Abstract
Latent TGFβ-binding protein 1 (LTBP-1) is a key regulator of TGFβ targeting and activation in the extracellular matrix. LTBP-1 is recognized as a major docking molecule to localize, and possibly to activate, TGFβ in the extracellular matrix. Despite this relevant function, the molecular mechanisms regulating Ltbp-1 transcription remain largely unknown. Previous results from our laboratory revealed that mouse embryonic fibroblasts (MEF) lacking dioxin receptor (AhR) had increased Ltbp-1 mRNA expression and elevated TGFβ activity, suggesting that AhR repressed Ltbp-1 transcription. Here, we have cloned the mouse Ltbp-1 gene promoter and analysed its mechanism of transcriptional repression by AhR. Reporter gene assays, AhR over-expression and site-directed mutagenesis showed that basal Ltbp-1 transcription is AhR-dependent. Chromatin immunoprecipitation (ChIP) and RNA interference (RNAi) revealed that AhR regulates Ltbp-1 transcription by a mechanism involving recruitment of co-activators such as CREB1 and co-repressors such as HDAC2 to the Ltbp-1 promoter. In AhR-expressing (AhR+/+) MEF cells, the recruitment of HDAC1, 2 and 4 correlated with decreased K8H4 acetylation and impaired binding of pCREBSer133 to the Ltbp-1 promoter, likely maintaining a constitutive repressed state. AhR−/− MEF cells had the opposite pattern of HDACs and pCREB1Ser133 binding to Ltbp-1 promoter, and therefore, over-expressed Ltbp-1 mRNA. In agreement, siRNA for HDAC2 increased Ltbp-1 expression and K8H4 acetylation in AhR+/+ but not in AhR−/− MEF cells. We suggest that HDAC2 binding keeps Ltbp-1 promoter repressed in AhR+/+ MEF cells, whereas in AhR-null MEF cells the absence of HDAC2 and the binding of pCREBSer133 allow Ltbp-1 transcription. Thus, epigenetics can contribute to constitutive Ltbp-1 repression by a mechanism requiring AhR activity.
Keywords: LTBP-1, dioxin receptor, CREB1, HDAC2, transcriptional regulation
Introduction
Transforming growth factor-βs (TGFβ) are dimeric, multifunctional cytokines, having a relevant role in cell homeostasis. They function by signalling through membrane-bound serine/threonine kinase coupled receptors.1 TGFβ is secreted from producing cells in a latent form that must be activated before the soluble molecule becomes competent for receptor binding2. Because altering TGFβ activity in the extracellular matrix (ECM) appears strongly associated to inheritable disorders and to human diseases such as cancer,3,4 a great effort is ongoing to understand its synthesis, association to latency proteins, secretion, localization and activation in the ECM. The initial step in the control of TGFβ activity occurs before secretion and involves the formation of a latent complex with the propeptide LAP. In most cells, however, the LAP-TGFβ latent complex will associate to a third protein called latent TGFβ binding protein (LTBP).5 This process will synthesize the large latent TGFβ complex (one molecule of LTBP bound to a dimer of TGFβ-LAP) that will be secreted outside the cell.2,6,7 Initially considered a protein to localize latent TGFβ to the ECM, increasing experimental evidence indicate that LTBP also contributes to the assembly, secretion and activation of the cytokine.5-10
Four forms of LTBP have been cloned and characterized from humans and rodents. They have differences in gene organization and structure, alternative splicing and use of alternative promoters that produce complex cell-specific patterns of expression.11-16 Two isoforms of LTBP-1, LTBP-1L (long) and LTBP-1S (short), have been cloned in humans12 and mice.17,18 LTBP-1L and LTBP-1S are transcribed from a single gene by the use of alternative promoters. After transcription, and during hnRNA processing, the Ltbp-1L transcript is formed by alternative splicing to an internal acceptor site in exon 1 of Ltbp-1S.12,17,18 Nevertheless, although the murine Ltbp-1L and Ltbp-1S genes have been cloned and their gene structure determined, the mechanisms regulating their transcription still remain largely unknown.
The aryl hydrocarbon (dioxin) receptor (AhR) is a transcription factor belonging to the basic-helix-loop-helix (bHLH) family of transcriptional regulators. Studies analysing the role of AhR in dioxin-induced toxicity and carcinogenesis have identified a battery of AhR-regulated genes. These include xenobiotic metabolizing enzymes such as cytochromes P450 1A1, 1A2, 1B1, 2S1, UDP-glucurono-syltranferase 1 and NAD(P)H: quinone acceptor oxidoreductase.19-23 The increasing relevance of AhR in cell physiology and in the control of endogenous processes24-28 has also prompted the search for new transcriptional targets. Microarray experiments have identified many genes that could be potentially regulated by AhR29-31 and previous studies have shown that constitutive expression of DNA polymerase kappa,32 N-myristoyltransferase 2,33 p2734 and Bax35 is AhR-dependent. These genes support AhR as a constitutive activator of gene expression, albeit this receptor can also repress transcription. Thus, AhR downregulates T-cadherin in smooth vascular cells,36 c-Myc in breast tumor cells and E2F-regulated genes in Hepa 1 cells.38 Mechanistically, AhR-dependent transcription involves receptor activation and translocation to the cell nucleus, dimerization with the bHLH protein aryl hydrocarbon receptor nuclear translocator (ARNT) and binding to xenobiotic responsive elements (XREs) located upstream in the promoter of target genes.39,40
AhR is functionally related to TGFβ. In vivo, liver tissue from AhR−/− mice had increased levels of TGFβ that were coincident with portal fibrosis.41-43 In cell culture, AhR−/− mouse embryonic fibroblasts (MEFs) produced high levels of active TGFβ that decreased their proliferation rates.44 AhR is also functionally linked to the control of Ltbp-1 expression. We have found that AhR−/− MEF over-expressed Ltbp-1 and secreted high levels of LTBP-1 protein.45 Furthermore, downregulation of Ltbp-1 by small interfering RNAs (siRNA) decreased the secretion of active TGFβ in AhR-null MEF cells and supported the contribution of LTBP-1 to TGFβ activation.46 Interestingly, AhR can also maintain lower TGFβ mRNA levels in fibroblast cells by a mechanism controlling RNA stability through the tristetraprolin RNA-binding protein (TTP).47 Altogether, these data support a role for AhR in TGFβ activation and identify Ltbp-1 as a yet uncharacterized AhR-regulated gene.
In this study, we have cloned the mouse Ltbp-1L gene promoter (hereafter Ltbp-1) and characterized its mechanism of transcriptional regulation by AhR. Promoter analysis revealed that AhR expression is required to maintain constitutive repression of the mouse Ltbp-1 gene. Epigenetics have a relevant role in the regulatory mechanism, since preferential binding of transcriptional repressors (e.g. histone deacetylase (HDAC)) versus activators e.g. pCREB1: (CREB, cAMP responsive element-binding protein)) to the Ltbp-1 promoter, in the presence of AhR, maintains repression and low levels of constitutive gene expression. We propose that AhR has a role in the control of constitutive Ltbp-1 transcription and, as consequence, in maintaining TGFβ activity.
Results
The mouse Ltbp-1 gene promoter contains XRE and CRE regulatory elements
We showed earlier that LTBP-1 is over-expressed in MEF cells and in liver tissue of AhR−/− mice. Since over-expression involved the LTBP-1 protein and the Ltbp-1 mRNA, we suggested that AhR repressed Ltbp-1 transcription.43,45,46
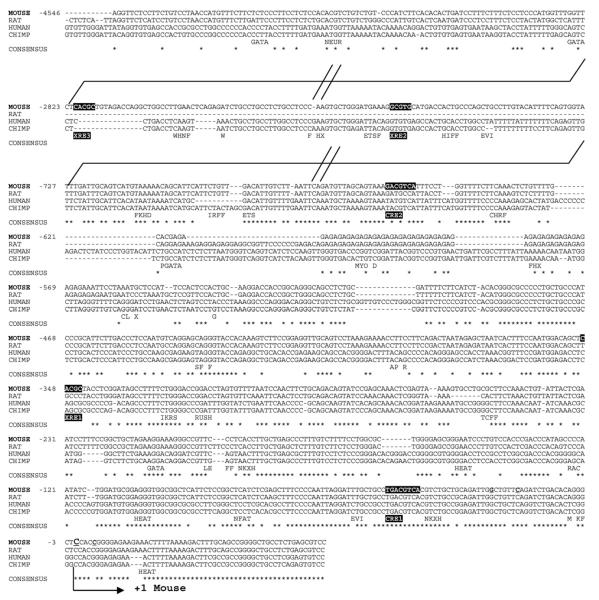
A 4725 bp genomic fragment containing 4547 bp of the Ltbp-1 mouse gene promoter was cloned in pGL2 Basic to generate the pGL2-LTBP-1 full-length construct. Transcription start sites were analysed by primer extension. Among the four putative sites found, C381 (numbered upstream from the translation start codon) appeared to be the predominant +1 position (underlined C in Fig. 1 and results not shown). Sequence alignment of the mouse Ltbp-1 promoter with those from rat, human and chimpanzee revealed variable degrees of homology among species: mouse was 87%, 55% and 30% homologous to rat, human and chimpanzee, respectively (Fig. 1). Consensus regulatory elements were identified also in the Ltbp-1 promoter. Three XRE sequences for AhR binding were found, one direct (XRE2, 5′GCGTG3′) and two inverse (XRE1 and XRE3, 5′CACGC3′). XRE1 was located in the proximal promoter (−347) whereas XRE2 (−2749) and XRE3 (− 2821) were located distal from +1. In addition, two cAMP-responsive elements (CRE), annotated as CRE1 (−49) and CRE2 (−657), were also identified (Fig. 1). Sequence alignment indicated that only the proximal XRE1 and CRE1 were conserved among mouse, human and chimpanzee. This conservation of XRE and CRE in the proximal Ltbp-1 promoter awaits further experiments.
Fig. 1.
Cloning the mouse Ltbp-1 promoter. A 4547 bp DNA fragment containing the mouse Ltbp-1L promoter was cloned and sequenced. Transcription start sites (underlined) were identified by primer extension and the major +1 labelled in bold upper case (cytosine located 381 upstream from ATG). Sequences from mouse, rat, human and chimpanzee were aligned using Clustal X software and conserved nucleotides are indicated by asterisks (*). XRE and CRE elements were located using MatInspector software and are highlighted in black. Only nucleotide regions relevant for this study are shown. For brevity, gaps have been introduced into the sequence shown (broken lines). The complete mouse Ltbp-1 promoter sequence reported in this study has been deposited in GenBank with accession number EF635913.
Constitutive Ltbp-1 repression is AhR-dependent
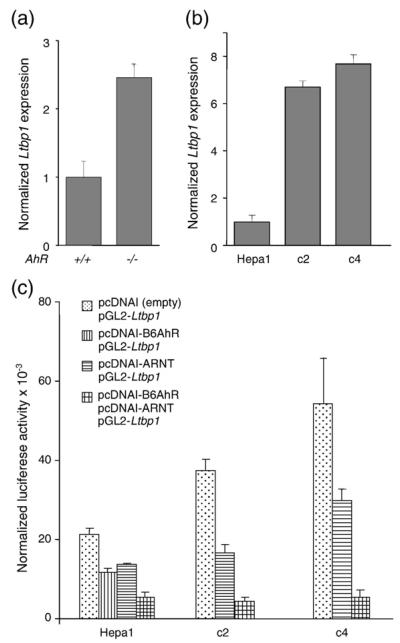
Real-time RT-PCR showed that, in agreement with our previous results, AhR−/− MEF cells over-expressed Ltbp-1 mRNA (Fig. 2a). To confirm that AhR was able to downregulate Ltbp-1 expression and to inhibit Ltbp-1 promoter activity, we used the mouse hepatoma Hepa 1 cell line and its variant clones c2 (less than 10% wild type AhR expression) and c4 (lacking ARNT expression). In agreement with the results obtained in AhR−/− MEF cells, c2 cells, with low levels of AhR, had high constitutive expression of Ltbp-1 mRNA. ARNT-lacking c4 cells had also increased Ltbp-1 mRNA expression (Fig. 2b). We then determined the effect on Ltbp-1 promoter activity of over-expressing AhR and ARNT using the pGL2-LTBP-1 full-length construct (Fig. 2c). Transfection of c2 cells with AhR (pcDNAI-B6AhR) or c4 cells with ARNT (pcDNAI-ARNT) decreased basal luciferase activity in either cell line. Co-transfection of AhR and ARNT in c2 or c4 cells had additive effect further decreasing luciferase activity. Similar results were found after transfection of these constructs in wild type Hepa 1 cells. In agreement with their constitutively elevated Ltbp-1 mRNA expression, basal luciferase activity was also higher in c2 and c4 than in Hepa 1 cells. Thus, AhR and ARNT are required to maintain constitutive repression of the mouse Ltbp-1 gene.
Fig. 2.
AhR is required to maintain constitutive repression of Ltbp-1. (a) Ltbp-1 mRNA expression was measured in AhR+/+ and AhR−/− MEF cells by real-time PCR and data were normalized by the expression of β-Actin. Data are shown as mean±SE from three experiments in triplicate. (b) Hepa 1 and its variant clones c2 and c4 were analysed for Ltbp-1 mRNA expression by real-time PCR. Data were normalized by β-Actin in the same samples. (c) The effect of AhR and/or ARNT over-expression on Ltbp-1 promoter activity was determined by luciferase reporter assays using the full-length pGL2-LTBP-1 construct. Co-transfections were performed with the expression vectors pcDNAI-B6AhR and pcDNAI-ARNT. In some experiments, both expression vectors were co-transfected with the luciferase reporter. The experiments were done in triplicate in at least three different cultures for each cell line. The data are shown as mean ± SE.
XRE sequences are involved in the constitutive repression of Ltbp-1
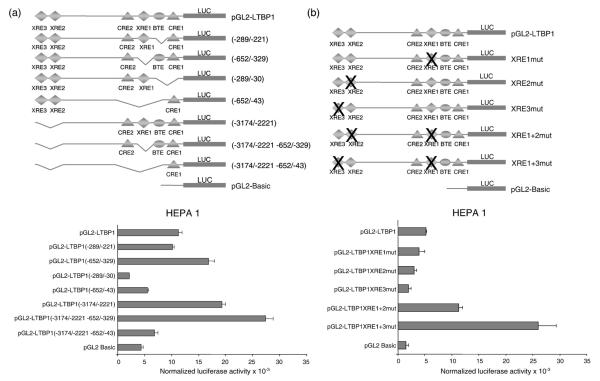
It is known that XRE and CRE elements are involved in AhR-mediated transcription of target genes.48,49 To analyse the importance of XRE and CRE on Ltbp-1 promoter activity, we have produced constructs containing deletions of these conserved elements and performed luciferase activity assays in Hepa 1 cells (Fig. 3a, upper). Two main patterns of regulation were found: (i) deletions that increased luciferase activity and lacked XRE1 (−652/−329), XRE2 and 3 (−3174/−2221) or all three XRE sequences (−3174/−2221 −652/−329), having the deletion of all three XREs an additive effect on luciferase activity (Fig. 3a, lower); (ii) deletions that decreased luciferase activity and lacked CRE1 (−289/−30) or CRE2 (−652/−43) (Fig. 3a, lower). Therefore, regions containing XREs had repressive activity whereas those having CRE elements activated Ltbp-1 transcription. To analyse the contribution of XRE sequences per se on the regulation of the Ltbp-1 promoter, site-directed mutagenesis was done (Fig. 3b, upper panel and Table 1). Single mutation of XRE1, XRE2, or XRE3 did not increase luciferase activity above basal levels (Fig. 3b, lower). However, the combined mutation of XRE1+2 or XRE1+3 increased luciferase activity markedly, with the double mutant XRE1+3 having the largest effect. Because XRE is the canonical binding site for AhR, these data suggest that this receptor participates in the molecular mechanism maintaining Ltbp-1 repression. In addition, the additive effect observed after deletion or mutation of proximal and distal XRE sequences suggests that AhR could be part of a protein complex maintaining constitutive Ltbp-1 repression. However, using ChIP assays we have failed to immunoprecipitate AhR bound to XREs in the Ltbp-1 promoter (using experimental conditions that detected AhR bound to the mouse Cyp1a1 promoter as positive control; data not shown). Since our data suggest strongly that AhR and XREs are relevant for Ltbp-1 repression, we think that the existence of a multimeric protein complex on the Ltbp-1 promoter could mask AhR, making it difficult for the antibody to reach its target in vivo.
Fig. 3.
XRE and CRE sequences regulate the constitutive repression of the mouse Ltbp-1 promoter. (a) Deletion constructs were produced from the full-length Ltbp-1 promoter vector pGL2-LTBP-1. DNA regions containing XRE or CRE elements were deleted as indicated in Materials and Methods. Each construct was transfected in Hepa 1 cells and luciferase activity determined and normalized by β-galactosidase activity (lower). (b) Site-directed mutagenesis was used to analyse the role of XRE sequences in Ltbp-1 repression, (crosses mark mutated XREs). Single and double XRE mutant constructs were transfected in Hepa 1 cells as indicated above. At least four experiments were done in triplicate. The data are shown as mean ± SE.
Table 1.
Site-directed mutagenesis of XRE elements present in the mouse Ltbp-1 promoter
| XRE1mut | XRE1 | |
| Wild type | 5′CCAATGGACAGCTCACGCTACCTCGGATAGCCTTTTCTGG3′ | |
| BamHI | ||
| Mutant 77.8 °C | 5′CCAATGGACAGCTCAGGATCCCTCGGATAGCCTTTTCTGG3′ | |
| 3′GGTTACCTGTCGAGTCCTAGGGAGCCTATCGGAAAAGACC5′ | ||
| XRE2mut | XRE2 | |
| Wild type | 5′CAAGTGCTGGGATGAAAGGCGTGCATGACCACTGCCCA3′ | |
| BamHI | ||
| Mutant 78.1 °C | 5′CAAGTGCTGGGATGAAAGGATCCCATGACCACTGCCCA3′ | |
| 3′GTTCACGACCCTACTTTCCTAGGGTACTGGTGACGGGT5′ | ||
| XRE3mut | XRE3 | |
| Wild type | 5′CTGTCCTGGAACTCACGCTGTAGACCAGGCTGGCCTTGAAC3′ | |
| EcoRI | ||
| Mutant 78.3 °C | 5′CTGTCCTGGAACTGAATTCTGTAGACCAGGCTGGCCTTGAAC3′ | |
| 3′GACAGGACCTTGACTTAAGACATCTGGTCCGACCGGAACTTG5′ |
Wild type XRE1, XRE2 and XRE3 are highlighted in bold. Base changes introduced into each XRE consensus sequence are underlined. For site-directed mutagenesis, the mutations introduced generated restriction sites for the endonucleases indicated.
CREB1 binding to the mouse Ltbp-1 promoter is AhR-dependent
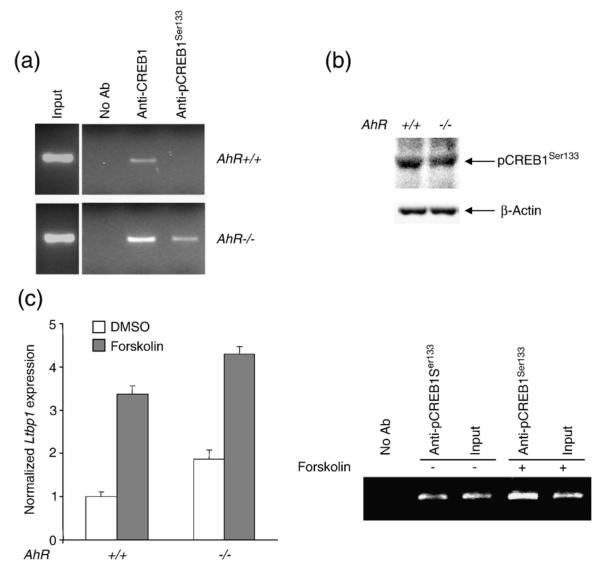
CREB and AhR are functionally related in the control of gene expression.48,49 Because deletion of CRE elements decreased Ltbp-1 promoter activity (Fig. 3a), we have analysed the contribution of CREB1 to the control of Ltbp-1 expression. ChIP assays showed increased binding of CREB1 to the Ltbp-1 promoter in AhR−/− than in AhR+/+ MEF cells (Fig. 4a). Consistently, the phosphorylated, active pCREB1Ser133 was also recruited more efficiently to the Ltbp-1 promoter in AhR-null MEF cells (Fig. 4a). The differences in pCREB1Ser133 binding were not due to changes in the amount of phosphorylated protein as determined by Western blotting (Fig. 4b). Treatment of AhR+/+ and AhR−/− and AhR−/− MEF cells with forskolin, which activates pCREB1Ser133 by raising the concentration of cAMP, increased Ltbp-1 mRNA levels in both genotypes (Fig. 4c), suggesting that pCREB1Ser133 had a role in regulating constitutive transcription of Ltbp-1. In agreement, the increase in Ltbp-1 expression induced by forskolin was concomitant with enhanced pCREB1Ser133 binding to the promoter (Fig. 4c). Thus, Ltbp-1 expression was positively regulated by CREB1 activation and pCREB1Ser133 binding to its promoter. Furthermore, decreased pCREB1Ser133 binding to the Ltbp-1 promoter in AhR+/+ MEF cells could contribute to their lower constitutive Ltbp-1 expression.
Fig. 4.
CREB1 is differentially recruited to the Ltbp-1 promoter in AhR+/+ and AhR−/− MEF cells. (a) The binding of CREB1 and activated pCREB1Ser133 to the Ltbp-1 promoter was analysed by ChIP in AhR+/+ and AhR−/− MEF cells. Positive controls were performed using the input fractions whereas negative controls were made in the absence of antibody. (b) pCREB1Ser133 protein levels were analysed in AhR+/+ and AhR−/− MEF cells by Western blotting. β-Actin was used to determine equal loading. (c) AhR+/+ and AhR−/− MEF cells were left untreated (DMSO) or treated with 10 μM forskolin for 6 h and Ltbp-1 mRNA expression determined by real-time PCR. Ltbp-1 mRNA expression was normalized by β-Actin. The binding of pCREB1Ser133 to the Ltbp-1 promoter in forskolin-treated AhR+/+ MEF cells was analysed by ChIP as in panel a. The data are shown as mean ± SE. The experiments were repeated three times with similar results.
HDAC2 recruitment to the mouse Ltbp-1 promoter is AhR-dependent
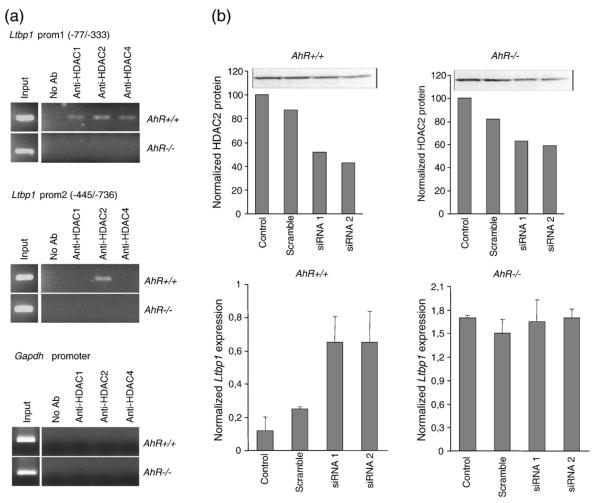
HDACs differentially affect CREB-regulated genes by inducing or repressing transcription.50 For instance, chromium repressed gene expression by preventing HDAC1 release and CREB-binding protein (CBP) recruitment to the proximal Cyp1a1 promoter51. On the basis of these data, we then analysed whether Ltbp-1 expression and CREB1 binding concurred with recruitment of HDACs to the Ltbp-1 promoter in MEF cells. As shown in Fig. 5a, HDAC1, −2, and −4 were recruited to the proximal Ltbp-1 promoter in AhR+/+ but not in AhR −/− MEF cells (upper panel). HDAC2 was also bound to more distal regulatory regions but only in AhR-expressing MEF cells (middle panel). Negative control experiments for HDAC1, −2 and −4 binding were performed using the mouse Gapdh promoter (lower panel). The relevance of HDACs on the regulation of Ltbp-1 was analysed by RNA interference (RNAi) for HDAC2 due to its prevalent binding on the Ltbp-1 promoter. Decreasing HDAC2 protein levels by two different siRNA molecules (Fig. 5b, upper panel) increased Ltbp-1 mRNA expression in AhR+/+ but not in AhR−/− MEF cells (Fig. 5b, lower panel).
Fig. 5.
HDAC2 is differentially recruited to the Ltbp-1 promoter in AhR+/+ and AhR−/− MEF cells. (a) The binding of HDAC1, −2 and −4 to the Ltbp-1 promoter was analysed by ChIP in AhR+/+ and AhR−/− MEF cells. Proximal (−77/−333) and more distal (−445/−736) regions were amplified by PCR. Positive controls included input fractions, whereas negative controls were done in the absence of antibodies. (b) HDAC2 protein was downregulated in AhR+/+ and AhR−/− MEF cells by transfecting two different siRNA molecules (siRNA 1 and siRNA 2). The effect of these siRNAs on the levels of HDAC2 protein in AhR+/+ and AhR−/− MEF cells is shown in the upper panels. The effect of HDAC2 siRNAs on Ltbp-1 mRNA expression in AhR+/+ and AhR−/− MEF cells was determined by real-time PCR and normalized by β-Actin (lower panels). Scramble siRNA was used as a control for RNAi specificity. The experiments were repeated four times with similar results. The data are shown as mean ± SE.
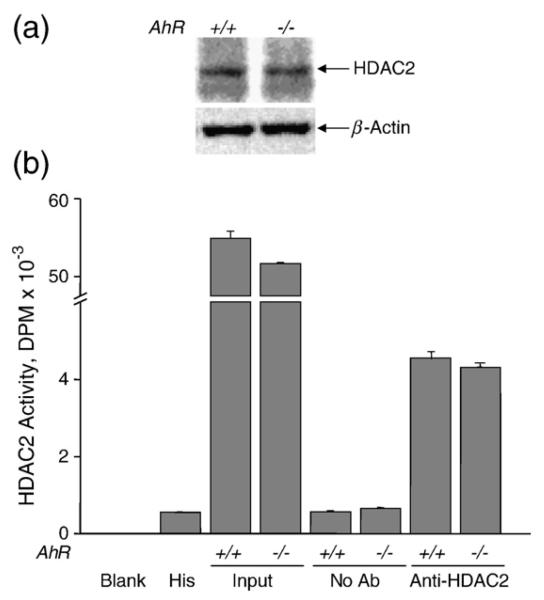
The difference in HDAC2 binding to the Ltbp-1 promoter in AhR+/+ and AhR−/− MEF cells was not due to changes in protein expression as determined by Western blotting (Fig. 6a). Additionally, the increase in Ltbp-1 mRNA expression produced by HDAC2 siRNA in AhR+/+ but not in AhR−/− MEF cells was not due to differences in HDAC2 deacetylase activity between both genotypes (Fig. 6b). Thus, HDAC1, −2 and −4 constitutively bind to the Ltbp-1 proximal promoter in AhR+/+ but not in AhR−/− MEF cells. Among these HDACs, HDAC2 appears relevant to modulate AhR-dependent repression of Ltbp-1 because HDAC2 downregulation by siRNA restored Ltbp-1 mRNA levels in AhR+/+ but not in AhR−/− MEF cells, which had undetectable HDAC2 binding to the promoter.
Fig. 6.
AhR genotype does not alter HDAC2 expression or deacetylase activity. (a) AhR+/+ and AhR−/− MEF cells were analysed for HDAC2 protein expression by Western blotting. β-Actin was used to determine equal loading. (b) HDAC2 deacetylase activity was measured in HDAC2 immunoprecipitates. HDAC2 was immuno-precipitated with a specific antibody and its deacetylase activity analysed in vitro using [3H]acetylated chicken erythrocyte histones as substrate. HDAC2 activity was quantitfied as DPM. Positive controls were done using input extracts. Negative controls included immunoprecipitates in absence of antibody (No Ab) and enzymatic reactions containing histones but not protein (His). The experiment was done in duplicate in two different AhR+/+ and AhR−/− MEF cultures. The data are shown as mean±SE.
HDAC2 recruitment modulates K8H4 acetylation in the proximal Ltbp-1 promoter
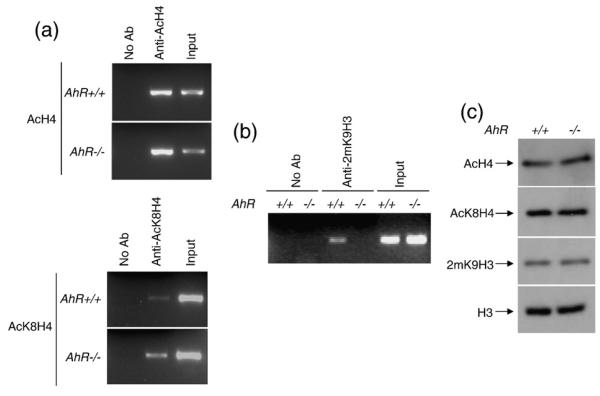
We next analysed whether HDAC2 recruitment to the proximal Ltbp-1 promoter modified histone acetylation marks associated to the inhibition of gene transcription. Acetylation status of specific marks was determined by ChIP assays. Histone H4 acetylation (AcH4) was increased in AhR−/− with respect to AhR+/+ MEF cells (Fig. 7a, upper panel). Systematic analyses of acetylable Lys residues in the histone H4 N-terminal tail revealed a specific increase in Lys8 acetylation (AcK8H4) in the proximal promoter of Ltbp-1 in AhR−/− MEF cells (Fig. 7a, lower panel). We investigated Lys9 dimethylation in histone H3 (2mK9H3) as an additional marker associated with gene repression. In agreement with their lower Ltbp-1 transcription, 2mK9H3 was present in the proximal Ltbp-1 promoter in AhR+/+ but not in AhR−/− MEF cells (Fig. 7b). Differences in AcH4, AcK8H4 and 2mK9H3 between AhR+/+ and AhR−/− MEF cells were not due to altered protein expression as determined by Western blotting (Fig. 7c). Thus, AhR+/+ MEF cells constitutively repress Ltbp-1 transcription by a coordinated mechanism in which impaired pCREBSer133 binding is associated with HDAC2 recruitment, decreased K8H4 acetylation and increased K9H3 dimethylation in the proximal promoter.
Fig. 7.
AhR+/+ MEF cells have acetylation and methylation markers in the Ltbp-1 promoter consistent with a repressed state. (a) Acetylation status of histone H4 (AcH4) (upper panel) and Lys8 of histone H4 (AcK8H4) (lower panel) in the proximal Ltbp-1 promoter was determined in AhR+/+ and AhR−/− MEF cells by ChIP. Positive controls corresponded to input fractions, whereas negative controls were done in the absence of antibody. (b) Methylation of Lys 9 of histone H3 (2mK9H3) was determined in AhR+/+ and AhR−/− MEF cells by ChIP. Positive and negative controls were performed as described above. (c) Protein levels of AcH4, AcK8H4 and 2mK9H3 were determined in AhR+/+ and AhR−/− MEF cells by Western blotting. Histone H3 was used as loading control. The experiments were repeated three times in independent AhR+/+ and AhR−/−MEF cultures.
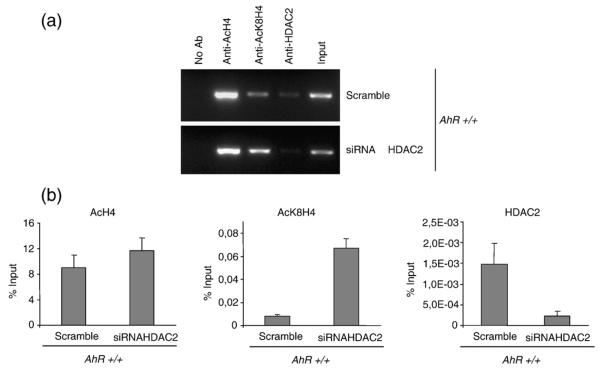
To determine the relevance of HDAC2 in decreasing K8H4 acetylation in the proximal Ltbp-1 promoter, HDAC2 was downregulated by siRNA and AcH4 and AcK8H4 levels analysed in AhR+/+ MEF cells by ChIP (Fig. 8a). Results were also quantified as the percentage of the input fraction (Fig. 8b). HDAC2 siRNA affected AcH4 acetylation only marginally. However, it significantly increased AcK8H4 in AhR+/+ MEF cells. Thus, HDAC2 binding to the Ltbp-1 promoter in AhR+/+ MEF cells maintained low levels of K8H4 acetylation. These data support a coordinated mechanism of recruitment of co-activators and co-repressors to the Ltbp-1 promoter that depends on AhR. Proteomic analyses are underway to identify differential protein complexes on the Ltbp-1 promoter in AhR+/+ and AhR−/− MEF cells.
Fig. 8.
HDAC2 downregulation by RNAi increases AcK8H4 levels in the Ltbp-1 promoter in AhR+/+ MEF cells. (a) AhR+/+ MEF cells were transfected with siRNAs for HDAC2 and ChIP assays performed for AcH4, AcK8H4 and HDAC2. Scramble RNA was transfected as control for RNAi specificity. Positive controls were performed using input fractions. Negative controls were done in the absence of antibody (No Ab). (b) Quantitative real-time PCR to measure the effect of HDAC2 siRNA on AcH4 and AcK8H4 in the proximal Ltbp-1 promoter. Levels of AcH4, AcK8H4 and HDAC2 measured by real-time PCR were compared to a calibration curve made with known amounts of input fraction. The experiment was repeated in duplicate using two different AhR+/+ MEF cultures with similar results. The data are shown as mean ± SE.
Discussion
AhR has been characterized extensively as a transcription factor that regulates the expression of XRE-containing genes.27,40 Classical AhR-regulated genes include those coding for xenobiotic metabolizing enzymes such as cytochromes P450, transferases and oxidoreductases.21,23,39,52 The study of AhR within a physiological context (i.e. xenobiotic-unrelated) has identified new target genes constitutively regulated by this receptor, including cell-cycle inhibitor p27,34 pro-apoptotic protein Bax35 and the modifying enzyme N-myristoyltransferase.33 Interestingly, recent reports have shown that AhR represses gene transcription. This repressive activity can be xenobiotic-dependent, as for T-cadherin in vascular smooth muscle cells,36 or constitutive and xenobiotic-independent, as for c-Myc in mammary tumor cells37 and TGFβ in mouse fibroblasts.47
Earlier, we reported that Ltbp-1 mRNA was constitutively over-expressed in AhR-null MEF cells,43,45,46 and that AhR knock-down increased the levels of active TGFβ in MEF cells,44,45 primary hepatocytes53 and liver tissue.43 Because Ltbp-1 is an important regulator of TGFβ activity, and the mechanism controlling Ltbp-1 synthesis (i.e. transcription) is largely unknown, we have cloned the mouse Ltbp-1 gene promoter to characterize its mechanism of transcriptional regulation by AhR.
Experiments performed in MEF cells and in mouse hepatoma Hepa 1 cells supported the hypothesis that AhR is required to maintain constitutive repression of Ltbp-1. In fact, MEF and Hepa 1 cells lacking AhR had constitutive over-expression of Ltbp-1 mRNA, and AhR re-expression was able by itself to restore repression on the Ltbp-1 promoter. Among the consensus sequences present in the Ltbp-1 promoter, three XREs and two CRE elements were identified. Deletion of these regulatory elements differentially affected Ltbp-1 promoter activity: whereas XRE sequences contributed to repress the Ltbp-1 promoter, CRE elements maintained its constitutive transcriptional activity. These results again suggested that AhR is a component of the regulatory complex repressing Ltbp-1 transcription. Site-directed mutagenesis on XRE elements produced interesting results. While mutation of each XRE did not significantly affect Ltbp-1 promoter activity, combined mutation of one proximal (XRE1) and one distal (XRE2 or XRE3) element released repression and increased promoter activity. Furthermore, an additive effect was observed after deletion or mutation of proximal and distal XRE sequences, suggesting that XRE1, −2 and −3 cooperate in maintaining Ltbp-1 repression. However, we could not immunoprecipitate AhR bound to the XRE sequences in the Ltbp-1 promoter. Considering the evidence supporting the involvement of AhR and XREs in Ltbp-1 repression, and since XREs are the canonical binding sites for AhR, we think that AhR is included in a multiprotein regulatory complex that masks recognition by its antibody in ChIP assays in vivo. Altogether, a consistent set of data supports the role of AhR in constitutive Ltbp-1 repression: (i) Ltbp-1 mRNA was constitutively over-expressed in AhR−/− MEF cells; (ii) Hepa 1 cells with a very low level of AhR (c2) had increased constitutive Ltbp-1 expression; (iii) Hepa 1 cells lacking the dimerization protein ARNT (c4) had Ltbp-1 over-expression; (iv) re-expression of AhR in c2 or ARNT in c4 Hepa 1 cells restored repression and decreased constitutive Ltbp-1 mRNA levels and (v) deletion or combined mutation of XRE sequences released repression on the Ltbp-1 promoter.
Considering that the level of conservation was higher in the proximal Ltbp-1 promoter, and since it contains XRE and CRE elements, we performed further analyses in this region. The proximal Ltbp-1 promoter contains two CRE elements whose deletion decreased promoter activity. CRE elements are recognized by CREB, a DNA-binding protein that is a co-activator of AhR in the regulation of the Cyclooxygenase-2 gene,48 and an intermediate molecule to recruit CREB-binding protein CBP to the Cyp1a1 promoter.49 Studies in osteoblastic cells have shown that parathyroid hormone induces Ltbp-1 by increasing cAMP levels, which suggested the involvement of CREB.54 Thus, CREB could participate in the regulation of constitutive Ltbp-1 expression in MEF cells. Forskolin, through adenyl cyclase, increases gene expression by inducing CREB phosphorylation at Ser133.55 Consistent with these previous data, forskolin treatment increased Ltbp-1 mRNA expression in AhR+/+ and AhR−/− MEF cells and promoted pCREBSer133 binding to the Ltbp-1 promoter, indicating that CREB signalling was involved in Ltbp-1 regulation. ChIP experiments demonstrated that AhR−/− MEF cells had higher levels of active pCREBSer133 bound to the Ltbp-1 promoter than AhR+/+ MEFs. These results support the suggestion that AhR+/+ MEF cells, having diminished binding of pCREBSer133 to the Ltbp-1 promoter also had lower constitutive Ltbp-1 transcription, and sustain that Ltbp-1 over-expression in AhR−/− MEF cells can be due to increased pCREBSer133 recruitment to the promoter. In agreement with our hypothesis, previous studies have shown that the transcriptional induction of the mouse C/EBPβ gene by TCDD is AhR-dependent and requires the recruitment of protein kinase A-activated CREB to the C/EBPβ promoter, despite the fact that such a mechanism does not seem to involve AhR binding to the proximal XRE sequence located downstream from the CREB-binding site.56,57 Thus, certain endogenous genes can be regulated by AhR through mechanisms that differ from the classical AhR/XRE pathway.
Several studies have linked histone deacetylases (HDACs) to the inhibition of CREB-dependent transcription. It was shown that HDAC1 induced pCREB dephosphorylation and inhibited its transcriptional activity.58 Certain nuclear receptors also use similar epigenetic mechanisms to regulate gene expression. For example, glucocorticoid receptor directly inhibited CREB-dependent CBP activity and recruited HDAC2 to the histone acetylase complex in the GM-Csf promoter.59 A recent report has shown that the CRE elements located in the promoter of the Interleukin-2 and c-Fos genes were important to regulate HDAC1 binding and their level of transcriptional activity.60 On the basis of these data, we have analysed if CREB1 binding to the Ltbp-1 promoter correlated to HDACs recruitment and whether this mechanism was dependent on AhR expression. Indeed, the absence of binding of pCREBSer133 coincided with recruitment of HDAC1, −2 and −4 in the proximal Ltbp-1 promoter in AhR+/+ MEF cells. As expected, AhR−/− MEF cells did not show significant recruitment of these HDACs to the Ltbp-1 promoter. HDAC2 had stronger and prevalent binding on the Ltbp-1 promoter than HDACs 1 and 4. The relevance of HDAC2 in maintaining Ltbp-1 repression was based on the fact that RNAi relieved repression of Ltbp-1 but only in AhR+/+ MEF cells having HDAC2 bound to the promoter. HDAC2 has some overlapping activity with HDAC1 and possibly with other HDACs. However, the effect of simultaneous RNAi for HDAC1, −2 and −4 could not be measured, since co-transfection with the corresponding siRNAs severely compromised the viability of MEF cells. Thus, at least HDAC2 contributes to the constitutive repression of Ltbp-1 in AhR+/+ MEF cells. In agreement, a similar mechanism has been proposed for the inhibition of benzo-[a]-pyrene-dependent (B[a]P) activation of Cyp1a1 by chromium. Chromium inhibited B[a]P-induced release of HDAC1 from the Cyp1a1 promoter and blocked recruitment of p300.51 The lower level of binding of pCREBSer133 to the Ltbp-1 promoter in AhR+/+ MEF cells can be explained by earlier studies showing that HDAC1 inhibits pCREBSer133 phosphorylation in cAMP-stimulated HEK293 cells.58 Thus, HDAC2 bound to the Ltbp-1 promoter could contribute to the in situ dephosphorylation of pCREBSer133 and to decreased Ltbp-1 expression in AhR+/+ MEF cells.
HDACs bound to promoter regions have been associated with deacetylation of specific lysine residues in histones and with gene repression. In A549 cells, glucocorticoid receptor inhibited GM-Csf expression by a mechanism in which recruitment of HDAC2 blocked interleukin-1β-dependent acetylation of K8H4 and K12H4.59 In agreement, increased binding of HDAC2 to the Ltbp-1 promoter in AhR+/+ MEF cells concurred with decreased K8H4 acetylation and lower Ltbp-1 gene expression. Moreover, HDAC2 has a role in K8H4 deacetylation since its down-regulation by RNAi restored K8H4 acetylation in the Ltbp-1 promoter. The regulation of the Ltbp-1 promoter is complex, as the methylation marker associated with gene repression 2mK9H3 was also increased in proximal promoter regions in AhR+/+ but not in AhR−/− MEF cells.
Thus, AhR expression determines a pattern of protein recruitment to the mouse Ltbp-1 promoter, in which constitutive gene repression involves HDAC2 binding, K8H4 deacetylation, K9H3 dimethylation and CREB1Ser133 dephosphorylation. More studies need to be done to precisely define the structure of the protein complexes involved. Current work is focused on defining the differential composition of these protein complexes in AhR+/+ versus AhR−/− MEF cells. A relevant aspect of the mechanism proposed here is whether the different upstream promoter regions of Ltbp-1 are able to regulate Ltbp-1 expression in a cell-specific manner. This is plausible, since previous studies have shown that the upstream promoter regions of Ltbp-1L differentially regulate reporter gene expression in WI-38 lung fibroblasts as compared to HA human amniotic and HT-1080 fibrosarcoma cells.12 Furthermore, the analysis of deletion reporter constructs of the Ltbp-1L promoter revealed the presence of a negative regulatory element that repressed constitutive Ltbp-1L expression in WI-38 but not in the SV40-transformed WI-38/VA13 cell line.12 From the toxicological point of view, Ltbp-1 could have an important role in regulating the increase in TGFβ activity induced by the carcinogen TCDD. In this context, several reports have shown that treatment with TCDD enhances TGFβ activation in the ECM of the thymic epithelium,61 in fibrotic myocardium in marmosets,62 and in the malignant transformation occurring after successive divisions of TCDD-damaged cells.63
This study provides evidence for the contribution of AhR to an epigenetic mechanism regulating the constitutive repression of the mouse Ltbp-1 gene. In the presence of AhR, HDAC2 binding to the Ltbp-1 promoter would decrease histone acetylation and diminish pCREBSer133 recruitment, thus lowering constitutive Ltbp-1 transcription. In the absence of AhR, decreased HDAC2 binding to the Ltbp-1 promoter could allow increased histone acetylation and pCREBSer133 recruitment, resulting in elevated constitutive Ltbp-1 mRNA expression. Defining the molecular mechanisms of Ltbp-1 transcriptional regulation is important owing its role in TGFβ secretion, localization and activation in the ECM. Although the precise location of AhR in this regulatory mechanism is not completely defined, our study implicates AhR in the epigenetic control of gene repression under normal cellular conditions.
Materials and Methods
Chemicals and reagents
Dulbecco's modified Eagle's medium (DMEM), cell media supplements and SuperScript II reverse transcriptase were from Life Technologies. Heat-inactivated fetal bovine serum (FBS) was from Hyclone. Taq DNA polymerase was from Ecogen. SYBR Green I was obtained from Molecular Probes, and QTaq DNA polymerase mix was from Becton-Dickinson. Antibodies against the following proteins were used: CREB1, pCREB1Ser133, AcH4 and AcK8H4 from Upstate Biotechnology, HDAC1, HDAC2, HDAC4, H3 and 2mK9H3 from AbCAM and rabbit and mouse IgG-HRP from Pierce and Santa Cruz Biotechnology, respectively. Antibodies against mouse AhR were obtained from Biomol and ABR.
Cell culture
AhR-null and wild type control mice were produced as described.42 Experiments involving mice were done following the Regulation for Animal Care and Use set forth by the University of Extremadura. MEF cultures were established from 14.5 dpc AhR+/+ and AhR−/− mouse embryos as described.64 MEF cells were cultured at 37 °C in a 5% (v/v) CO2 atmosphere in DMEM supplemented with 10% (v/v) FBS, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. MEFs at second passage were used in all experiments. Mouse hepatoma Hepa 1 cells and its variant clones c2 (expressing less than 10% wild type levels of AhR) and c4 (lacking ARNT expression)65 were cultured at 37 °C in a 5% CO2 atmosphere in alpha-MEM containing 5% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
Cloning the mouse Ltbp-1L gene promoter
The mouse Ltbp-1L promoter was isolated by PCR from genomic DNA using the bacterial artificial chromosome (BAC) clone RPCI23 as template (BACPAC Resources CHORI, Children's Hospital Oakland Research Institute). Amplification of a 4725 bp DNA fragment containing the 5′ upstream region of Ltbp-1L was performed using the primers forward:
5′ CGACGCGTCGAGGTTCTCCTTCTGTCCTAACC 3′
and reverse:
5′ CGACGCGTGGCGCGAGCTGCTCAAGGTGAGGGTC 3′
MluI sites are underlined. PCR was carried out for 40 cycles in 50 μl of reaction mixture containing: 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 μM each primer, 2.5 units of Taq polymerase and 200 ng of DNA template. PCR conditions were: initial denaturation at 94 °C for 1 min, denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and extension at 72 °C for 1 min. The PCR fragment obtained was digested with MluI and cloned into the pGL2 Basic vector to generate the full-length construct pGL2-Ltbp-1. The cloned Ltbp-1 promoter was sequenced in both strands by chromosome walking using fluorescent dideoxyterminator chemistry in an automatic ABI Prism sequencer (Applied Biosystems). Consensus regulatory sequences were identified in the Ltbp-1 promoter using MatInspector software (Genomatix). Sequence alignment was performed using CLUSTAL X (1.83) software.
Transcription start sites were identified in the mouse Ltbp-1L promoter by primer extension using total and poly (A)+ RNA as described.12 The same oligonucleotides were used to extend the Ltbp-1L mRNA and to sequence the upstream region of the gene: forward:
5′ CGAGCTGCTCAAGGTGAGGGTC 3′
and reverse:
5′ CCCGCCATCGCGGGCAGTCTC 3′.
Deletion mutants and site-directed mutagenesis
The transcriptional activity of the XRE and CRE elements in the Ltbp-1 promoter was analysed by producing deletion mutants fused to the luciferase reporter gene. Deletions were located and named with respect to the transcription start site. Constructs pGL2-Ltbp-1 (−289/−30), pGL2-Ltbp-1 (−652/−43) and pGL2-Ltbp-1 (−4565/−430) were obtained by digesting pGL2-Ltbp-1 with PstI, AatII and KpnI, respectively, followed by self-ligation of the linearized DNA. pGL2-Ltbp-1 (−4565/−430, −289/−30) was subcloned from pGL2-Ltbp-1 (−4565/−430) by digesting with PstI and self-ligation. pGL2-Ltbp-1 (−3174/−2221) was obtained from pGL2-Ltbp-1 by digesting with BstXI and NsiI and making blunt-ended with Klenow DNA polymerase. From this latter construct, pGL2-Ltbp-1 (−4565/ −430, −652/−43) was obtained by digesting with AatII and self-ligation. Constructs lacking only XRE1 (pGL2-Ltbp-1 (−652/−329)) or BTE (pGL2-Ltbp-1 (−289/−221)) were made as follows: pGL2-Ltbp-1 was digested with AatII and PstI to delete the region containing both sites. Then, BTE was re-introduced in pGL2-Ltbp-1 (−652/−329) and XRE1 in pGL2-Ltbp-1 (−289/−221) by PCR. Oligonucleotides used were: for pGL2-Ltbp-1 (−652/−329) forward:
5′ TCGGATAGCCTTTTCTGGGACC 3′
and reverse:
5′ GACGTGACGTCAGGCAGCAAATC 3′
and for pGL2-Ltbp-1 (−289/−221): forward:
5′ AAAACTGCAGCTGCTAGAAGGAAAGGGC 3′
and reverse:
5′ TGAACAGCCAATCTGCAGCAGACG 3′
(AatII and PstI sites underlined). pGL2-Ltbp-1 (−3174/ −2221, −652/−43) was cloned from pGL2-Ltbp-1 (−652/ −329) by digesting with BstXI and NsiI, and self-ligation.
Site-directed mutagenesis of XRE1, XRE2 and XRE3 was done using the QuickChange II XL kit (Stratagene) following the manufacturer's instructions. XREs were mutated by introducing restriction sites for the endonucleases BamHI (XRE1 and 2) or EcoRI (XRE3) using the oligonucleotides shown in Table 1. Double mutants for XRE1 and XRE2 or XRE1 and XRE3 were obtained by digesting each single mutant with AatII. Next, the 609 bp fragment released from the XRE1mut (e.g. containing the mutated XRE1) was cloned into AatII-digested XRE2mut and XRE3mut. Constructs were sequenced to confirm site-directed mutagenesis.
Real time RT-PCR
Total RNA was purified from Hepa 1 and MEF cells using the RNeasy kit (Qiagen) following the manufacturer's instructions. The purity of the RNA was estimated by measuring the A260 nm/A280 nm ratio. After denaturation at 70 °C for 10 min, 10 μg of RNA was reverse transcribed for 2 h at 42 °C using 2.5 μM random hexamers and a reaction mixture containing 2.5 mM each dNTP, 10 mM DTT, 10 U RNase inhibitor and 150 U SuperScript II reverse transcriptase. RNA was then degraded by incubation for 10 min at 70 °C in 35 mM NaOH. After neutralization with HCl, synthesized cDNAs were ethanol-precipitated, washed in 80% (v/v) ethanol, dried and dissolved in DEPC-treated water. Real time PCR was carried out using SYBR Green I, QTaq DNA Polymerase Mix and 5 μl of RT template in a DNA Engine Opticon 2 System (MJ Research). PCR conditions were: initial denaturation at 95 °C for 10 min and 30 cycles of denaturation at 95 °C for 30 s, annealing at 56.5 °C for 30 s (60 °C for AcH4, HDAC2 and AcK8H4) and extension at 72 °C for 30 s. A fusion curve was performed after each PCR reaction by heating from 60 °C to 95 °C at a rate of 0.2 degC/s. As internal control, β-actin mRNA was amplified and used to normalize gene expression.46 Real-time PCR for AcH4, AcK8H4 and HDAC2 after chromatin immunoprecipitation (ChIP) was quantified as percentage of input from a titration curve made with known concentrations of input DNA. Negative controls were also made in the absence of cDNA.
SDS-PAGE and Western blotting
MEF cells were washed in PBS and lysed at 4 °C in buffer containing 50 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2 mM EGTA, 270 mM sucrose, 10 mM β-glycerophosphate, 50 mM sodium fluoride, 1% (v/v) Triton X-100, 0.1 mM sodium orthovanadate, 1% (v/v) β-mercaptoethanol and 4 μg/ml Complete Protease Inhibitor Cocktail (Roche). Lysates were centrifuged and protein concentration determined in the supernatants using Coomassie Plus protein assay reagent and BSA as standard. Aliquots of 15–20 μg protein were denatured, separated by SDS/8% PAGE and transferred to nitrocellulose membranes. Membranes were blocked for 2 h at room temperature in TBS-T (50 mM Tris-HCl (pH 7.5), 10 mM NaCl and 0.5% (v/v) Tween 20) containing 5% (w/v) non-fat milk and incubated overnight at 4 °C with the corresponding primary antibodies. Following washing in TBS-T, blots were incubated with HRP-coupled secondary antibodies for 1 h at room temperature. After washing in TBS-T, SuperSignal chemiluminescence substrate (Pierce) was added and the blots were exposed and developed using a Molecular Imager FX System (Bio-Rad Labs).
Transient transfections and luciferase activity assays
Hepa 1 cells were grown at 4 × 104 cells/well and its variants c2 and c4 clones at 6 × 104 cells/well in 24-well plates. Hepa 1 cells were transfected with 400 ng of each pGL2-LTBP-1-LUC construct using Lipofectamine + Plus Reagent (Invitrogen) in 250 μl of alpha-MEM medium containing 5% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin. Cultures were co-transfected with 20 ng of pCMVβGal vector (Clontech) to normalize luciferase activity. Six hours after transfection, fresh medium was added to the cultures for an additional 48 h. Cells were then lysed with Reporter lysis buffer (Promega) and luciferase activity determined in a microtiter plate lumino-meter (Dynex). β-Galactosidase activity was also measured and used to normalize luciferase activity. pGL2 Basic (empty vector) served to determine background luciferase activity.
To analyse the effects of AhR and ARNT over-expression on Ltbp-1 promoter activity, Hepa 1, c2 or c4 cell lines were co-transfected with 100 ng of the expression vectors pcDNAI-B6AhR or pcDNAI-ARNT and 400 ng of full-length pGL2-LTBP-1. Luciferase activity was measured and normalized as indicated above.
RNAi for HDAC2
siRNAs specific for HDAC2 were designed, synthesized (Qiagen) and characterized as described.66 AhR+/+ and AhR−/− MEF cells were plated at 14 × 104 cells/35 mm plate for 24 h. Then, cells were transfected with (50 nM) one of the two siRNAs for HDAC2 synthesized. The following sequences were used: Hs-Hdac2-1 sense:
r(GGUCAAUAAGACCAGAUAA)dTdT
and antisense:
r(UUAUCUGGUCUUAUUGACC)dGdT)
and Hs-Hdac2-2 sense:
r(GGGUUGUUUCAAUCUAACA)dTdT
and antisense:
r(UGUUAGAUUGAAACAACCC)dAdG).
Transfections were done using Oligofectamine Reagent (Invitrogen) as indicated by the manufacturer. In brief, 2.5 μl of Oligofectamine and 2.5 μlof Hs-Hdac2-1 or Hs-Hdac2-2 were added to 500 μl of Opti-MEM I medium. The resulting solutions were mixed gently and incubated for 1 h at room temperature. These transfection mixtures were then added for 6 h to MEF cells previously maintained in 500 μl of Opti-MEM I medium. After 48 h, cells were lysed to obtain RNA or protein, or fixed for chromatin immunoprecipitation (ChIP) assays.
HDAC2 immunoprecipitation and associated deacetylase activity
AhR+/+ and AhR−/− MEF cells were washed in PBS and lysed for 30 min at 4 °C in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) Nonidet NP-40 and 5 mM EDTA. Extracts were centrifuged and the protein concentration was determined in the supernatants as described above. Aliquots (3 mg) of protein were pre-cleared by incubation for 1 h at 4 °C with 40 μl of protein G/agarose (Upstate Biotechnology). Pre-cleared extracts were then rotated gently overnight at 4 °C with 12 μg of anti-HDAC2 antibody. Samples were rotated again for 1 h at 4 °C with 40 μl of fresh protein G/agarose. After brief centrifugation, agarose beads were washed with high-salt buffer (50 mM Tris-HCl (pH 7.4), 450 mM NaCl, 1% Nonidet NP-40, 5 mM EDTA). An aliquot of total extract was used as a positive control (input) and fresh protein G/agarose beads served as a negative control.
To determine HDAC2 deacetylase activity, washed beads were resuspended in 100 μl of buffer (25 mM Tris-HCl (pH 7.5), 10% glycerol, 1 mM EDTA, 50 mM NaCl) containing 60 μgof[3H]acetylated chicken erythrocyte histones. After incubation for 2 h at 37 °C, 37.5 μl stop solution (1 M HCl, 0.4 M acetic acid) was added and acetylated histones were extracted by the addition of 700 μl of ethyl acetate. Following centrifugation, equal volumes of the organic phase were measured using a Beckman LS-3801 scintillation counter.
Chromatin immunoprecipitation (ChIP)
ChIP assays were used to determine in vivo binding of transcription factors to the Ltbp-1 promoter and to analyse specific modifications (acetylation or methylation) of nucleosomal histones H3 and H4. Protein-DNA interactions were crosslinked in MEF and Hepa 1 cultures by the direct addition of 1% (v/v) formaldehyde (Sigma) during 15 min at room temperature. Cross-linking was stopped by incubation in 0.125 M glycine for 5 min and cells were washed and harvested in PBS containing 4 μg/ml Complete protease inhibitor. MEF and Hepa 1 cells were then lysed on ice for 10 min in 50 mM Tris-HCl (pH 8.1), 1% (w/v) SDS, 10 mM EDTA and 4 μg/ml Complete Protease Inhibitor Cocktail. Lysates were sonicated for 20 min in a Bioruptor (Diagenode) set at full power with 0.5 min sonication/ 0.5 min resting. Under these sonication conditions, DNA was fragmented in a range of 200–700 bp as determined by agarose gel electrophoresis. Next, samples were centrifuged and diluted 1:10 in ChIP buffer (6.7 mM Tris-HCl (pH 8.1), 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl and 4 μg/ml Complete Protease Inhibitor Cocktail). Diluted extracts were precleared by incubation for 30 min at 4 °C with protein A/agarose containing salmon sperm DNA. Following centrifugation, supernatants were incubated overnight at 4 °C with the corresponding antibodies. Then, fresh aliquots of protein A/agarose were added and incubation for an additional hour at 4 °C. Agarose beads were washed with the following buffers: low ionic strength (120 mM Tris-HCl (pH 8.1), 0. 1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl), high ionic strength (120 mM Tris-HCl (pH 8.1), 0. 1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl), LiCl buffer (10 mM Tris-HCl (pH 8.1), 0.25 M LiCl, 1% nonidet P40, 1% deoxycholate, 1 mM EDTA) and TE (10 mM Tris-HCl (pH 8.1), 1 mM EDTA). Protein-DNA complexes were separated from the agarose beads by three elution steps in 0.1 M NaHCO3 containing 1% SDS. DNA was then released from proteins by incubation for 4 h at 65 °C in 200 mM NaCl followed by incubation for 1 h at 45 °C in 40 mM Tris-HCl (pH 6.5), 10 mM EDTA and 4 μg/ml Complete Protease Inhibitor Cocktail. DNA was extracted with phenol/chloroform and precipitated with ethanol. PCR for the Ltbp-1 promoter was performed using the oligonucleotides: promoter region (−77/−333) forward:
5′ AGCTCACGCTACCTCGGATA 3′
and reverse:
5′ GAGATGAGCCGGAGAATGAG 3′
and promoter region (−445/−736) forward:
5′ TCATGGACTTTTGATTGCAG 3′
and reverse:
5′ CATTGGAGGGTCAAGAATGC 3′.
Amplification was done for 35 cycles in 25 μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.5 μM each primer, 2.5 U Taq polymerase and 3 μl of DNA template. Cycling conditions were: initial denaturation at 94 °C for 5 min, denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 1 min. PCR products were visualized in agarose gels stained with ethidium bromide. Extracts incubated in the absence of antibody (input) were used as positive controls. Fresh protein A/ agarose was used as a negative control. In some experiments, crosslinking was performed using 2% formaldehyde or 1% formaldehyde plus 2 mM disuccinyl glutarate or 10 mM dimethyl apidimate.
Statistical analyses
Analysis of variance (ANOVA) was done with Instat software program (GraphPAD Sotfware, version 1.11). Data are shown as mean ± SE.
Acknowledgements
This work has been supported by grants SAF2002-00034 and SAF2005-00130 from the Spanish Ministry of Education and Sciences, and from the Junta de Extremadura 2PR04A060 (to P.M.F-.S) and by US NIEHS grants 5R01 ES06273 and P30 ES06096 (to A.P.). A.G.D. and J.M.C.-G. were fellows from the Spanish Ministry of Education and Sciences and from the Junta de Extremadura, respectively. We are very grateful to Dr Santiago Ropero for assistance with HDAC2 deacetylase activity experiments.
Abbreviations used
- AhR
aryl hydrocarbon (dioxin) receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- ChIP
chromatin immunoprecipitation
- CREB
cAMP responsive element-binding protein
- ECM
extracellular matrix
- HDAC
histone deacetylase
- LTBP-1
latent transforming growth factor-binding protein 1
- MEF
mouse embryonic fibroblasts
- TGFβ
transforming growth factor β
- RNAi
RNA interference
- siRNA
small interfering RNA
Footnotes
Data Bank accession number
The mouse Ltbp-1L promoter sequence has been deposited in GenBank with accession number EF635913.
References
- 1.Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 2.Saharinen J, Hyytiainen M, Taipale J, Keski-Oja J. Latent transforming growth factor-beta binding proteins (LTBPs) – structural extracellular matrix proteins for targeting TGF-beta action. Cytokine Growth Factor Rev. 1999;10:99–117. doi: 10.1016/s1359-6101(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 3.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 4.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nature Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 5.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J. Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 6.Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J. Latency, activation, and binding proteins of TGF-beta. Microsc. Res. Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence DA. Latent-TGF-beta: an overview. Mol. Cell Biochem. 2001;219:163–170. doi: 10.1023/a:1010819716023. [DOI] [PubMed] [Google Scholar]
- 8.Dallas SL, Rosser JL, Mundy GR, Bonewald LF. Proteolysis of latent transforming growth factor-beta (TGF-beta)-binding protein-1 by osteoclasts. A cellular mechanism for release of TGF-beta from bone matrix. J. Biol. Chem. 2002;277:21352–21360. doi: 10.1074/jbc.M111663200. [DOI] [PubMed] [Google Scholar]
- 9.Oklu R, Hesketh R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem. J. 2000;352:601–610. [PMC free article] [PubMed] [Google Scholar]
- 10.Taipale J, Keski–Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- 11.Giltay R, Kostka G, Timpl R. Sequence and expression of a novel member (LTBP-4) of the family of latent transforming growth factor-beta binding proteins. FEBS Lett. 1997;411:164–168. doi: 10.1016/s0014-5793(97)00685-6. [DOI] [PubMed] [Google Scholar]
- 12.Koski C, Saharinen J, Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-beta binding protein-1 (LTBP-1) in a cell type-specific manner. J. Biol. Chem. 1999;274:32619–32630. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- 13.Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, et al. Identification and characterization of LTBP-2, a novel latent transforming growth factor-beta-binding protein. J. Biol. Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- 14.Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new latent transforming growth factor-beta-binding protein, LTBP-4. J. Biol. Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji T, Okada F, Yamaguchi K, Nakamura T. Molecular cloning of the large subunit of transforming growth factor type beta masking protein and expression of the mRNA in various rat tissues. Proc. Natl Acad. Sci. USA. 1990;87:8835–8839. doi: 10.1073/pnas.87.22.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor-beta binding protein gene (LTBP-3) J. Biol. Chem. 1995;270:10147–10160. doi: 10.1074/jbc.270.17.10147. [DOI] [PubMed] [Google Scholar]
- 17.Noguera I, Obata H, Gualandris A, Cowin P, Rifkin DB. Molecular cloning of the mouse Ltbp-1 gene reveals tissue specific expression of alternatively spliced forms. Gene. 2003;308:31–41. doi: 10.1016/s0378-1119(03)00463-3. [DOI] [PubMed] [Google Scholar]
- 18.Weiskirchen R, Moser M, Gunther K, Weiskirchen S, Gressner AM. The murine latent transforming growth factor-beta binding protein (Ltbp-1) is alternatively spliced, and maps to a region syntenic to human chromosome 2p21-22. Gene. 2003;308:43–52. doi: 10.1016/s0378-1119(03)00464-5. [DOI] [PubMed] [Google Scholar]
- 19.Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 20.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 21.Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and Ah gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 23.Rivera SP, Saarikoski ST, Hankinson O. Identification of a novel dioxin-inducible cytochrome P450. Mol. Pharmacol. 2002;61:255–259. doi: 10.1124/mol.61.2.255. [DOI] [PubMed] [Google Scholar]
- 24.Marlowe JL, Puga A. Aryl hydrocarbon receptor, cell cycle regulation, toxicity, and tumorigenesis. J. Cell. Biochem. 2005;96:1174–1184. doi: 10.1002/jcb.20656. [DOI] [PubMed] [Google Scholar]
- 25.Elferink CJ. Aryl hydrocarbon receptor-mediated cell cycle control. Prog. Cell Cycle Res. 2003;5:261–267. [PubMed] [Google Scholar]
- 26.Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 27.Bock KW, Kohle C. Ah receptor: dioxin-mediated toxic responses as hints to deregulated physiologic functions. Biochem. Pharmacol. 2006;72:393–404. doi: 10.1016/j.bcp.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Barouki R, Coumoul X, Fernandez-Salguero PM. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 29.Frericks M, Meissner M, Esser C. Micro-array analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol. Appl. Pharmacol. 2007;220:320–332. doi: 10.1016/j.taap.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Frericks M, Temchura VV, Majora M, Stutte S, Esser C. Transcriptional signatures of immune cells in aryl hydrocarbon receptor (AHR)-proficient and AHR-deficient mice. Biol. Chem. 2006;387:1219–1226. doi: 10.1515/BC.2006.151. [DOI] [PubMed] [Google Scholar]
- 31.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- 32.Ogi T, Mimura J, Hikida M, Fujimoto H, FujiiKuriyama Y, Ohmori H. Expression of human and mouse genes encoding polkappa: testis-specific developmental regulation and AhR-dependent inducible transcription. Genes Cells. 2001;6:943–953. doi: 10.1046/j.1365-2443.2001.00478.x. [DOI] [PubMed] [Google Scholar]
- 33.Kolluri SK, Balduf C, Hofmann M, Gottlicher M. Novel target genes of the Ah (dioxin) receptor: transcriptional induction of N-myristoyltransferase 2. Cancer Res. 2001;61:8534–8539. [PubMed] [Google Scholar]
- 34.Kolluri SK, Weiss C, Koff A, Gottlicher M. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13:1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nature Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- 36.Niermann T, Schmutz S, Erne P, Resink T. Aryl hydrocarbon receptor ligands repress T-cadherin expression in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2003;300:943–949. doi: 10.1016/s0006-291x(02)02970-4. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Liu D, Murray TJ, Mitchell GC, Hesterman EV, Karchner SI, et al. The aryl hydrocarbon receptor constitutively represses c-myc transcription in human mammary tumor cells. Onco-gene. 2005;24:7869–7881. doi: 10.1038/sj.onc.1208938. [DOI] [PubMed] [Google Scholar]
- 38.Marlowe JL, Knudsen ES, Schwemberger S, Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 2004;279:29013–29022. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- 39.Denison MS, Whitlock JP., Jr Xenobioticinducible transcription of cytochrome P450 genes. J. Biol. Chem. 1995;270:18175–18178. doi: 10.1074/jbc.270.31.18175. [DOI] [PubMed] [Google Scholar]
- 40.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 41.Peterson TC, Hodgson P, Fernandez-Salguero P, Neumeister M, Gonzalez FJ. Hepatic fibrosis and cytochrome P450: experimental models of fibrosis compared to AHR knockout mice. Hepatol. Res. 2000;17:112–125. doi: 10.1016/s1386-6346(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 43.Corchero J, Martin-Partido G, Dallas SL, Fernandez-Salguero PM. Liver portal fibrosis in dioxin receptor-null mice that overexpress the latent transforming growth factor-beta-binding protein-1. Int. J. Expt. Pathol. 2004;85:295–302. doi: 10.1111/j.0959-9673.2004.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elizondo G, Fernandez-Salguero P, Sheikh MS, Kim GY, Fornace AJ, Lee KS, Gonzalez FJ. Altered cell cycle control at the G(2)/M phases in aryl hydrocarbon receptor-null embryo fibroblast. Mol. Pharmacol. 2000;57:1056–1063. [PubMed] [Google Scholar]
- 45.Santiago-Josefat B, Mulero-Navarro S, Dallas SL, Fernandez-Salguero PM. Overexpression of latent transforming growth factor-{beta} binding protein 1 (LTBP-1) in dioxin receptor-null mouse embryo fibroblasts. J. Cell Sci. 2004;117:849–859. doi: 10.1242/jcs.00932. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Duran A, Mulero-Navarro S, Chang X, Fernandez-Salguero PM. LTBP-1 blockade in dioxin receptor-null mouse embryo fibroblasts decreases TGF-beta activity: role of extracellular proteases plasmin and elastase. J. Cell. Biochem. 2006;97:380–392. doi: 10.1002/jcb.20637. [DOI] [PubMed] [Google Scholar]
- 47.Chang X, Fan Y, Karyala S, Schwemberger S, Tomlinson CR, Sartor MA, Puga A. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol. Cell Biol. 2007;27:6127–6139. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang F, Bleich D. Transcriptional regulation of cyclooxygenase-2 gene in pancreatic beta-cells. J. Biol. Chem. 2004;279:35403–35411. doi: 10.1074/jbc.M404055200. [DOI] [PubMed] [Google Scholar]
- 49.Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol. Cell Biol. 2005;25:5317–5328. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J. Biol. Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- 51.Wei YD, Tepperman K, Huang MY, Sartor MA, Puga A. Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin. J. Biol. Chem. 2004;279:4110–4119. doi: 10.1074/jbc.M310800200. [DOI] [PubMed] [Google Scholar]
- 52.Whitlock JP, Jr, Okino ST, Dong L, Ko HP, Clarke-Katzenberg R, Ma Q, Li H. Cytochromes P450 5: induction of cytochrome P4501A1: a model for analyzing mammalian gene transcription. FASEB J. 1996;10:809–818. doi: 10.1096/fasebj.10.8.8666157. [DOI] [PubMed] [Google Scholar]
- 53.Zaher H, Fernandez-Salguero PM, Letterio J, Sheikh MS, Fornace AJ, Jr, Roberts AB, Gonzalez FJ. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 1998;54:313–321. doi: 10.1124/mol.54.2.313. [DOI] [PubMed] [Google Scholar]
- 54.Kwok S, Qin L, Partridge NC, Selvamurugan N. Parathyroid hormone stimulation and PKA signaling of latent transforming growth factor-beta binding protein-1 (LTBP-1) mRNA expression in osteoblastic cells. J. Cell. Biochem. 2005;95:1002–1011. doi: 10.1002/jcb.20453. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 56.Liu PC, Dunlap DY, Matsumura F. Suppression of C/EBPalpha and induction of C/EBPbeta by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mouse adi-pose tissue and liver. Biochem. Pharmacol. 1998;55:1647–1655. doi: 10.1016/s0006-2952(98)00012-4. [DOI] [PubMed] [Google Scholar]
- 57.Vogel CF, Sciullo E, Park S, Liedtke C, Trautwein C, Matsumura F. Dioxin increases C/EBPbeta transcription by activating cAMP/ protein kinase. A. J. Biol. Chem. 2004;279:8886–8894. doi: 10.1074/jbc.M310190200. [DOI] [PubMed] [Google Scholar]
- 58.Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, et al. Attenuation of a phosphorylation-dependent activator by an HDACPP1 complex. Nature Struct. Biol. 2003;10:175–181. doi: 10.1038/nsb895. [DOI] [PubMed] [Google Scholar]
- 59.Ito K, Barnes PJ, Adcock IM. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell Biol. 2000;20:6891–6903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tenbrock K, Juang YT, Leukert N, Roth J, Tsokos GC. The transcriptional repressor cAMP response element modulator alpha interacts with histone deacetylase 1 to repress promoter activity. J. Immunol. 2006;177:6159–6164. doi: 10.4049/jimmunol.177.9.6159. [DOI] [PubMed] [Google Scholar]
- 61.Nottebrock C, Riecke K, Kruse M, Shakibaei M, Stahlmann R. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the extracellular matrix of the thymus in juvenile marmosets (Callithrix jacchus) Toxicology. 2006;226:197–207. doi: 10.1016/j.tox.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 62.Riecke K, Grimm D, Shakibaei M, Kossmehl P, Schulze-Tanzil G, Paul M, Stahlmann R. Low doses of 2,3,7,8-tetrachlorodibenzo-p-dioxin increase transforming growth factor beta and cause myocardial fibrosis in marmosets (Callithrix jacchus) Arch. Toxicol. 2002;76:360–366. doi: 10.1007/s00204-002-0338-6. [DOI] [PubMed] [Google Scholar]
- 63.Yang JH, Vogel C, Abel J. A malignant transformation of human cells by 2,3,7,8-tetrachlorodibenzo-p-dioxin exhibits altered expressions of growth regulatory factors. Carcinogenesis. 1999;20:13–18. doi: 10.1093/carcin/20.1.13. [DOI] [PubMed] [Google Scholar]
- 64.Santiago-Josefat B, Pozo-Guisado E, Mulero-Navarro S, Fernandez-Salguero PM. Proteasome inhibition induces nuclear translocation and transcriptional activation of the dioxin receptor in mouse embryo primary fibroblasts in the absence of xenobiotics. Mol. Cell Biol. 2001;21:1700–1709. doi: 10.1128/MCB.21.5.1700-1709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Legraverend C, Hannah RR, Eisen HJ, Owens IS, Nebert DW, Hankinson O. Regulatory gene product of the Ah locus. Characterization of receptor mutants among mouse hepatoma clones. J. Biol. Chem. 1982;257:6402–6407. [PubMed] [Google Scholar]
- 66.Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nature Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]