes an ATP-dependent conformational change that promotes oligomerization and interaction with IPS-1 via homotypic CARD-CARD binding. TRAF3 and FADD can form a complex with IPS-1 that signals to TBK1 and IKKi to phosphorylate and activate IRF3 and IRF7 for translocation to the nucleus and activation of genes including Ifna and Ifnb. The IPS-1/TRAF3/FADD complex also activates caspases 8 and 10 leading to IKK complex activation and release of inhibitory IκB from the NF-κB complex. NF-κB can then move into the nucleus to transactivate cytokine genes. Satoh et al. show that NF-κB and IRF activation are severely impaired in Lgp2−/− cells. Although MDA5 dimerization has not been demonstrated, signaling from MDA5 (not shown) is expected to be similar to RIG-I signaling.
At least three protein families are involved in the recognition of virus-specific components, typically double-stranded (ds) or single-stranded (ss) RNA and DNA, by innate immune cells: the Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD)-like receptors (NLRs), and the retinoic acid-inducible gene I (RIG-I)-like receptor (RLRs). RLRs, the focus of this commentary, are members of the DExD/H box-containing RNA helicase family and are responsible for cytoplasmic virus recognition and ensuing signaling leading to type I interferon (IFN) and inflammatory cytokine production. Two RLRs, RIG-I and MDA5, sense specific viruses including influenza A, Sendai, and vesicular stomatitis viruses in the case of RIG-I, and encephalomyocarditis and murine hepatitis viruses in the case of MDA5, through recognition of 5′-triphosphorylated and uncapped ssRNA or dsRNA (2 –4), species not found among endogenous self-RNA. RIG-I can also detect 5′-triphosphorylated dsRNA transcribed from AT-rich dsDNA, or DNA from bacteria or DNA viruses, by DNA-dependent RNA polymerase III (Pol III) (5, 6).
The third RLR, LGP2, contains helicase and C-terminal domains (CTDs) homologous in sequence and structure to those of RIG-I and MDA5 (7, 8). LGP2 binds with higher affinity than either RIG-I or MDA5 to dsRNA and ssRNA via the CTD, which folds to form a basic RNA-binding groove bounded on one side by a critical RNA-binding loop (9 –12). However, LGP2 lacks the two caspase recruitment domains (CARDs) present at the N termini of RIG-I and MDA5 that mediate homotypic interactions involved in the assembly of a complex with the essential RLR adapter IFN-β promoter stimulator 1 (IPS-1; also known as MAVS, VISA, and Cardif) and other downstream molecules. The absence of any CARD in LGP2, together with in vitro data demonstrating that transient overexpression of LGP2 inhibited IFN-stimulated response element (ISRE)-dependent reporter gene transcription upon Newcastle disease virus or Sendai virus infection (7, 8, 13), prompted the view that LGP2 functions as a negative regulator of signaling initiated by RIG-I and MDA5, which sense these paramyxoviruses.
However, the function of LGP2 in virus recognition and signaling has remained controversial. In support of the proposed inhibitory role of LGP2 in RIG-I signaling, mouse embryonic fibroblasts (MEFs) from mice with a targeted deletion of LGP2 produced increased amounts of IFN-β and active NF-κB in response to transfected poly-I:C relative to wild-type MEFs (14). In addition, Lgp2−/− mice displayed enhanced resistance to intranasal infection with a lethal inoculum of vesicular stomatitis virus (VSV), known to be sensed by RIG-I. In contrast, Lgp2−/− mice showed increased susceptibility and decreased serum levels of type I IFNs and interleukin (IL)-6 in response to i.p. infection with encephalomyocarditis virus (EMCV), which is detected by MDA5.
Using Lgp2−/− and Lgp2K30A/K30A mice (both independently generated, the latter homozygous for a mutation in the helicase ATP-binding site), Satoh et al. address the role of LGP2 in resistance to viruses sensed by RIG-I and/or MDA5. LGP2 functions specifically in conventional dendritic cells (cDCs), and both mutant alleles negatively influenced IFN-β production by cDCs responding to EMCV, Mengo virus, VSV, Sendai virus, Japanese encephalitis virus, and reovirus, although responses to influenza virus or to transfected poly-I:C were normal. Similar results were found for MEFs, in which reconstitution with wild-type LGP2 restored EMCV-induced IFN-β production. Lgp2−/− and Lgp2K30A/K30A mice displayed increased susceptibility to EMCV infection in vivo, showing undetectable serum levels of IFN-β after infection. These findings are bolstered by experiments demonstrating impaired activation of ISRE and NF-κB response elements, as well as reduced STAT1 phosphorylation, in Lgp2−/− cells following EMCV infection. Together, these comprehensive investigations indicate a positive role for LGP2 in virus recognition.
LGP2 works upstream of RIG-I and MDA5 to potentiate viral RNA-induced signaling.
The data of Satoh et al. differ from those of Venkataraman et al. (14) with respect to viral susceptibility of LGP2-deficient mice and the response of Lgp2−/− cells to poly-I:C stimulation. Satoh et al. also observe vaginal atresia in adult Lgp2−/− females, and a reduced frequency of Lgp2−/− mice born from heterozygous crosses than the expected Mendelian frequency. No developmental defects were reported for Lgp2−/− mice by Venkataraman et al., suggesting critical genetic differences distinguish the two knockout strains that may similarly account for the disparity in responses to viral infection and exogenous RNA treatment. Interestingly, Lgp2K30A/K30A mice showed no developmental defects, indicating that the function of LGP2 in development may be distinct from its role in viral sensing.
To probe the mechanism by which LGP2 participates in virus recognition and signaling, Satoh et al. use the Lgp2K30A allele to show that the LGP2 ATPase domain is critical for IFN-β production and viral resistance. The Lgp2K30A allele was equivalent to the Lgp2 deletion allele in its effect on antiviral responses in vitro and in vivo. Importantly, overexpression of the CARDs of either RIG-I or MDA5 in Lgp2−/− MEFs activated the IFN-β promoter, suggesting that LGP2 functions upstream of RIG-I and MDA5. Satoh et al. propose that LGP2 may function to modify viral RNA by removing viral RNA-binding proteins, altering the conformation of RNA or perhaps modulating the intracellular localization of viral RNA, thereby facilitating recognition by RIG-I or MDA5 (Fig. 1). Consistent with this hypothesis, Lgp2−/− cells display intact IFN-β production in response to transfected poly-I:C that lacks complex secondary and ribonucleoprotein-induced structure. Whether the LGP2 helicase domain actually needs to unwind RNA to mediate antiviral responses was not investigated, but neither RIG-I nor MDA5 requires helicase activity for dsRNA-induced IFN induction (15, 16). With the exception of influenza, LGP2 deficiency appeared to affect MDA5-dependent responses to viruses more severely than RIG-I-dependent responses, possibly because on its own, MDA5 has weaker RNA-binding activity than RIG-I (9). Thus, LGP2 may specifically aid MDA5 in viral RNA recognition by promoting recruitment or conformational alteration of viral RNAs.
Significantly, the findings of several groups probing the mechanism of LGP2 action are consistent with Satoh and colleagues’ hypothesis that LGP2 structurally modifies or recruits viral RNA to facilitate detection by RIG-I and/or MDA5. Previously, direct binding of LGP2 to dsRNA was proposed to sequester viral RNA from RIG-I/MDA5, preventing signaling. Whereas this may be true in the case of LGP2 overexpression, which has consistently been found to inhibit RIG-I- and MDA5-mediated antiviral responses, the results of Satoh et al. suggest that LGP2 binds to RNA to mediate conformational changes that promote recognition by RIG-I and MDA5. LGP2 has also been shown to interact with RIG-I via its CTD (17). The RIG-I-LGP2 interaction was hypothesized to prevent RIG-I from oligomerizing with itself and with IPS-1, thereby blocking downstream signaling. However, such an interaction may represent recruitment by LGP2 of RIG-I to its ligand. Finally, LGP2 was shown to interact with IPS-1, potentially blocking binding by IKKi, an activator of IFN regulatory factor 3 (IRF3) and IRF7 (13). The findings of Satoh et al. suggest that LGP2 interactions with RIG-I and IPS-1 may serve to bring sensor and signaling molecules into close proximity to stimulate downstream signaling.
Satoh et al. convincingly show that LGP2 works upstream of RIG-I and MDA5 to potentiate viral RNA-induced signaling. The precise action of LGP2 in facilitating such signaling is not yet known, but future exploration will be important to address several key questions. For example, what types of structural changes in RNA and in RNA sensors are mediated by LGP2? What ATP-dependent activities are required for LGP2 viral sensing? And what countermeasures do viruses take to evade detection through the LGP2 → RIG-I/MDA5 axis? In short, many questions remain; viruses must know the answers already.
Footnotes
The authors declare no conflict of interest.
See companion article on page 1512 .
References
- 1.Satoh T, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2009;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 3.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 5.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 8.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 9.Takahasi K, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: Identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murali A, et al. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J Biol Chem. 2008;283:15825–15833. doi: 10.1074/jbc.M800542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pippig DA, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, et al. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komuro A, Horvath CM. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J Virol. 2006;80:12332–12342. doi: 10.1128/JVI.01325-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 15.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J Biol Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]