Abstract
Senescence is an irreversible cell-cycle arrest that is elicited by a wide range of factors, including replicative exhaustion. Emerging evidences suggest that cellular senescence contributes to ageing and acts as a tumour suppressor mechanism. To identify novel genes regulating senescence, we performed a loss-of-function screen on normal human diploid fibroblasts. We show that downregulation of the AMPK-related protein kinase 5 (ARK5 or NUAK1) results in extension of the cellular replicative lifespan. Interestingly, the levels of NUAK1 are upregulated during senescence whereas its ectopic expression triggers a premature senescence. Cells that constitutively express NUAK1 suffer gross aneuploidies and show diminished expression of the genomic stability regulator LATS1, whereas depletion of NUAK1 with shRNA exerts opposite effects. Interestingly, a dominant-negative form of LATS1 phenocopies NUAK1 effects. Moreover, we show that NUAK1 phosphorylates LATS1 at S464 and this has a role in controlling its stability. In summary, our work highlights a novel role for NUAK1 in the control of cellular senescence and cellular ploidy.
Keywords: LATS1, NUAK1, senescence
Introduction
Normal human diploid cells have a finite replicative potential, mainly because of telomere erosion. When they reach this critical point they become senescent (Hayflick, 1965). Senescent cells remain metabolically active, show characteristic changes in cell morphology, physiology, and gene expression, and typically show a senescence-associated β-galactosidase (SA-β-gal) activity (Dimri et al, 1995) and the appearance of senescent-associated heterochromatin foci (Narita et al, 2003). A phenotypically similar end point can be reached in response to different cellular stresses, notably inappropriate oncogenic signalling or oxidative stress. The fact that normal cells undergo senescence in response to oncogenes has led to the hypothesis that senescence, similar to apoptosis (Lowe et al, 2004; Dimri, 2005), is an intrinsic genetic programme with tumour-suppressive properties (Braig et al, 2005; Chen et al, 2005; Collado et al, 2005; Dimri, 2005).
A corollary of the above observations is that oncogenes and tumour suppressor alterations can compromise the senescence programme, thus favouring cell immortalisation and cancer. This is exemplified by our current understanding of the Rb and p53 tumour suppressor networks, which are pivotal in controlling the occurrence of senescence and tumour development (Sherr, 1996; Sionov and Haupt, 1999; Hanahan and Weinberg, 2000; Yamasaki, 2003; Campisi, 2005). Interference with the Rb or p53 pathway through mutation, overexpression, or depletion of various components of these pathways or by overexpression of oncogenic proteins (including the oncoviral proteins E6 and E7) results in bypassed senescence and extension of the replicative lifespan of normal human cells (Jacobs et al, 1999; Carnero et al, 2000; Lundberg et al, 2000; Ohtani et al, 2001; Brummelkamp et al, 2002; Shvarts et al, 2002; d'Adda di Fagagna et al, 2003; Gil et al, 2004; Herbig et al, 2004).
Aneuploidy caused by a weak mitotic spindle checkpoint or by cytokinetic defects results in premature senescence (Baker et al, 2004; Takahashi et al, 2006). To our knowledge, however, nobody has ever reported having increased the replicative potential of normal cells by enforcing genomic stability safeguards. Strong enforcement of genomic stability safeguards, such as by LATS1 or BubR1, by constitutive expression can even inhibit cell growth (Yang et al, 2001; Shin et al, 2003; Iida et al, 2004). Whether slightly enhanced genomic stability can increase the replicative potential of normal human cells thus remains an open question.
To identify new genes regulating the replicative potential of normal human cells, we have performed a loss-of-function genetic screen, using a retroviral shRNA library. This screen has revealed that the level of NUAK1 has an effect on the replicative potential of normal human cells and this might be due to the level of aneuploidie. This suggests that the maintenance of the normal DNA content can increase the replicative potential of human cells.
Results
NUAK1 depletion prevents senescence
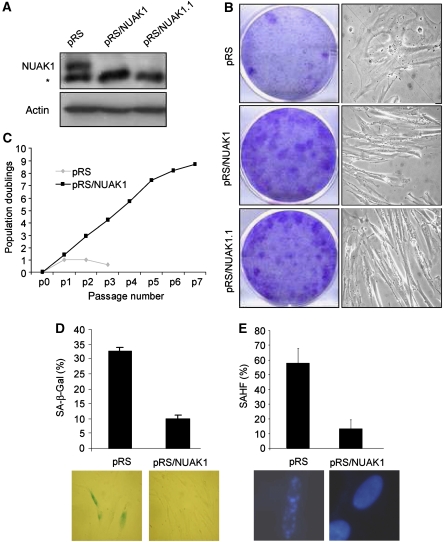
WI-38 normal human diploid fibroblasts (HDFs) approaching senescence were infected with an shRNA retroviral library (Berns et al, 2004). After puromycin selection, the cells were split every week for 3 weeks. Cells infected with the empty (control) vector stopped growing, whereas some cells infected with library pools continued to grow. Genomic DNA was extracted from these cells and an shRNA construct targeting NUAK1 conferring extension of the cellular replicative lifespan was identified, along with shRNAs directed against PLA2R (Augert et al, 2009), topoisomerase 1 (Humbert et al, 2009), and another shRNA that is still under characterisation. To analyse the role of NUAK1 in replicative senescence, we generated two independent retroviral shRNA vectors, each targeting NUAK1. We then verified the ability of these shRNAs to downregulate endogenous NUAK1 protein levels (Figure 1A). When HDFs were infected with either of the two NUAK1 knockdown shRNAs, they reversed senescence, as monitored using colony assay and by their loss of senescent morphology (Figure 1B). This effect was further confirmed using growth curve analysis in which we observed an extension of replicative lifespan of approximately eight population doublings in NUAK1 knockdown cells (Figure 1C). This increase in replicative potential was accompanied with a decrease in the number of senescent cells, as observed by SA-β-Gal staining or SAHF analysis in WI-38 cells infected with pRS/NUAK1 when compared with cells infected with an empty vector (Figure 1D and E). Ultimately, the NUAK1 knockdown cells entered a senescence state. Indeed, the cells then stopped growing, showed an SA-β-Gal staining and the typical senescent morphology, and were immortalised by hTert expression (Supplementary Figure S1).
Figure 1.
NUAK1 downregulation extends the replicative lifespan. (A) WI-38 cells were infected with control pRS, pRS/NUAK1, or pRS/NUAK1.1 (these constructs encode two different shRNAs directed against NUAK1) and selected by puromycin. Cell extracts were prepared and resolved using SDS–PAGE. * denotes a non-specific band. The efficiency of the two shRNAs to downregulate endogenous NUAK1 protein levels was monitored using immunoblot analysis with an anti-NUAK1 antibody. (B) WI-38 cells were infected with a retroviral vector (pRS control, pRS/NUAK1, or pRS/NUAK1.1) and selected with puromycin. Selected cells were seeded at low density and stained after 2 weeks with crystal violet. Alternatively, transmitted images were taken 5 days after seeding to monitor any change in cellular morphology. (C) WI-38 cells were infected with pRS or pRS/NUAK1 and selected with puromycin. The cells were split every 5 days and the cell numbers were counted. Cumulative population doublings were showed at each passage. (D, E) Infected and selected WI-38 cells were analysed for SA-β-Gal activity or SAHF positivity. Analysis was performed when control cells entered senescence.
Collectively, these results indicate that the downregulation of NUAK1 expression results in replicative lifespan extension.
Constitutive NUAK1 expression triggers premature senescence
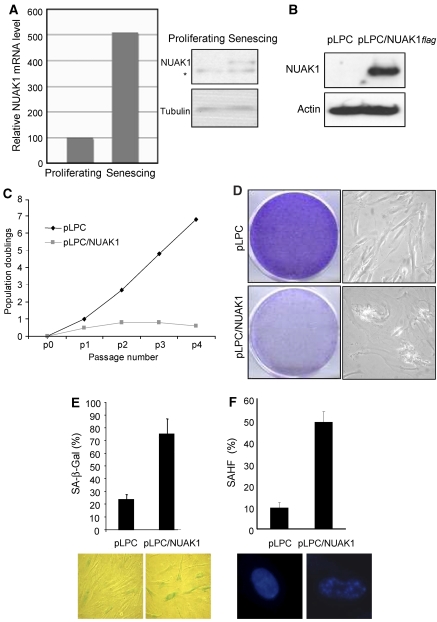
As downregulation of NUAK1 extends the replicative lifespan, we next analysed whether the endogenous NUAK1 level increases during replicative senescence. To verify this, we measured the NUAK1 transcript level using Q–PCR and protein level using immunoblot analysis in proliferating- versus senescing-cultured HDFs. NUAK1 mRNA and protein increased strongly in late-passage HDFs when compared with early-passage ones (Figure 2A).
Figure 2.
NUAK1 overexpression induces premature senescence. (A) RNA and protein were prepared from proliferating and senescing WI-38 cells. After reverse-transcription, real-time quantitative PCR was used to determine NUAK1 mRNA expression. Relative NUAK1 mRNA levels are shown. Alternatively, NUAK1 protein levels were measured by immunoblot analysis using an anti-NUAK1 antibody. * denotes a non-specific band. (B) WI-38 cells were infected with the pLPC or pLPC/NUAK1flag retroviral vector. After puromycin selection, cells were used to perform different assays. Cell extracts were prepared and resolved using SDS–PAGE. Constitutive expression of flag-tagged NUAK1 levels was checked using immunoblot analysis with an anti-flag-tag antibody. Actin protein levels were used as a loading control. (C) Cells were split and counted every 5 days. Cumulative population doublings were calculated and showed after each passage of pLPC-or pLPC/NUAK1flag-infected cells. (D) Cells were seeded at low density and left to grow for 5 days and phase contrast images were taken to monitor cell morphology. Alternatively, after 2 weeks, cells were fixed with PFA and stained with crystal violet. Representative plates are shown. (E, F) SA-β-Gal and SAHF assays were performed to analyse senescence of pLPC-infected (control) and pLPC/NUAK1 flag-infected HDFs.
To confirm that NUAK1 modulates the occurrence of senescence, we analysed the effect of NUAK1 overexpression. Early-passage HDFs were infected with a retroviral vector expressing a flag-tagged human NUAK1 or with a control retroviral vector. Ectopic expression of NUAK1 was checked using western blotting with an anti-flag tag antibody (Figure 2B). The growth potential of NUAK1-transduced cells was examined by means of growth curves and a colony formation assay. NUAK1 expression was found to block cell growth and induce a characteristic senescent morphology (Figure 2C and D). This growth arrest was due to premature senescence induction, as evidenced by the increased proportion of SA-β-Gal-positive and SAHF-positive cells among the NUAK1-overexpressing cells, as compared with control cells (Figure 2E and F). These results reveal that both downregulation and overexpression of NUAK1 have a major effect on the cellular replicative lifespan by regulating senescence.
NUAK1-induced senescence requires activation by LKB1 kinase
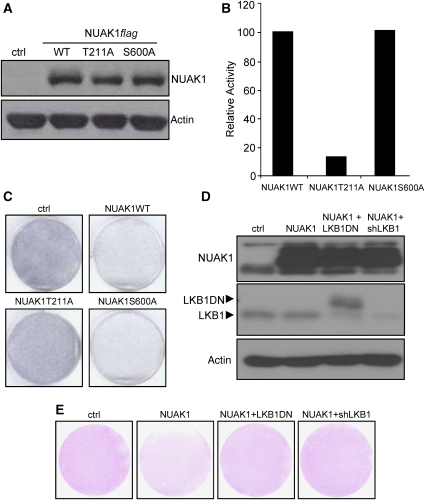
Phosphorylation of NUAK1 at T211 by the LKB1 kinase has been shown to activate its kinase activity (Lizcano et al, 2004). The phosphorylation of NUAK1 at S600 by Akt kinase could also regulate NUAK1 (Suzuki et al, 2003b). We generated retroviral vectors encoding T211A or S600A mutation and checked their expression using immunoblot (Figure 3A). Interestingly, whereas NUAK1 S600A mutant kept its ability to phosphorylate an AMARA peptide, the NUAK1 T211A mutant lost it (Figure 3B). Accordingly, only the NUAK1 S600A mutant was still able to block cell growth, whereas the NUAK1 T211A mutant was not (Figure 3C). We next wondered whether or not activation of NUAK1 through the T211 involved LKB1 kinase (Lizcano et al, 2004). To evaluate this, we stably expressed in WI-38 cells a control vector, or an NUAK1 vector, or an NUAK1 and an LKB1 dominant-negative (LKB1DN) vectors, or an NUAK1 and an shRNA directed against LKB1 (shLKB1) encoding vectors. We first verified the expression of all the proteins (Figure 3D). The growth-inhibitory effect of NUAK1 was largely reverted when LKB1 activity was inhibited by either expression of a dominant-negative form or by its knockdown (Figure 3E). These results indicate that the effect of NUAK1 oversenescence required its phosphorylation by LKB1.
Figure 3.
NUAK1 activation by LKB1 is required for NUAK1-induced senescence. (A) WI-38 cells were infected with the indicated constructs and puromycin selected. Cellular extracts were prepared and analysed using immunoblotting. Wild type and mutants of NUAK1 expression levels were checked with an anti- flag-tag antibody. Anti-actin antibody was used to monitor protein loading. (B) HEK 293 cells were transfected with the indicated vectors. After immunoprecipitation with an anti-flag-tag antibody, NUAK1 kinase activity towards the AMARA peptide was measured. The activity was normalised to 100% to NUAK1-transfected cells. (C) After infection and selection, the cells were seeded at low density. After 2 weeks, the cells were fixed and stained by crystal violet. (D) WI-38 cells were infected with ctrl or NUAK1 encoding vectors (neo resistance) together with ctrl or LKB1DN or shLKB1 encoding vectors (puro selection) and selected. Cellular extracts were prepared and analysed using immunoblotting. NUAK1 expression was analysed using an anti-flag-tag antibody, and LKB1 expression was analysed using an anti- LKB1 antibody. Anti-actin antibody was used to monitor protein loading. (E) After infection and selection as in D, cells were seeded at low density. The cells at 2 weeks after they were fixed and stained by crystal violet are shown. A full-colour version of this figure is available at The EMBO Journal Online.
NUAK1-induced senescence requires the kinase activity of NUAK1
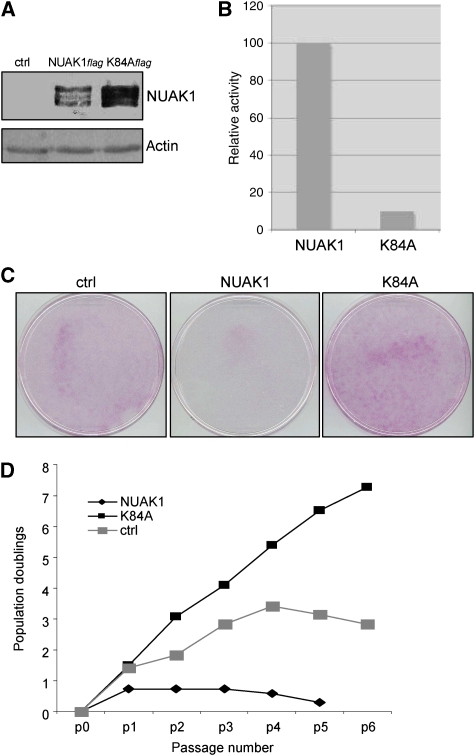
To gain insight into the mechanism by which NUAK1 controls the occurrence of senescence, we examined whether the NUAK1 kinase activity mediates this control. We generated a retroviral vector encoding a kinase-dead mutant (K84A) (Lizcano et al, 2004; Suzuki et al, 2006), and verified its expression using immunoblotting (Figure 4A). We next tested whether this mutant had lost its kinase activity towards the AMARA peptide, a known NUAK1 substrate. As expected, the kinase-dead mutant was unable to phosphorylate a serine residue in the AMARA peptide (Figure 4B). To examine the effect of this mutant on senescence, we compared the ability of NUAK1-expressing and NUAK1K84A-expressing cells to form colonies. As expected, NUAK1 expression was found to inhibit colony formation when compared with control cells (Figure 4C). Interestingly, the kinase-dead mutant K84A was unable to block colony formation and even seemed to favour cell growth (Figure 4C). To further confirm these effects we performed growth curve analysis. The NUAK1K84A-expressing cells showed an extended replicative lifespan when compared with control infected cells, suggesting that it might act as a dominant-negative form (Figure 4D). Nevertheless, the cells entered the senescence state after few additional passages, a senescence that was avoided by hTert constitutive expression (Supplementary Figure S2). We conclude that the kinase activity of NUAK1 is required for NUAK1-induced senescence.
Figure 4.
NUAK1 kinase activity is required for senescence induction. (A) WI-38 cells were infected with a control vector or with a vector expressing wild-type or K84A NUAK1 mutant. After puromycin selection, cell extracts were prepared and analysed using immunoblotting. Wild-type and mutant NUAK1 expression levels were checked with an anti- flag-tag antibody. Anti-actin antibody was used to monitor protein loading. (B) HEK 293 cells were transfected with a vector encoding wild-type or K84A mutant NUAK1. After immunoprecipitation with an anti-flag-tag antibody, NUAK1 kinase activity towards the AMARA peptide was measured. The activity was normalised to 100% to NUAK1-transfected cells. (C) WI-38 cells were infected by the indicated constructs and selected with puromycin. The cells were next seeded at low density, allowed to grow, and stained with crystal violet as in Figure 2. (D) WI-38 cells were infected with the indicated vectors and selected with puromycin. Growth was monitored up to 6 passages and curves were drawn. A full-colour version of this figure is available at The EMBO Journal Online.
NUAK1 induces gross aneuploidies
To identify the pathways and the target through which NUAK1 controls senescence, we first examined the status of the two main senescence-regulating pathways: the p53 and the Rb pathways. Quite surprisingly, p53 and p21 proteins levels were not modified by NUAK1 knockdown or overexpression. In addition, E6 expression was not able to revert NUAK1-induced senescence (Supplementary Figure S3A–C). Rb was found hyper- or hypo-phosphorylated in NUAK1 overexpressed or knockdown cells (Supplementary Figure S3A and B). However, the inhibition of the Rb pathway by E7 was not sufficient to revert NUAK1-induced senescence (Supplementary Figure S3C). The modification observed on Rb is thus insufficient to explain the effect of NUAK1 and could thus be only a mark of the proliferative state.
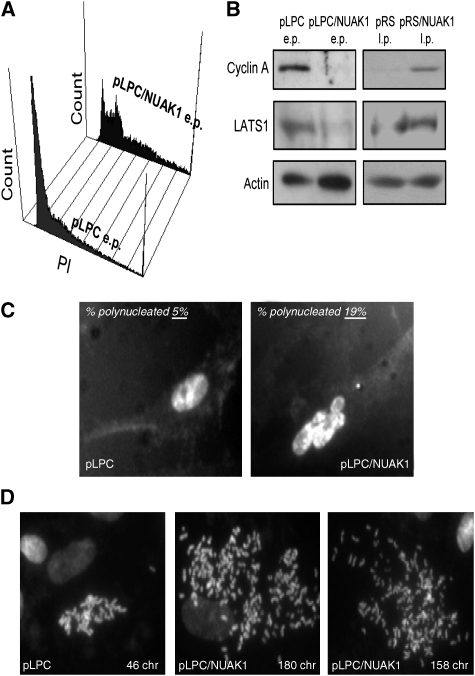
As genomic instability is reported to cause premature senescence (Baker et al, 2004; Takahashi et al, 2006), we next examined whether NUAK1 might regulate senescence by affecting the DNA content. We first performed FACS-based cell-cycle analysis of control and NUAK1-overexpressing cells. NUAK1-expressing cells shifted from a mainly 2n DNA content (G1 phase) to a 3–4n DNA content (Figure 5A). In addition, numerous NUAK1-expressing cells had more than 4n DNA content according to the FACS analysis (Figure 5A). Interestingly, cells expressing an shRNA directed against NUAK1 show a normal DNA content profile in contrast with control senescent cells that show a shift of cells in 3–4n DNA content (Supplementary Figure S4). We then wondered whether NUAK1-expressing cells or control senescent cells showing 3–4n DNA content might be blocked in late S-G2/M. This possibility was ruled out on the basis of the observation that cyclin A, a cyclin accumulating in the late S-G2/M phases, was downregulated in NUAK1-expressing cells or in control senescing cells (Figure 5B). From these experiments we conclude that NUAK1-expressing cells or control senescent cells have an aberrant DNA content.
Figure 5.
NUAK1 regulates the DNA content. (A) At 10 days after infection with a pLPC or a pLPC/NUAK1flag vector and puromycin selection, WI-38 cells were fixed with ethanol, stained with propidium iodide, and analysed using FACS to determine the DNA content. (B) Cell extracts were prepared from early-passage (e.p.) pLPC-infected and pLPC/NUAK1-infected and late-passage (l.p.) pRS- and pRS/NUAK1-infected WI-38 cells. Levels of the cyclin A and LATS1 proteins were checked using immunoblotting and actin level was used as a loading control. (C) WI-38 cells were fixed with PFA and stained with Hoechst. Percentages of polynucleated cells were estimated and Hoechst-stained images are shown. (D) Chromosome spreading experiments. At 7 days after infection and selection, cells were treated with colcemid. Cell images with normal and aberrant chromosome numbers are shown.
A decrease in LATS1 and subsequent block of cytokinesis has been implicated in senescence (Takahashi et al, 2006). The LATS1 level was found to be lower in NUAK1-expressing cells and higher in NUAK1-depleted cells than in respective control cells (Figure 5B). Microscopic analysis confirmed that NUAK1-expressing cells had greater proportion of polynucleated cells when compared with control cells (30 versus 8%; Figure 5C). Interestingly, 5–10% of the mitotic NUAK1-expressing cells contained more than 100 chromosomes, whereas control cells never showed such an aberrant number of chromosomes (Figure 5D). As this assay required cell entry into mitosis, we think that we underestimated the number of NUAK1-expressing cells with a high chromosome number as the majority of these cells were not growing. LATS1 downregulation might allow cells to undergo aberrant mitosis without cytokinesis (Yang et al, 2004). This would explain the aberrant DNA content observed upon constitutive NUAK1 expression. In contrast, downregulation of NUAK1 seems to lead to sustained levels of LATS1 to thus avoid the appearance of aberrant DNA content when compared with control senescing cells.
LATS1 inhibition induces premature senescence
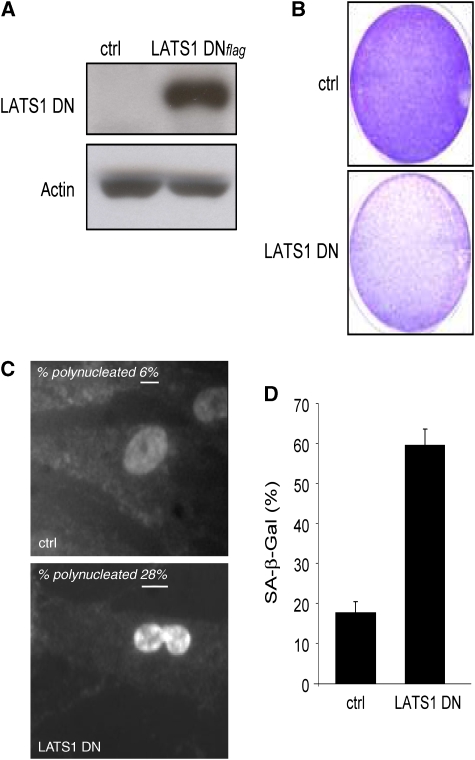
We next examined whether LATS1 inactivation alone might be sufficient to produce the phenotype induced by ectopic NUAK1 expression (i.e., aberrant DNA content and senescence). We infected HDFs with a dominant-negative form of LATS1 (LATS1 DN) (Takahashi et al, 2006). LATS1 DN expression was confirmed using immunoblot analysis (Figure 6A). A growth arrest was observed when compared with control infected cells, as judged using colony formation assays (Figure 6B). Similar to constitutive NUAK1 expression, the LATS1 DN expression induced gross aneuploidies (Figure 6C) and the appearance of SA-β-Gal activity (Figure 6D). In addition, LATS1 DN expression in shNUAK1-expressing cells was still able to block cell growth, suggesting that LATS1 might be a downstream target of NUAK1 (Supplementary Figure S5). Thus, inactivation of LATS1 mimics the effect of NUAK1 on aberrant DNA content appearance and senescence.
Figure 6.
LATS1 downregulation induces gross aneuploidies and premature senescence. (A) WI-38 cells were infected with a control vector or with a vector encoding a dominant-negative form of LATS1. After puromycin selection, cell extracts were prepared and expression of LATS1 DN was checked by immunoblotting using anti-flag-tag antibody. Actin level was used as a loading control. (B) Infected cells were seeded at low density. After 10 days of culture, the cells were stained with crystal violet. (C) The nuclei of infected cells were stained with Hoechst dye. Percentages of polynucleated cells were estimated and Hoechst-stained images are shown. (D) SA-β-Gal assays were performed on cells infected with the LATS1DN-encoding construct and on control cells. A full-colour version of this figure is available at The EMBO Journal Online.
NUAK1 interacts, phosphorylates, and controls LATS1 levels
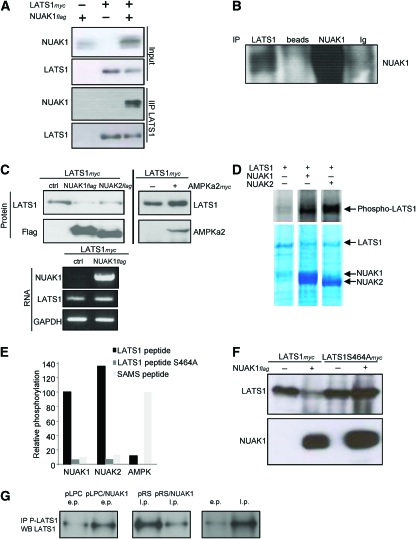
As a deficiency of LATS1 causes aneuploidies and premature senescence, we next analysed the relationships between NUAK1 and LATS1by performing co-immunoprecipitation assays on extracts from cells co-expressing NUAK1 and LATS1. Interestingly, NUAK1 was detected in the LATS1 immunoprecipitate (Figure 7A). We next repeated the immunoprecipitation experiment with endogenous proteins. Once again, NUAK1 protein was found in the LATS1 immunoprecipitate (Figure 7B).
Figure 7.
NUAK1 regulates LATS1 levels through phosphorylation at S464. (A) HEK 293 cells were transfected with the NUAK1flag, LATS1myc-encoding vectors, or with both. Immunoprecipitation was performed with an anti-flag antibody at 1 day after transfection and proteins of interest were monitored using immunoblotting with the indicated antibodies. (B) WI-38 cells closed to senescence were used to prepare cell extracts. Immunoprecipitation was performed with LATS1 or NUAK1 antibodies. IgG or beads alone were used as controls. The immunoprecipitates were analysed by immunoblotting using an anti-NUAK1 antibody. (C) HEK 293 cells were co-transfected with the LATS1myc-encoding vector and either with control vector or the NUAK1flag- or NUAK2flag- or AMPKα2myc-encoding vectors. At 1 day after transfection, protein or RNA extracts were prepared. Protein expression was analysed using immunoblotting with the indicated antibodies. RNA expression was analysed using RT–PCR. (D) HEK 293T cells were transfected with flag-tagged LATS1, NUAK1, and NUAK2. They were affinity purified on M2-flag resin and eluted with flag-peptide. Eluted LATS1 protein was phosphorylated alone or with NUAK1 or with NUAK2 and separated on SDS–PAGE. LATS1 phosphorylation by NUAK1 and NUAK2 (Phosphoimage, top) and the protein levels (Coomassie stain) are shown. (E) HEK 293 cells were transfected with vectors encoding either NUAK1flag- or NUAK2flag-encoding vectors and immunoprecipitated with a flag resin. Recombinant AMPK was prepared and activated as described in the Materials and methods section. The ability of these three kinases to phosphorylate the LATS1 peptide, LATS1 S464A mutant peptide, or SAMS peptide was measured. (F) HEK 293 cells were transfected with the LATS1myc- or LATS1S464Amyc-encoding vector or with or without NUAK1 flag encoding vector. At 2 days after transfection, cell extracts were prepared and the protein levels were monitored using immunoblotting with the indicated antibodies. (G) A total of 5 million WI-38 cells were used in each condition to prepare cell extracts (e.p. for early passage and l.p. for late passage). Immunoprecipitations were performed using the anti-phosphoLATS1 antibody. Half of each IP was used for the immunoblot using a LATS1 antibody. A full-colour version of this figure is available at The EMBO Journal Online.
We next transiently co-expressed NUAK1 and LAST1 in HEK 293 cells to analyse the effect of NUAK1 on LATS1 protein levels. Interestingly, LATS1 protein levels decreased when co-expressed with NUAK1, whereas LATS1 mRNA levels remained unchanged (Figure 7C). NUAK2, which belongs to the same sub-family of NUAK1 in the ARK family, was also found to regulate the LATS1 protein levels, whereas AMPKα2, which does not belong to the same sub-family, lacked this ability (Figure 7C). Interestingly, expression of the NUAK1 kinase-dead mutant reverted the effect of NUAK1 WT expression on LATS1 protein levels (Supplementary Figure S6). These results also confirmed that NUAK1 kinase-dead mutant behaved as an NUAK1 dominant-negative form (Figure 4C and D).
As NUAK1 interacts and regulates LATS1 levels, we next wanted to know whether purified NUAK1 protein would be able to phosphorylate full-length LATS1 protein. Interestingly, NUAK1 and its closest member NUAK2 were both able to phosphorylate LATS1 (Figure 7D). We then synthesised a LATS1 peptide containing a consensus ARK motif around the S464 (predicted to be phosphorylated in the Swiss Prot database) and found that NUAK1 as well as NUAK2, but not AMPK, were able to phosphorylate it (Figure 7E). Mutation of S464 to A in the LATS1 peptide completely abolished phosphorylation by NUAK1 and NUAK2 (Figure 7E), confirming that NUAK1 can specifically phosphorylate S464.
Importantly, we also found that NUAK1 expression had no effect on the protein levels of S464 to A mutant form of LATS1 protein (Figure 7F). To further confirm this phosphorylation and its significance during the senescence regulated by NUAK1, we generated a phospho-specific antibody directed against the phosphor S464 of LATS1. As expected, this antibody was unable to recognise the S464A LATS1 mutant (Supplementary Figure S7). Interestingly, we observed an increase in the endogenous LATS1 S464 phosphorylation during replicative or NUAK1-induced senescence and a decrease in pRS/NUAK1-infected cells during escape of replicative senescence (Figure 7G). Thus, NUAK1 is able to regulate the LATS1 protein levels directly through phosphorylation at S464.
Discussion
In a search for new genetic events that are involved in senescence of normal human cells, we have isolated NUAK1 as a modulator of senescence. A total of 13 proteins belong to the ARK (AMP-activated protein kinase-related kinase) family. They can be classified into five subfamilies: AMP-activated protein kinase (AMPK), salt-induced kinase (SIK), microtubule-affinity-regulating kinase (MARK), brain-specific kinase (BRSK), and SNF1-like kinase 1 (NUAK). These proteins show various biological activities, from controlling cell polarity (MARK proteins) to sensing metabolic changes (AMPK proteins) (Drewes et al, 1997; Kahn et al, 2005). Some of them are also involved in basic processes, such as controlling cell death and proliferation (Blazquez et al, 2001; Meisse et al, 2002; Inoki et al, 2003; Kimura et al, 2003; Li et al, 2003; Jones et al, 2005). Apart from the evidence suggesting that NUAK1 may participate in inducing invasion and metastasis (Suzuki et al, 2003a; Kusakai et al, 2004a, 2004b), little is known about its biological function. In this study we show that NUAK1 downregulation increases the replicative potential of HDFs, whereas NUAK1 constitutive expression decreases their replicative potential by inducing premature senescence.
Importantly, AMPKα2 modulates the replicative potential of MEFs through the p53 pathway (Jones et al, 2005). Our results suggest that NUAK1 regulates senescence by a mechanism not involving p53, as neither the overexpression nor the knockdown of NUAK1 affected p53 activity in WI-38 cells (Supplementary Figure S3). WI-38 cells are immortalised by expressing the hTert enzyme (Augert et al, 2009) or show replicative senescence delay by expressing E6 or E7 oncoproteins (Supplementary Figure S3D). Interestingly, NUAK1 expression blocks growth of E6-expressing cells as well as of hTert-immortalised cells, showing that NUAK1 is not acting through the hTert/telomere length/p53 pathway (Supplementary Figure S3C). Altogether, these results suggest that NUAK1 is acting independently of the hTert pathway, or downstream of telomere shortening signalling through an alternate p53-independent pathway. AMPK and NUAK1 have different target specificities, as AMPK is unable to downregulate the level of LATS1 or to phosphorylate S464 of the LATS1 peptide. In contrast, NUAK2, which belongs to the same sub-family as NUAK1, phosphorylates full-length LATS1 protein or LATS1 peptide and decreases LATS1 levels. This similarity between NUAK1 and NUAK2 was further confirmed as NUAK2 was able to induce premature senescence and downregulation of LATS1 in WI-38 cells (Supplementary Figure S8). Nevertheless, the mechanism of NUAK2 regulation seems to be different from that evoked by NUAK1. This was supported by the observation that unlike NUAK1 mRNA levels that increased during replicative senescence, there was no significant change in NUAK2 mRNA levels during replicative senescence (Supplementary Figure S9). In addition, when we induced senescence through RASV12 expression, we only observed slight variations in NUAK1 and NUAK2 mRNA levels (Supplementary Figure S9). Taking all these observations together, we conclude that mainly NUAK1 is involved in the regulation of the replicative senescence.
A decrease in LATS1 levels is known to induce premature senescence and genomic instability through blocking cytokinesis (Takahashi et al, 2006). Our FACS analyses and metaphase spread experiments show that NUAK1 expression induces gross aneuploidies, causing a strong increase in the DNA content per cell. This phenotype correlates with decreased LATS1. Furthermore, premature senescence and aberrant DNA content can be induced by the expression of a dominant-negative form of LATS1. Interestingly, in MCF10a immortal cells, NUAK1 was also found to induce a strong aneuploidy (Supplementary Figure S10A). Nevertheless, in these cells the aneuploidy provoked cell death instead of senescence (Supplementary Figure S10B and C), resulting in a decreased ability to form colonies (Supplementary Figure S10D). Altogether, these results suggest that depending of the cell type the aneuploidy induced by NUAK1 might have different consequences: senescence or cell death.
Hara and collaborators (Takahashi et al, 2006) strongly suggest a function for LATS1 in regulating senescence. These researchers propose that there is no return to proliferation in human senescent cells (in contrast with mouse cells) because of an irreversible cytokinesis blockage and aneuploidy (Takahashi et al, 2006). Interestingly, the knockdown of NUAK1 after 4 days of its constitutive expression does not allow the cells to re-proliferate, suggesting that NUAK1 also induces an irreversible cell-cycle arrest (Supplementary Figure S11). Hara and collaborators (Takahashi et al, 2006) have implicated LATS1 downregulation in irreversible cell-cycle arrest in normal human cells. In their elegant system, they looked at the role of aberrant DNA content accumulation after senescence induction. Our results also suggest a function for LATS1 and aberrant DNA content in the regulation of senescence, but in our system LATS1 and aberrant DNA content accumulation could also contribute to the initiation of senescence. Indeed, NUAK1 downregulation can preserve cells from gross aneupoidies and extend their replicative lifespan, whereas NUAK1 upregulation induces gross aneuploidies and senescence. According to our data, LATS1 participates in both initiation and enforcement of senescence but the molecular mechanism involved in the downregulation of LATS1 may be different. Indeed, we did not detect any effect of the proteasome inhibitor MG-132 over LATS1 downregulation by NUAK1 (Supplementary Figure S12) and we also found that the mutation of S464 to A effects LATS1 levels in contrast with the findings of Takahashi et al (Takahashi et al, 2006). It is thus possible that there are two different mechanisms acting through different upstream kinases, resulting in LATS1 downregulation, aneuploidy, and senescence. Nevertheless, the experimental systems that are used are different and hence make it difficult to derive a definitive conclusion by comparing these results.
In conclusion, our paper joins emerging evidence in suggesting that aberrant DNA content might be, similar to aberrant DNA replication, DNA damage, or oxidative stress, one of the triggers contributing to elicit senescence, and that NUAK1 modulation of LATS1 levels is critical for the process.
Materials and methods
Cell culture
WI-38 cells were grown in DMEM (Invitrogen). HEK 293, 293T (ATCC), and the packaging cell line 293 GP (Clontech) were grown in RPMI (Invitrogen). Both media were supplemented with 10% FCS (Hyclone) and gentamycin (Invitrogen).
Vectors
Pools of shRNAs cloned in the pRetroSUPER vector were used for screening (Berns et al, 2004). The shRNA sequences inserted into the pRetroSUPER vector to generate the pRS/NUAK1 and pRS/NUAK1.1 constructs are, respectively, 5′-CATCCTCTGATTCTAGGTG-3′ and 5′-GAAGTTATGCTTTATTCAC-3′. NUAK1flag cDNA was excised from pcDNA3.1/NUAK1flag (Suzuki et al, 2003b) and inserted into the pLPC retroviral vector (pLPC/NUAK1flag) or in LNCX2 retroviral vector (LNCX2/NUAK1flag). NUAK2 cDNA was TOPO cloned essentially with a 5′ flag tag as described (Lizcano et al, 2004). pBabe-flag-LKB1 DN was described elsewhere (Shaw et al, 2004). The shRNA sequence inserted into the pRS to generate the pRS/LKB1 is 5′-CGGGACTGACGTGTAGAACAA-3′.
The single mutants NUAK1 K84A (kinase dead) were prepared by using pLPC/NUAK1flag as template with the Mutagenesis kit (Stratagene) as instructed by the manufacturer. The primers used K84A forward 5′-GGTTGCTATAAGATCCATTCGTAAGGACAAGCTTAAGGATGAACAAG-3′, K84A reverse 5′-CTTGTTCATCCTAAGCTTTGTCCTTACGAATGGATCTTATAGCAACC-3′, T211A forward 5′-TAAGTTCTTACAAGCGTTTTGTGGGAGTC-3′, T211A reverse 5′-GACTCCCACAAAACGCTTGTAAGAACTTA-3′, S600A forward 5′-CCAGCGCATCCGCGCCTGCGTCTCTGCAG-3′ and S600A reverse 5′-CTGCAGAGACGCAGGCGCGGATGCGCTGG-3′. The vectors encoding LATS1 flag, LATS1 DN flag, and LATS1myc have been described elsewhere (Hirota et al, 2000; Yang et al, 2004; Takahashi et al, 2006).
Infection and genetic screening
Retrovirus was produced by transfection of 293 GP packaging cells (Clontech) according to the manufacturer's recommendations. For screening, WI-38 cells close to senescence were infected with the pRS control vector or with a library pool. Cells were initially selected with 500 ng/ml puromycin and later cultured in the presence of 200 ng/ml puromycin. WI-38 cells were split every week for 3 weeks until clones appeared. Genomic DNA was then prepared and nested PCRs were performed to identify the positive shRNA. The primer pairs used were: PCR1 reverse 5′-GAGACGTGCTACTTCCATTTGTC-3′ and forward 5′-CCCTTGAACCTCCTCGTTCGACC-3′, PCR2 reverse 5′-TGTGAGGGACAGGGGAG-3′ and forward 5′-ACCTCCTCGTTCGACCC-3′. A total of 20 PCR cycles were performed with the PCR1 primer pair. Afterwards, 10% of the PCR product was subjected to 20 additional PCR cycles with the PCR2 primer pair. The resulting PCR products were cloned with the TOPO TA cloning kit (Invitrogen) and finally sequenced.
Growth curves, colony formation assays, senescence-associated β-galactosidase staining, and senescent-associated heterochromatin formation
After infection and puromycin selection, 90 000 cells were seeded per well in a six-well plate. Every 5–8 days, the cells were split and 90 000 cells were seeded per well into six-well plates. The number of population doublings was calculated at each passage. For colony formation assays, 60 000 cells were seeded and left to grow for 2 weeks. The cells were then fixed with 4% paraformaldehyde and stained with a crystal violet solution. SA-β-Gal and SHAF analyses were performed as described Dimri et al (1995) and Narita et al (2003), at 2 or 3 days after seeding 90 000 cells per well in six-well plates.
Cell-cycle analysis
For cell-cycle analysis, the cells were fixed in ice-cold 70% ethanol, washed in PBS, and treated with 10 μg/ml RNaseA for 30 min at 37 °C. Propidium iodide (Sigma) was added to the samples (final concentration: 10 μg/ml) before the analysis of at least 5 × 103 cells with an Epics Elite Cytometer (Coulter).
Quantitative RT–PCR
RNA was extracted from cells with the help of the RNeasy kit from Invitrogen. cDNAs were made from RNA polyA with Superscript II according to the manufacturer's recommendations (Invitrogen). Q-PCR was performed with the following primers: NUAK1 forward 5′-GACATGGTTCACATCAGACGA-3′, NUAK1 reverse 5′-CAATAGTGCACAGCAGAGACG-3′, Control RPS14 forward 5′-GACCAAGACCCCTGGACCT-3′ and Control RPS14 reverse 5′-GAGTGCTGTCAGAGGGGATG-3′.
Immunoblotting
Immunoblot analyses were performed as described in Bernard et al (2003). Membranes were incubated with the antibodies directed against the following antigens: flag tag (F3165, Sigma), cyclin A (H-432, Santa Cruz Biotechnology), NUAK1 (Abgent), LATS1 (A300–477A, Bethyl, or G-16, Santa Cruz Biotechnology), LKB1 (sc-32245, Santa Cruz Biotechnology), and actin (A5316, Sigma). Antibody against the phospho S464 of LATS1 was prepared by injecting a phospho S464 peptide (H2N—IPV RSN S464 FN NPL G—CONH2). Rabbit phospho-specific antibody was purified by its ability to bind the phospho peptide but not the non phosphorylated peptide (Euromedex). The nitrocellulose membranes were then incubated with the corresponding secondary antibodies (Amersham) and the signal revealed using the ECL kit (PerkinElmer Life Sciences).
Phosphorylation assay
HEK 293T cells were transfected with flag-tagged NUAK1, NUAK2, or LATS1 DNA by means of either calcium phosphate or jetPEI. After 36–48 h, the cells were washed three times with ice-cold PBS and collected by scraping into 0.7 ml lysis buffer containing 25 mM HEPES pH 7.5, 1% Triton X-100, protease inhibitors (Roche; 1 tablet/50 ml), phosphatase inhibitors (50 mM sodium fluoride and 5 mM sodium pyrophosphate), 100 mM NaCl, and 1 mM DTT and kept on ice. Harvested cells were dispersed by four passages through a 21-G needle and were kept on ice for approximately 20 min. The lysate was clarified by centrifuging at 14 000 r.p.m. for 20 min and the supernatant was collected. The clarified lysate was incubated with M2-flag resin (50 μl/500 μl lysate) overnight at 4 °C. The protein-bound resin was washed twice with lysis buffer containing 150 mM NaCl and once with lysis buffer. Flag-resin-bound NUAK1 was eluted with 100 μl elution buffer (25 mM HEPES pH 7.5, 1% Triton X-100, protease and phosphatase inhibitors, 10% glycerol, and 300 ng/μl flag peptide) by shaking at 4 °C for 3–6 h. In vitro kinase assays with the NUAK1 and AMPK kinases (purified and activated with CamKKbeta as described by Sanders et al, 2007) were carried out with AMARA (AMARAASAAALRRR), SAMs (HMRSAMSGLHLVKRR), LATS1 (PNIPVRSNS464FNNPLGPRRR), and LATS1S464A (PNIPVRSNA464FNNPLGPRRR) peptides, with a 25 μl mixture containing 50 mM HEPES, pH 7.5 and 1% Triton X-100, 1 mM DTT, 10 mM MgCl2, 1 mM EDTA, 0.1 mM [32P]-ATP, and 1 mM peptide. The reaction mixtures were incubated at 30 °C for 20 min. Transfer of [32P]-phosphate to the peptide substrate was measured by placing 20 μl of the reaction mixture onto P81 phosphocellulose paper and the paper was washed in 1 mM phosphoric acid three times and subjected to scintillation counting.
NUAK1 and NUAK2 phosphorylation of full-length LATS1: Flag peptide eluted LATS1 protein was incubated at 37 °C in 25 mM HEPES pH 7.5, 1% Triton X-100, protease and phosphatase inhibitors, 10% glycerol and 0.5 μl 32P ATP (6000 Ci/mmol, Perkin-Elmer) either alone or with flag peptide eluted NUAK1 or NUAK2 for 20 min in a 30-μl reaction. Reactions were stopped with SDS–PAGE sample buffer and separated on NUPAGE (Invitrogen) gels in Tris-Glycine buffer. Gels were stained with Simply Blue stain (Invitrogen) and destained with water, dried, and phosphor imaged.
Chromosome spreading
The cells were incubated for 1 h in a Karyomax Colcemid (Invitrogen Corporation), trypsinised, and incubated in a 60 mM KCl hypotonic buffer. The cells were fixed with freshly made methanol/acetic acid solution (3:1 v/v), spread onto frozen slides, and then air-dried. DNA was stained with 1 μg/ml Hoechst 33258 for 3 min. The chromosomes were observed and counted under an epifluorescence microscope (Zeiss, axioplan 2) with a Hoechst filter.
Supplementary Material
Supplementary Figures S1–S12
Supplementary Figure Legends
Review Process File
Acknowledgments
We thank the members of the Laboratory for helpful discussions. We also thank Virginie Glippa and Julie Bertout for technical assistance. We thank H Esumi for the NUAK1 cDNA, E Hara and H Saya for the LATS1-encoding vector. This work was carried out with the support of the ‘Association pour la Recherche sur le Cancer', the ‘Fondation pour la Recherche Médicale Nord Pas de Calais', the ‘Comité du Pas de Calais de la Ligue Nationale contre le Cancer', the RTRS Fondation Synergie Lyon Cancer, and the Medical Research Council, UK.
Footnotes
The authors declare that they have no conflict of interest.
References
- Augert A, Payre C, de Launoit Y, Gil J, Lambeau G, Bernard D (2009) The M-type receptor PLA2R regulates senescence through the p53 pathway. EMBO Rep 10: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM (2004) BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749 [DOI] [PubMed] [Google Scholar]
- Bernard D, Pourtier-Manzanedo A, Gil J, Beach DH (2003) Myc confers androgen-independent prostate cancer cell growth. J Clin Invest 112: 1724–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437 [DOI] [PubMed] [Google Scholar]
- Blazquez C, Geelen MJ, Velasco G, Guzman M (2001) The AMP-activated protein kinase prevents ceramide synthesis de novo and apoptosis in astrocytes. FEBS Lett 489: 149–153 [DOI] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA (2005) Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, van Lohuizen M, Bernards R (2002) TBX-3, the gene mutated in Ulnar-Mammary syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem 277: 6567–6572 [DOI] [PubMed] [Google Scholar]
- Campisi J (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM, Beach DH (2000) p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol 2: 148–155 [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Paolo Pandolfi P (2005) Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M (2005) Tumour biology: senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- Dimri GP (2005) What has senescence got to do with cancer? Cancer Cell 7: 505–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E (1997) MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89: 297–308 [DOI] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D (2004) Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM (2004) Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 14: 501–513 [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H (2000) Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol 149: 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert N, Martien S, Augert A, Da Costa M, Mauen S, Abbadie C, de Launoit Y, Gil J, Bernard D (2009) A genetic screen identifies topoisomerase 1 as a regulator of senescence. Cancer Res 69: 4101–4106 [DOI] [PubMed] [Google Scholar]
- Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, Saya H (2004) Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene 23: 5266–5274 [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG (2005) AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25 [DOI] [PubMed] [Google Scholar]
- Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino K, Hara K, Kemp BE, Witters LA, Mimura O, Yonezawa K (2003) A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells 8: 65–79 [DOI] [PubMed] [Google Scholar]
- Kusakai G, Suzuki A, Ogura T, Kaminishi M, Esumi H (2004a) Strong association of ARK5 with tumor invasion and metastasis. J Exp Clin Cancer Res 23: 263–268 [PubMed] [Google Scholar]
- Kusakai G, Suzuki A, Ogura T, Miyamoto S, Ochiai A, Kaminishi M, Esumi H (2004b) ARK5 expression in colorectal cancer and its implications for tumor progression. Am J Pathol 164: 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang P, Robinson M, Lawrence TS, Sun Y (2003) AMPK-beta1 subunit is a p53-independent stress responsive protein that inhibits tumor cell growth upon forced expression. Carcinogenesis 24: 827–834 [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 23: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G (2004) Intrinsic tumour suppression. Nature 432: 307–315 [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Hahn WC, Gupta P, Weinberg RA (2000) Genes involved in senescence and immortalization. Curr Opin Cell Biol 12: 705–709 [DOI] [PubMed] [Google Scholar]
- Meisse D, Van de Casteele M, Beauloye C, Hainault I, Kefas BA, Rider MH, Foufelle F, Hue L (2002) Sustained activation of AMP-activated protein kinase induces c-Jun N-terminal kinase activation and apoptosis in liver cells. FEBS Lett 526: 38–42 [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D (2007) Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J 403: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC (2004) The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 101: 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ (1996) Cancer cell cycles. Science 274: 1672–1677 [DOI] [PubMed] [Google Scholar]
- Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM, Lee HS, Yoo SH, Chung DH, Sung YC, McKeon F, Lee CW (2003) Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 4: 483–497 [DOI] [PubMed] [Google Scholar]
- Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley GQ, Spits H, Bernards R (2002) A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19ARF-p53 signaling. Genes Dev 16: 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov RV, Haupt Y (1999) The cellular response to p53: the decision between life and death. Oncogene 18: 6145–6157 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Esumi H (2003a) ARK5 suppresses the cell death induced by nutrient starvation and death receptors via inhibition of caspase 8 activation, but not by chemotherapeutic agents or UV irradiation. Oncogene 22: 6177–6182 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kusakai G, Kishimoto A, Lu J, Ogura T, Lavin MF, Esumi H (2003b) Identification of a novel protein kinase mediating Akt survival signaling to the ATM protein. J Biol Chem 278: 48–53 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Ogura T, Esumi H (2006) NDR2 acts as the upstream kinase of ARK5 during insulin-like growth factor-1 signaling. J Biol Chem 281: 13915–13921 [DOI] [PubMed] [Google Scholar]
- Takahashi A, Ohtani N, Yamakoshi K, Iida S, Tahara H, Nakayama K, Nakayama KI, Ide T, Saya H, Hara E (2006) Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat Cell Biol 8: 1291–1297 [DOI] [PubMed] [Google Scholar]
- Yamasaki L (2003) Role of the RB tumor suppressor in cancer. Cancer Treat Res 115: 209–239 [DOI] [PubMed] [Google Scholar]
- Yang X, Li DM, Chen W, Xu T (2001) Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene 20: 6516–6523 [DOI] [PubMed] [Google Scholar]
- Yang X, Yu K, Hao Y, Li DM, Stewart R, Insogna KL, Xu T (2004) LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat Cell Biol 6: 609–617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S12
Supplementary Figure Legends
Review Process File