Abstract
GATA transcription factors interact with FOG proteins to regulate tissue development by activating and repressing transcription. FOG-1 (ZFPM1), a co-factor for the haematopoietic factor GATA-1, binds to the NuRD co-repressor complex through a conserved N-terminal motif. Surprisingly, we detected NuRD components at both repressed and active GATA-1/FOG-1 target genes in vivo. In addition, while NuRD is required for transcriptional repression in certain contexts, we show a direct requirement of NuRD also for FOG-1-dependent transcriptional activation. Mice in which the FOG-1/NuRD interaction is disrupted display defects similar to germline mutations in the Gata1 and Fog1 genes, including anaemia and macrothrombocytopaenia. Gene expression analysis in primary mutant erythroid cells and megakaryocytes (MKs) revealed an essential function for NuRD during both the repression and activation of select GATA-1/FOG-1 target genes. These results show that NuRD is a critical co-factor for FOG-1 and underscore the versatile use of NuRD by lineage-specific transcription factors to activate and repress gene transcription in the appropriate cellular and genetic context.
Keywords: FOG-1, chromatin, haematopoiesis, NuRD, transcription
Introduction
Lineage-specific gene expression patterns are established by the combinatorial action of transcription factors and their co-factors. Maturation of a given cell lineage requires the activation of gene expression and the simultaneous repression of genes that mark earlier stages of development or alternative cell fates. GATA-1 is a haematopoietic zinc-finger transcription factor that activates all known erythroid and megakaryocyte (MK)-specific genes (Crispino, 2005; Ferreira et al, 2005). In addition to activating gene expression, GATA-1 can function as transcriptional repressor (Welch et al, 2004). Genes directly repressed by GATA-1 include Gata2 (Grass et al, 2003), Kit (Munugalavadla et al, 2005; Jing et al, 2008) and Myc (Rylski et al, 2003).
The regulation of most, but not all, GATA-1-dependent genes requires binding to FOG-1 (ZFPM1), a multi-type zinc-finger protein whose expression in the haematopoietic system is restricted largely to erythroid cells and MKs (Tsang et al, 1997). FOG-1 contains nine zinc fingers, four of which are capable of binding GATA-1. The GATA-1/FOG-1 interaction is direct and requires the N-terminal zinc finger of GATA-1, and at least one of the four GATA-1-binding zinc fingers of FOG-1 (Tsang et al, 1997; Fox et al, 1999). Point mutations in GATA-1 that disrupt FOG-1 binding impair erythroid and MK development in engineered mice and human patients, establishing the importance of this interaction in vivo (Crispino et al, 1999; Nichols et al, 2000).
Chromatin immunoprecipitation (ChIP) experiments revealed that FOG-1 colocalizes with GATA-1 at genes in which GATA-1 functions as an activator and repressor (Wang et al, 2002; Letting et al, 2004; Pal et al, 2004; Jing et al, 2008). How FOG-1 cooperates with GATA-1 during activation and repression of transcription is unclear. Biochemical approaches to address this question revealed interactions of FOG-1 with several protein complexes that modulate GATA-1/FOG-1 activity, including TACC3, CtBP2 and NuRD (Fox et al, 1999; Garriga-Canut and Orkin, 2004; Hong et al, 2005; Rodriguez et al, 2005). TACC3 is capable of competing with GATA-1 for FOG-1 binding and might sequester FOG-1 in the cytoplasm thus inhibiting erythroid maturation (Garriga-Canut and Orkin, 2004). CtBP2 might link FOG-1 to other co-repressor molecules, but it remains uncertain to what extent this interaction contributes to FOG-1 function in vivo (Katz et al, 2002). Affinity purification of proteins associated with the N-terminal repression domain of FOG-1 identified a high-affinity interaction with NuRD (Hong et al, 2005). The composition of the NuRD complex can vary (Bowen et al, 2004), but in the context of FOG-1 contains the ATPase Mi-2β, MTA-1, MTA-2, p66, RbAp46 (RBBP7), RbAp48 (RBBP4), MBD3 and the histone deacetylases HDAC1 and HDAC2 (Hong et al, 2005). NuRD bears features of a classical co-repressor complex because of the presence of histone deacetylases and its association with diverse transcriptional repressors (Bowen et al, 2004; Denslow and Wade, 2007; Manavathi and Kumar, 2007).
The interaction between FOG-1 and NuRD is mediated by a small, conserved motif contained within the N-terminal 12 amino acids of FOG-1 that is also present in FOG-2 (Hong et al, 2005; Roche et al, 2008). Other molecules known to repress transcription, including Bcl11A, Bcl11B, SALL1-4, EBFAZ and Evi3, bear an identical motif at their N-termini (for reference see Hong et al, 2005). For FOG and SALL molecules, this motif has been shown to be necessary and sufficient for NuRD binding, and point mutations that disrupt this interaction impair transcriptional repression in transfection-based assays (Lin et al, 2004; Hong et al, 2005; Lauberth and Rauchman, 2006; Roche et al, 2008). Truncated forms of SALL1 found in patients with Townes–Brock syndrome, a dominantly inherited developmental disorder characterized by multiple birth defects, maintain the minimal repression motif suggesting that the disease is caused by interference with the normal SALL1/NuRD interaction (Kiefer et al, 2003). Moreover, the zinc-finger transcriptional co-repressor ZFHX1B bears a similar motif near its N-terminus that also mediates NuRD binding (Verstappen et al, 2008). Mutations or deletions affecting this region are associated with rare forms of Mowat–Wilson syndrome, a disorder associated with mental retardation (Zweier et al, 2006; Verstappen et al, 2008). These examples illustrate the importance of NuRD recruitment by select transcription factors to ensure normal development. So far, no germ line mutations within the NuRD-binding motif of FOG-1 or FOG-2 have been described in human patients.
The association of FOG-1 with NuRD immediately suggested a mechanism by which GATA-1/FOG-1 repress transcription. Indeed, NuRD is detectable in vivo at genes that are repressed by GATA-1 in a FOG-1-dependent manner (Hong et al, 2005; Rodriguez et al, 2005). However, further exploration suggested broader functions for NuRD during haematopoietic gene transcription. Here, we report that NuRD is present not only at repressed, but also active FOG-1-regulated genes in erythroid cells and MKs. Surprisingly, transfection assays using GATA-1/FOG-1-dependent reporter genes showed that NuRD is directly required for transcriptional activation. To assess the in vivo function of the GATA-1/FOG-1/NuRD axis during haematopoietic development, we generated mice with a triple point mutation in FOG-1 that abrogates NuRD binding.
Homozygous mutant animals revealed defects in the erythroid and MK lineages, consistent with a function for NuRD in most of FOG-1 biology. Alterations in gene expression in mutant animals included the failure to activate and repress select GATA-1/FOG-1-regulated genes. The dual function of NuRD during transcriptional activation and repression suggests that the classification of NuRD as co-repressor might not do justice to its versatile function in gene expression.
Results and discussion
NuRD broadly occupies active and repressed GATA-1/FOG-1 target genes
Earlier ChIP experiments that were performed in cells expressing a conditional form of GATA-1 suggested that the Mi-2β subunit of NuRD is recruited to select sites at the Gata2 and Kit genes upon their repression by GATA-1, consistent with NuRD serving as a GATA-1/FOG-1 co-repressor (Hong et al, 2005; Rodriguez et al, 2005). Here, we sought to extend these findings by (1) examining a total of three subunits of NuRD that would more reliably reflect the presence of the NuRD complex, (2) analysing multiple regulatory regions of the Gata2 and Kit genes through which GATA-1 represses their expression, (3) studying additional genes that are repressed by GATA-1 and (4) investigating genes that are directly activated by GATA-1.
ChIP experiments were carried out in the erythroid cell lines G1E and G1E-ER4. G1E-ER4 cells were derived from the GATA-1-deficient cell line G1E that lacks an intact GATA-1 gene and is developmentally arrested at the proerythroblast stage (Weiss et al, 1997). G1E-ER4 cells contain a stably integrated form of GATA-1 fused to the ligand-binding domain of the oestrogen receptor (GATA-1-ER). Treatment with oestradiol leads to activation of GATA-1 followed by erythroid maturation along with the activation and repression of GATA-1 target genes (Weiss et al, 1997). This system faithfully recapitulates erythroid maturation and has been invaluable for numerous studies of erythroid biology and transcriptional regulation.
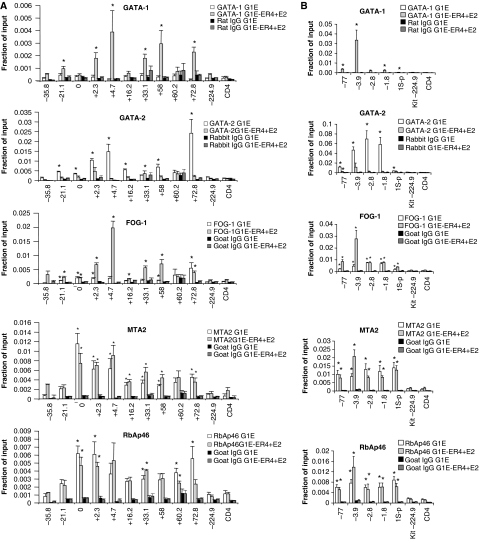
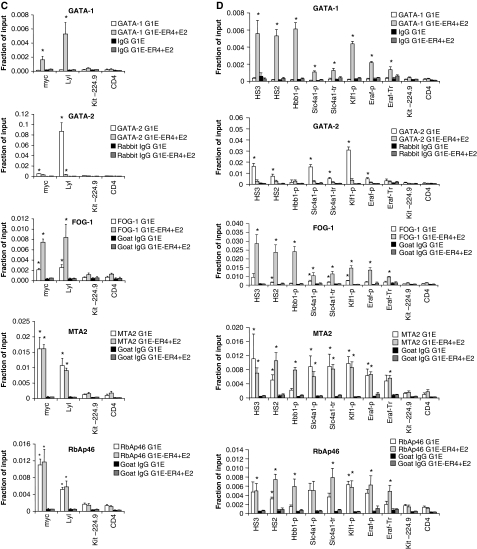
We began by examining FOG-1 and NuRD occupancy across ∼100 kilobases (kb) of the Kit gene that is expressed in immature erythroid cells driven in part by transcription factor GATA-2. During terminal erythroid maturation, GATA-1 activation leads to loss of GATA-2 binding and repression of Kit in a FOG-1-dependent manner (Jing et al, 2008). We and others earlier identified sites of high GATA-1 occupancy at multiple positions including −114.5, +4.7, +58.1 and +72.8 kb with respect to the transcription start site (TSS) (Munugalavadla et al, 2005; Jing et al, 2008). Thus, the study of the Kit locus under dynamic conditions is well suited to examine possible spatial and temporal correlations between FOG-1 and NuRD occupancy. In parental G1E cells lacking GATA-1, FOG-1 was detected at +72.8 kb and at low levels at additional sites of the locus (Figure 1A). GATA-1 and FOG-1 were near background levels at control regions −224.9 kb and the silent CD4 gene. Low levels of FOG-1 might reflect the presence of GATA-2 that also binds FOG-1 (Figure 1A) (Jing et al, 2008). In G1E-ER4 cells treated with oestradiol, FOG-1 levels remained high at +72.8 kb and increased at sites of high GATA-1 occupancy (Figure 1A). When we examined the distribution along the Kit locus of two NuRD components, MTA-2 and RbAp46, three findings were especially noteworthy. First, the levels of MTA-2 and RbAp46 closely correlated with each other and tended to be high at sites with high FOG-1 occupancy (Figure 1A). Second, both proteins showed occupancy significantly above background throughout the Kit locus including sites with little or no GATA-1/FOG-1 binding (Figure 1A). This might result from spreading along the chromatin fibre and is consistent with the ability of NuRD to associate with chromatin in a non-targeted manner (Li et al, 2002). Third, surprisingly, both MTA-2 and RbAp46 were detected at the active Kit locus before its repression by GATA-1, but not at control regions (Figure 1A). To examine whether NuRD occupies other genes repressed by GATA-1, we chose the Gata2 gene that contains several known GATA-1-binding sites (−77, −3.9, −2.8 and −1.8 kb with respect to the TSS) (Grass et al, 2003, 2006; Martowicz et al, 2005), and the Myc and Lyl1 genes (Rylski et al, 2003; Johnson et al, 2007). Notably, at all three genes, MTA-2 and RbAp46 occupancy was high before repression and changed very little on GATA-1-induced silencing (Figure 1B and 1C), mirroring our observations at the Kit locus.
Figure 1A-B.
FOG-1 and NuRD proteins occupy active and repressed GATA-1 target genes. ChIP at the repressed GATA-1 target genes Kit (A), Gata2 (B), Myc and Lyl1
Figure 1C-D.
(C) using antibodies anti-GATA-1, GATA-2, FOG-1, MTA2, RbAp46 and the corresponding control IgG, in G1E and G1E-ER4 cells after treatment with oestradiol for 24 h. (D) FOG-1 and NuRD ChIP at sites in which GATA-1 functions as an activator. HS2, DNase hypersensitive sites 2. The regions at 224.9 kb upstream of the Kit promoter and in the first intron of CD4 serve as negative control. The results are averages of 4–10 independent experiments. Error bars represent s.e.m. Statistical significance was determined by comparing the occupancy at each site and negative control regions; *P<0.05.
These results indicated that the presence per se of NuRD is not prohibitory to active transcription, and prompted us to explore whether NuRD also occupies genes that are directly activated by GATA-1. Indeed, at three sites of the β-globin locus in which GATA-1 occupancy is high, including DNase 1 hypersensitive sites (HS) 2 and 3 of the locus control region and the Hbb-b1 promoter (Horak et al, 2002; Johnson et al, 2002; Letting et al, 2003; Im et al, 2005), significant levels of MTA-2 and RbAp46 were observed before activation of GATA-1 when compared with the controls at Kit-224.9 and CD4 (Figure 1D). Surprisingly, NuRD levels remained high or even increased on activation of Hbb-b1 transcription by GATA-1 (Figure 1D). High levels of NuRD at active genes seems to be a general phenomenon, as similar results were found at the GATA-1-activated target genes Klf1, Band3 (Slc4a1) and Eraf (Figure 1D).
We next examined the occupancy of Mi-2β, one of the defining subunits of NuRD. Using two independently derived antibodies, A301-081A and A301-082A, Mi-2β was found at both GATA-1-activated and repressed genes in a manner very similar to that of RbAp46 and MTA-2 (Supplementary Figure S1). However, we noticed a discrepancy between these results and those earlier obtained with different Mi-2β antibodies that had detected strongly inducible increases in Mi-2β occupancy at positions at the Gata2 and Kit genes (Hong et al, 2005; Rodriguez et al, 2005). To resolve this issue, we introduced HA-tagged Mi-2β into G1E-ER4 cells through retrovirus. ChIP analysis with HA antibodies showed that the spatial distribution of HA–Mi-2β across the Kit locus closely resembled that of RbAp46 and MTA-2 (Supplementary Figure S2A). We also observed a modest, but reproducible, two-fold increase of HA–Mi-2β near the +4.7 kb region of Kit and −2.8 kb of Gata2 in addition to similar increases at several other sites (Supplementary Figure S2A, B and C). In addition, at HS2, HS3, Hbb-b1, Klf1, Eraf and Band3 (Slc4a1), HA–Mi-2β was present in the absence of GATA-1 and slightly increased further following GATA-1 activation (Supplementary Figure S2D).
Two observations deserve comments: first, HA–Mi-2β displayed a more general trend towards increased occupancy on GATA-1 activation than RbAp46 and MTA-2. This might be a result of over-expression or of variation in the populations of the cells analysed. Second, at some sites, such as the Hbb-b1 promoter, changes in FOG-1 occupancy were more pronounced than those of NuRD proteins. This might be due to changes associated with transcriptional activation that either influence NuRD recruitment and/or epitope recognition, or to posttranslational modifications that regulate the FOG-1/NuRD interaction similar to what has been observed with SALL1 (Lauberth et al, 2007). Nevertheless, together, the above results indicate that at least three components of the NuRD complex are present at GATA-1/FOG-1-activated and repressed genes, and that occupancy of NuRD can be further augmented on gene activation. Therefore, a simple recruitment mechanism does not account for the distinction between activated versus repressed GATA-1/FOG-1 target genes.
We next examined whether NuRD occupies GATA-1/FOG-1-activated genes in cultured primary foetal liver-derived MKs. We found that MTA-2 and RbAp46 were enriched at all active genes examined, including αIIb (Itga2b), Mpl, Gp9, Gp1bα and Pf4 when compared with a control region or the silent erythroid GATA-1/FOG-1 target gene Hbb-b1 (Supplementary Figure S3). This indicates that NuRD recruitment to active GATA-1/FOG-1-regulated genes is not limited to erythroid cells, but is a general phenomenon.
NuRD as transcriptional co-activator
The presence of the Mi-2β component of NuRD has been observed at the actively transcribed CD4 locus, and loss of Mi-2β in gene-targeted mice resulted in reduced CD4 expression, suggesting that NuRD functions as an activator or anti-silencer at this gene (Williams et al, 2004; Naito et al, 2007). However, it remained unresolved whether Mi-2β acted in the context of the entire NuRD complex. Moreover, inferences as to a direct function of Mi-2β in transcription activation are complicated by the presence of increased levels of the related protein Mi-2α in mutant mice, and the possibility that changes in gene expression are an indirect consequence of Mi-2β loss (Williams et al, 2004). Similarly, Mi-2α was shown to activate transcription by MYB, but it was suggested that Mi-2α might function in the absence of other NuRD components in this context (Saether et al, 2007).
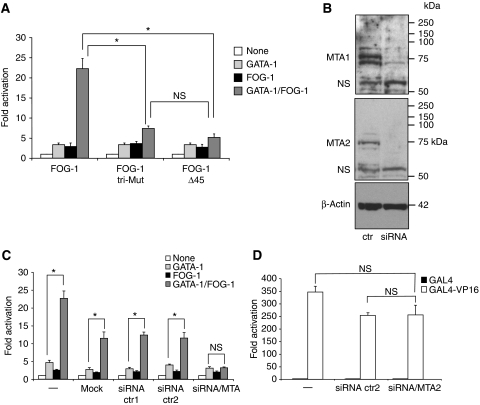
To examine a direct requirement for NuRD during transcriptional activation, we made use of an assay that measures transcriptional activation by FOG-1. Specifically, we and others have shown that FOG-1 augments the transcriptional activity of GATA-1 in reporter assays incorporating the MK-specific promoter of the αIIb gene that bears adjacent GATA and ETS elements (Gaines et al, 2000; Wang et al, 2002; Pang et al, 2006). Consistent with earlier results, FOG-1 increased the activity of GATA-1 when co-expressed with an αIIb-luciferase reporter gene in 3T3 fibroblasts (Figure 2A). Deletion of 45 amino acids comprising the NuRD-binding domain (Δ45) of FOG-1 strongly impaired activation by FOG-1 (Figure 2A). Moreover, a triple point mutation (R3G,R4G,K5A) that abolishes NuRD binding (Supplementary Figure S4) had the same effect as the deletion (Figure 2A). One concern was that the point mutations might compromise binding to unknown molecules that function as co-activators. Although numerous attempts failed to detect any non-NuRD polypeptides that interact with this domain of FOG-1, we measured FOG-1 activity under conditions in which key NuRD proteins were depleted. We chose the MTA proteins, as they bind to the repression domain at the N-terminus of FOG-1 (Hong et al, 2005). Twenty-four hours before transfection with the reporter construct, 3T3 cells were exposed to a mixture of siRNAs directed against MTA-1, MTA-2 and MTA-3 as described earlier (Roche et al, 2008) or with two different control siRNAs. Knockdown of >90% of control was achieved for MTA-1 and MTA-2 as determined by western blotting (Figure 2B). MTA-3 is not detectable by western blotting in 3T3 cells (Roche et al, 2008). Generally, mock transfection or transfection with two distinct control siRNAs lead to a slight reduction in overall reporter activity when compared with untreated cells (Figure 2C). However, loss of MTA proteins specifically and completely abrogated activation by FOG-1 (Figure 2C). Importantly, the FOG-1-independent activation by GATA-1 was not significantly affected. A second set of siRNAs against MTA-1, MTA-2 and MTA-3 produced a less pronounced, but consistent effect (Supplementary Figure S5). To further rule out non-specific effects of MTA knockdowns on high-level transcription, we examined the transcriptional activator GAL4–VP16 in the context of a reporter gene bearing 5 GAL4-binding sites. Knockdown of MTA proteins did not significantly impair GAL4–VP16-mediated activation (Figure 2D), further supporting a specific function for NuRD during FOG-1-dependent transcriptional activation.
Figure 2.
NuRD binding is required for transcriptional activation by FOG-1. Transient transfection of 3T3 cells with constructs expressing GATA-1, FOG-1 and a luciferase reporter gene driven by the αIIb promoter. FOG-1 mutants bearing triple point mutations (tri-mut) or a deletion of the NuRD-binding domain (Δ45) fail to substantially activate GATA-1-dependent transcription (A). Bars denote averages of five independent experiments. Error bars represent s.e.m. *P<0.05; NS: not significant. (B) Western blots of cells co-transfected with siRNAs against MTA-1 and MTA-2. NS, non-specific bands. (C) Transient transfections as in (A), but 24 h before transfection with GATA-1, FOG-1 and the reporter gene, cells were additionally transfected with MTA-1, MTA-2 and MTA-3 siRNAs, no siRNAs (mock), or two different control siRNAs (ctr1 and ctr2). Note that the exposure to an additional round of transfection leads to non-specific reduction in GATA-1/FOG-1 activity. The results are averages of 3–12 independent experiments. Error bars represent s.e.m. *P<0.05; NS: not significant. (D) 3T3 cells were treated with control siRNA (siRNA ctr2) or siRNA against MTA-1 and MTA-2 (siRNA/MTA) as in (C); 24 h later, GAL4–VP16 or GAL4 alone were co-transfected with a reporter gene driven by the thymidine kinase promoter and containing 5 GAL4-binding sites; n=3 (error bars denote s.e.m.; NS: not significant).
Together, these results show a direct requirement of NuRD for transcriptional activation. Moreover, as the N-terminus of FOG-1 binds stoichiometrically and with high affinity to the entire NuRD complex in vitro and in vivo (Hong et al, 2005), this suggests that the complex as a whole and not just a subset of NuRD proteins is associated with active transcription. The consistent presence of NuRD at both GATA-1/FOG-1-activated and repressed genes in vivo together with the requirement for the FOG-1/NuRD interaction for both transcriptional activation (Figure 2) and repression (Hong et al, 2005) in transfection assays suggests that NuRD can function distinctly depending on gene context. Whether this context determines the specific activity of NuRD or affects the recruitment or loss of additional components are questions that need to be addressed in future studies.
Methylation of lysine 4 of histone H3 (H3K4) leads to loss of NuRD binding to the N-terminal tail of histone H3 in vitro (Nishioka et al, 2002; Zegerman et al, 2002) providing a possible mechanism by which NuRD is displaced from active chromatin. However, our ChIP studies indicate that NuRD is not displaced from active genes. It is possible that instead of displacing NuRD, H3K4 methylation interferes with the way NuRD engages chromatin to deacetylate histones. Another possibility is that modification of FOG-1 alters the way it interacts with NuRD at active versus repressed genes. The transcriptional repressor SALL1 binds to NuRD through an N-terminal motif that is identical to that of FOG-1 (Lauberth and Rauchman, 2006). Phosphorylation of serine 2 leads to disruption of this interaction in vitro (Lauberth et al, 2007). Whether FOG-1 is phosphorylated or modified in any other way to regulate NuRD binding remains unknown, but deserves further investigation. Finally, it has become clear that the presence of HDACs per se does not necessarily specify transcriptional repression. In addition to histones, HDACs deacetylate nuclear factors and can influence transcription positively and negatively (Smith, 2008).
Disruption of the FOG-1/NuRD interaction in mice
We showed earlier that the K5A substitution in FOG-1 impaired NuRD binding and transcriptional repression, but did not completely abrogate it (Hong et al, 2005). Therefore, we also altered amino-acids R3 and R4 that contribute to the NuRD interaction. To verify that the triple mutation disrupts NuRD binding, we generated GST fusion proteins containing amino-acids 1–45 of FOG-1 and single (K5A) and triple (R3G, R4G, K5A) amino-acid substitutions. When exposed to in vitro translated MTA-1, MTA-2, RbAp48 or RbAp46, wild-type GST–FOG-1–45 associated efficiently with all of these proteins (Supplementary Figure S4A). The K5A substitution greatly diminished these associations, but residual binding was observed. However, the triple substitution completely abolished association with NuRD components (Supplementary Figure S4A). Correspondingly, when the N-terminus of FOG-1 was fused to the DNA-binding domain of GAL4, the single K5A mutation led to a significant loss of transcriptional repression towards a GAL4-regulated reporter gene in transfected cells. However, the triple mutation essentially abolished all of transcriptional repression (Supplementary Figure S5B). Finally, the triple mutation also abrogated the FOG-1/NuRD interaction in the context of full-length FOG-1 without diminishing FOG-1 protein levels (not shown).
To determine the function of the FOG-1/NuRD axis on GATA-1-regulated haematopoietic lineages in vivo and to evaluate the dual function of NuRD in gene expression under conditions in which FOG-1 is expressed at physiological levels, we engineered mice in which the FOG-1/NuRD interaction is disrupted. We used recombineering (Copeland et al, 2001) to generate a targeting vector to ‘knock-in' (ki) the triple mutation into the Fog-1 gene (Zfpm1; for details see Supplementary Figures S6A and S5B–D). Mice heterozygous for the ki mutations were bred to homozygosity and studied as outlined below. Generally, the triple point mutation did not significantly change FOG-1 protein levels in erythroid cells or MKs (Supplementary Figure S6E).
Defective erythropoiesis in homozygous mutant animals
Among 293 live born offspring from heterozygous crosses, 97 (33.1%) were wild type (+/+), 149 (50.9%) +/ki and only 47 (16%) ki/ki, consistent with embryonic or perinatal mortality of ki/ki mice (Supplementary Table S1). As the frequency of homozygous embryos was close to normal at E14.5, the predominant loss of ki/ki animals likely occurred between E14.5 and birth (Supplementary Table S1). The ki/ki animals that survived past their birth seemed normal with regard to appearance, weight and activity, and were fertile. The exact time points and cause(s) of death need to be investigated further, but might result from a combination of haematopoietic and cardiac defects. Nevertheless, these results show that the FOG-1/NuRD interaction is important for normal murine development.
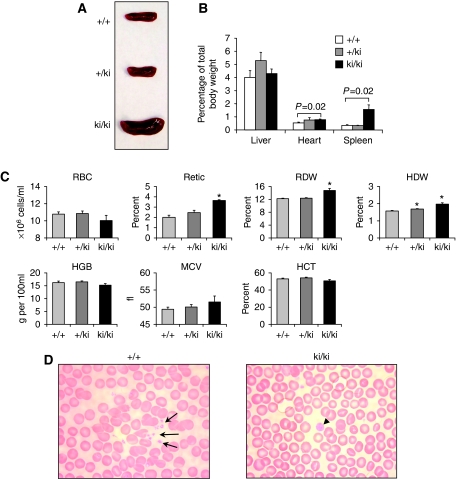
When 6–7-week-old mice were examined, marked splenomegaly was evident in all ki/ki animals with spleens weighing more than three times that of +/+ or +/ki littermates (Figure 3A and B), consistent with extramedullary haematopoiesis. Other organs, including heart and liver, were grossly normal in morphology and weight (Figure 3B). A modest increase in heart weight in ki/ki animals might be secondary to their anaemia (see below). Peripheral blood analysis of ki/ki mice revealed signs of a mild compensated anaemia as reflected in increased reticulocyte counts, red cell distribution width and haemoglobin distribution width, but no microcytosis (normal mean corpuscular volume). Other erythroid indices were within the normal range, including red blood cell count, haemoglobin concentration and haematocrit (Figure 3C). Smears of peripheral blood were generally unremarkable with regard to erythroid cells (Figure 3D). We also observed a two-fold increase in neutrophil granulocytes and monocytes in ki/ki animals, whereas the remaining white cell parameters were normal (Supplementary Table S2). The effects of the FOG-1 mutation on the myeloid lineages will be explored in future studies.
Figure 3.
(A) Spleens from 6-week-old mice with indicated genotype. (B) Organ weights plotted as per cent of total body weight (n=4). (C) Red blood cell indices of wild-type (+/+, n=15), heterozygous (+/ki, n=11) and homozygous mutant (ki/ki, n=10) animals. Error bars represent s.e.m. P-values were determined by two-tailed t-tests. *P<0.001. (D) Giemsa—Wright-stained peripheral blood smears from +/+ and ki/ki mice. Note scarcity of normal sized platelets and the presence of enlarged platelets.
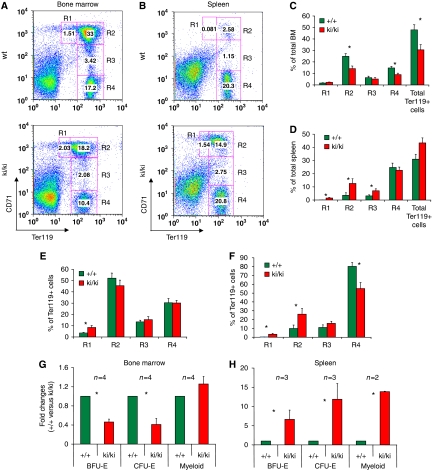
Histological sections of ki/ki bone marrow appeared normal with regard to overall cellularity (Supplementary Figure S6). We next examined the maturation of erythroid cells in the bone marrow by determining the Ter119/CD71 profile (Socolovsky et al, 2001) from 6-week-old mice. The ki/ki mice displayed a significant reduction in the total number of Ter119+ cells indicative of impaired erythropoiesis (Figure 4A). Moreover, the levels of Ter119 per cell were reduced ∼two-fold. However, the maturation from basophilic erythroblasts to orthochromatic erythroblasts was not significantly impaired (Figure 4A, C and E). When cultured under the appropriate conditions, the ki/ki bone marrow contained fewer erythroid progenitors as reflected in an ∼50% reduction in BFU-E and CFU-E colonies, whereas myeloid progenitors were normal in number (Figure 4G).
Figure 4.
Erythroid defects in ki/ki erythroid cells from bone marrows (A, C, E, G) and spleens (B, D, F, H). (A, B) Representative flow cytometric analysis using CD71 and Ter119 antibodies. R1, R2, R3 and R4 represent proerythroblasts, basophilic, polychromatic and othrochromatic erythroblasts, respectively (Socolovsky et al, 2001) and were plotted as per cent of total (C, D) or per cent of Ter119+ cells (E, F). Error bars represent s.e.m. n=7 (C, D) n=9 (D, F). *P<0.05. (G, H) Colony assays measuring BFU-E and CFU-E from bone marrows and spleens. The results were plotted as fold changed comparing +/+ with ki/ki-derived samples. N-values are indicated for each graph. *P<0.05.
The ki/ki spleens displayed a marked increase in the number of haematopoietic cells, including that the erythroid (Ter119+) compartment (Figure 4D) commensurate with the increase in spleen size, and a relative decrease of the white pulp (Supplementary Figure S6). The ki/+ spleens were normal (not shown). The severity of the phenotype varied significantly between ki/ki animals. Among the Ter119+ cells, the proerythroblast (R1), basophilic (R2) and polychromatic (R3) compartments were markedly expanded in ki/ki spleens when compared with wild type (Figure 4D and F). The ki/ki erythroid cells largely maintained their ability to progress normally throughout later stages of maturation, although the progression from basophilic to orthochromatic erythrobasts appeared to be somewhat impaired (Figure 4B and D). The number of BFU-E, CFU-E and myeloid progenitors in ki/ki spleens was dramatically increased consistent with splenic compensation (Figure 4H).
The analysis of ki/ki animals described above was performed between 6 and 8 weeks of age and their phenotypes were consistent. Interestingly, in a subset of animals aged over 1 year, the anaemia and splenomegaly worsened progressively. In these cases, the anaemia was accompanied by peripheral reticulocytosis (Supplementary Figure S8A, B) and a pronounced expansion of the precursor populations (R1+R2) relative to the mature cells in the bone marrow and spleens (Supplementary Figure S8C) and increased cell death (Supplementary Figure S8D) consistent with ineffective erythropoiesis. Although the causes underlying this age-related deterioration are unclear, the variation in penetrance among siblings is likely because of differences in their genetic background.
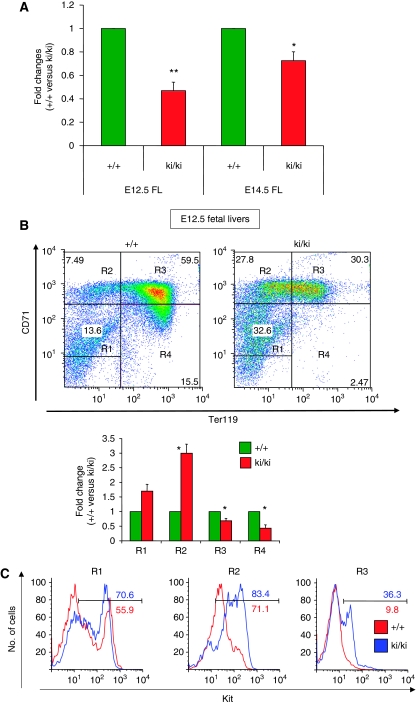
To investigate whether erythroid defects occur at earlier stages of development, we examined erythropoiesis in foetal livers. Notably, the total number of cells contained in ki/ki E12.5 foetal livers was reduced by 50% when compared with wild-type counterparts (Figure 5A). Moreover, ki/ki foetal liver erythroid cells displayed a delay in maturation as determined by Ter119/CD71 expression (Figure 5B). To examine the cause for the reduction in erythroid cell numbers, we measured cell cycle progression by propidium iodide staining and flow cytometry in erythroid populations at the proerythroblast and basophilic stages of maturation (Supplementary Figure S9A). A reduction in the fraction of cells in S/G2/M-phases was detected in both populations, indicating a significant delay in cell cycle progression in ki/ki erythroblasts (Supplementary Figure S9A). The fractions of apoptotic and dead cells were determined by staining the cells with annexinV and by measuring uptake of 7-amino-actinomycin D (7-AAD). The percentages of both apoptotic cells (AnnexinV+ 7-AAD− cells) and dead cells (7-AAD+ cells) were significantly increased in ki/ki foetal liver (Supplementary Figure S9B, C). The combination of the delay in cell cycle progression and decreased cell viability explains the reduced cell numbers found at E12.5. Together with the delay in erythroid maturation, this accounts for the peripheral anaemia in mutant embryos at this stage of development (not shown).
Figure 5.
Hypocellularity (A) and erythroid maturation defects in ki/ki erythroid cells from E12.5 foetal livers as revealed by flow cytometric analysis using CD71 and Ter119 antibodies (B). R1, R2, R3 and R4 represent immature erythroblasts, proerythroblasts, basophilic and polychromatic/orthrochromatic erythroblasts, respectively (Zhang et al, 2003) and were plotted as per cent of total cells. (C) R1, R2 and R3, populations from (B), were analysed by flow cytometry for surface expression of Kit. Note residual expression of kit in R2 to R4 populations, which might reflect a delay in kit silencing. Error bars represent s.e.m. n=4 E12.5 litters and n=3 E14.5 litters. *P<0.05. **P<0.005.
At E14.5, erythroid maturation, cell cycle progression and cell viability improved approximating those of normal littermates (Figure 5A and data not shown). Thus, the most severe erythroid defects occur transiently and early in development, but can surface again in aged specimens.
Defective megakaryopoiesis in homozygous mutant animals
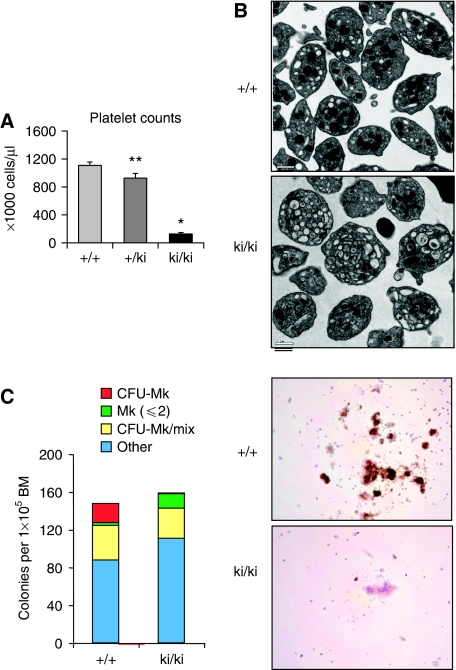
The most prominent change found on peripheral blood smears was a reduction in platelet numbers with the occasional presence of dramatically enlarged platelets in ki/ki animals (Figure 4D), consistent with macrothrombocytopaenia. Flow cytometry showed that the number of blood platelets was reduced to ∼11% of that found in +/+ or ki/+ animals (Figure 6A). Ultrastructural analysis revealed heterogeneity among ki/ki platelets (Figure 6B). Strikingly, the majority of platelets exhibited a paucity of α-granules and increased numbers of lysosomal-like vacuoles (Figure 6B and data not shown) resembling those found in mice and human patients with macrothrombocytopaenia owing to mutations in GATA-1 that impair binding to FOG-1 (Nichols et al, 2000; Freson et al, 2001; Chang et al, 2002). Despite defects in platelet numbers and appearance, the ki/ki animals had no overt bleeding diathesis indicating the presence of residual platelet function.
Figure 6.
(A) Platelet counts of wild-type (+/+, n=15), heterozygous (+/ki, n=11) and homozygous mutant (ki/ki, n=10) animals. Error bars represent s.e.m. *P<0.001; **P<0.05. (B) Electron micrographs of +/+ and ki/ki platelets. (C) Reduced MK potential in ki/ki mice as revealed by colony assays from bone marrows. ki/ki bone marrows produced markedly fewer CFU-Mk colonies in but increased numbers of single MKs. Right panel: acetyl-cholinesterase (AChE) stain of representative colonies.
In histological sections, the number of MKs in the ki/ki bone marrows appeared to be in the normal range with cells displaying an apparent decrease in cytoplasmic to nuclear ratio when compared with +/+ MKs (Supplementary Figure S6). As another measure of the maturation of MKs in the spleen and bone marrow, we determined their ploidy using propidium iodide staining. Surprisingly, we found virtually normal degrees of ploidy in ki/ki MKs, suggesting that the FOG-1/NuRD interaction is dispensable for endoreduplication (Supplementary Figure S10). When cultured in vitro in the presence of thrombopoietin, ki/ki bone marrows were unable to produce large acetyl-cholinesterase positive MK colonies (Figure 6C). However, the number of MK occurring in two-cell colonies or as single cells was increased. Thus, in vitro conditions uncovered a requirement for the FOG-1/NuRD interaction in the proliferative capacity of maturing MKs. This suggests that in vivo a compensatory mechanism might exist, perhaps triggered by the decreased platelet count, that leads to normal numbers of MKs.
Together, these results show an essential function of the FOG-1–NuRD interaction for normal MK development. Mice that are null for FOG-1 displayed a more dramatic phenotype with complete failure to produce any MKs (Tsang et al, 1998), indicating that FOG-1 performs functions in addition to interacting with NuRD.
Deregulation of gene expression in mutant erythroid cells and MKs
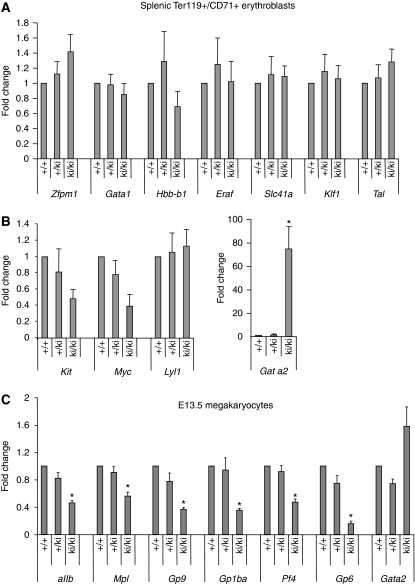
To examine the underlying causes for the defective erythroid development in ki/ki mice, we measured the expression of key erythroid GATA-1/FOG-1 target genes in stage-matched erythroid cells. Total mRNA was purified from splenic TER119+, CD71+ erythroid precursors from wild-type, +/ki and ki/ki littermates. Transcript levels were analysed by real-time RT–PCR and normalized to GAPDH. First, we examined the GATA-1/FOG-1-activated genes β-globin (Hbb-b1), Eraf (AHSP), Band3 (Slc4a1), Klf1, Tal1, Fog-1 (Zfpm1) and Gata1. We found their expression to be essentially unchanged (Figure 7), although Hbb-b1 levels showed a modest trend towards reduced expression in the ki/ki erythroid cells. This suggests that the FOG-1/NuRD interaction is dispensable for the activity of most activated GATA-1/FOG-1 target genes, consistent with the relatively normal maturation of splenic ki/ki erythroid cells. When Ter119+, CD71+ cells from bone marrow were examined, levels of Hbb-b1 and Hba-a1 were significantly reduced, whereas the remaining genes were expressed normally (Supplementary Figure S10). Of note, as Hbb-b1 and Hba-a1 are GATA-1/FOG-1 target genes, this implies a direct function of NuRD for their activation.
Figure 7.
Expression of GATA-1-activated (A) and repressed genes (B) in stage-matched (R3) +/+, +/ki and ki/ki erythroid cells as determined by real-time RT–PCR, normalized to GAPDH and plotted as fold change from +/+ samples. (C) mRNA levels of indicated genes in MKs cultured under serum-free conditions from E13.5 MKs determined by qRT–PCR. The results are averages of seven independent experiments. Error bars represent s.e.m. *P<0.05.
We next turned our attention to genes that are directly repressed by GATA-1 in a FOG-1-dependent (Kit, myc, Gata2) and -independent (Lyl1) manner. Strikingly, splenic ki/ki erythroid cells displayed a ∼75-fold increase in Gata2 expression, whereas the other genes were expressed at normal or near-normal levels (Figure 7). Likewise, in bone marrow erythroblasts (Supplementary Figure S11) and in E12.5 and E14.5 foetal liver erythroblasts (not shown), Gata2 expression was substantially elevated with normal or close to normal levels of Kit and Lyl. Gata2 is normally silenced as erythroid cells mature and accumulate GATA-1 (Weiss et al, 1994). Our results indicate that NuRD is required for the normal silencing of Gata2, but not of all repressed GATA-1 targets.
This gene-selective requirement of NuRD cannot be explained by differences in NuRD recruitment, as the occupancy of NuRD proteins was comparable at NuRD-dependent and -independent genes (Figure 1). NuRD-independent genes might have compensatory mechanisms in place that can substitute for NuRD during transcriptional repression. It is also possible that the subunit composition of NuRD varies between genes in a way that influences the activity of the complex. Finally, as the mere presence of NuRD does not reflect the activity of a gene, it is possible that gene-specific transcription factors modulate the catalytic activity of NuRD. The stimulation of histone acetyltransferase activity by nuclear factors has been observed earlier (e.g. Chen et al, 2001). In this manner, FOG-1 or neighbouring molecules might alter the specific activities of the HDACs or the ATPase of Mi-2β. For example, SCL and its associated proteins have been recently shown to be present at activating, but not repressive GATA elements (Tripic et al, 2009). Moreover, in the context of MK-specific genes, select Ets proteins such as Fli1 specify transcriptional activation by FOG-1(Wang et al, 2002). It is conceivable that co-operating transcription factors such as SCL and Fli1 modulate NuRD in a way to promote positive transcriptional outcomes. Perhaps, similar to our results, although the SWI/SNF complex can bind to DNA and chromatin without sequence specificity, a transcription activation domain contributes to targeting and stimulation of the catalytic activity of this complex to trigger nucleosome remodelling (Gutierrez et al, 2007).
Using a gene complementation assay, we earlier observed a delay in Kit silencing in erythroid cells expressing mutant FOG-1 (Hong et al, 2005). To examine whether this is the case in the ki/ki mice, Kit expression was measured by flow cytometry at maturation stage-matched erythroid cells from foetal livers. Interestingly, we observed a delay in silencing of Kit expression in ki/ki E12.5 foetal liver erythroblasts with substantially elevated fractions of cells in the R3 and R4 populations expressing Kit when compared with +/+ controls (Figure 5C). Elevated Kit expression was also observed in the R3 population by RT–PCR (not shown). Whether the delay in Kit repression is a direct consequence of failed FOG-1 function or an indirect consequence of the maturation delay remains uncertain. It is also possible that elevated GATA-2 levels in ki/ki erythroblasts contribute to sustained Kit expression (Jing et al, 2008). Forced Kit expression has been shown to antagonize GATA-1-induced cell cycle arrest in G1E cells (Munugalavadla et al, 2005). Therefore, additional mechanisms need to be invoked to explain the slowing of the cell cycle progression in ki/ki foetal liver erythroid cells.
GATA-1 and GATA-2 contribute to the expression of most if not all MK-specific genes. The function of FOG-1 during megakaryopoiesis is entirely dependent on GATA factors (Chang et al, 2002). To examine a requirement for FOG-1/NuRD during MK gene expression, we cultured primary MKs from E13.5 foetal livers under serum-free conditions in the presence of thrombopoietin for 5 days and prepared total RNA. Real-time RT–PCR was used to measure the expression of the MK-specific genes αIIb, Mpl, Gp9, GP1bα, Pf4 and Gp6. All of the genes analysed were reduced in ki/ki mice when compared with +/ki or +/+ counterparts. However, the extent of expression was affected differently. For example, Mpl expression was only reduced by ∼40% in ki/ki MKs, whereas Gp6 expression was reduced by almost six-fold (Figure 7C). Thus, there are differences in NuRD requirement among distinct genes. In contrast to mature erythroid cells, Gata2 expression is maintained throughout megakaryopoiesis. Yet, ki/ki MKs displayed increased levels of Gata2, suggesting that GATA-1 limits the expression of this gene through FOG-1 and NuRD. Culture of MKs under serum-free conditions for 5 days does not completely eliminate the presence of other haematopoietic cells. However, the contribution by such cells was comparable between cultures from +/+, +/ki and ki/ki mice (not shown), and hence does not explain the differences in gene expression. In addition, contribution by non-MKs does not account for the different degrees of reduced gene expression.
These result show that NuRD is required in vivo for the activation of MK-specific gene expression by FOG-1 through GATA-1 and likely also GATA-2. These findings are consistent with our experiments showing that NuRD is directly required for transcriptional activation in reporter assays using a promoter of a MK-expressed gene (Figure 2) and the presence of NuRD proteins at active megakaryocytic genes (Supplementary Figure S3). Thus, the classification of NuRD as co-repressor does not account for all of its functions during gene expression.
In concert, the work presented here reveals that NuRD has a function in several key functions of FOG-1 in the erythroid and MK lineages. In addition to its function in promoting the development of these two lineages, FOG-1 is also involved in restricting the formation of the GATA-1-dependent mast cell and eosinophil lineages in which FOG-1 is normally down-regulated. In fact, forced expression of FOG-1 in either cell type can block their differentiation (Querfurth et al, 2000; Cantor et al, 2008). Our preliminary work suggests that suppression of mast cell differentiation by FOG-1 requires NuRD as several mast cell-specific genes are deregulated not only in haematopoietic precursor cells, but also in mature erythroid cells and MKs from ki/ki mice (Gregory et al, in preparation). Moreover, colony assays revealed that MK-erythroid precursors have the potential to form mast cells (Gregory et al, in preparation). As up-regulation of Gata2 is the most salient difference in gene expression between +/+ and ki/ki haematopoietic cells, it will be interesting to determine the relative contributions of GATA-2 over-expression versus direct effects of the FOG-1 mutation on haematopoietic differentiation.
In the context of GATA-4, FOG-1 contributes to normal heart morphogenesis (Katz et al, 2003). Our ki/ki mice display defects in this functions as well (unpublished observations). This suggests that NuRD is required for normal FOG-1 function in most if not all FOG-1-dependent tissues. Yet, in all cell types investigated, the phenotypes caused by disrupted NuRD binding are less severe than those associated with complete loss of FOG-1. Given the interaction of FOG-1 with diverse protein partners, many of whom still remain to be discovered, this hardly comes as a surprise. On the other hand, it was unexpected to observe defects in erythroid development in ki/ki mice as expression of a mutant form of FOG-1 lacking the N-terminal 254 amino acids was capable of rescuing erythroid maturation of a FOG-1−/− cell line (Cantor et al, 2002). However, the erythroid phenotype in mutant animals is relatively mild and might be missed in a cell line-based system. Moreover, forced expression of proteins can obscure subtle defects in their functions.
A key advantage of our approach to selectively disrupt a specific molecular interaction between a nuclear factor and its co-regulator is that it allows examining this interaction in vivo under conditions of normal protein expression. An alternative approach is to knock out a component of NuRD itself. Indeed, conditional deletion of Mi-2β produced broad defects in haematopoiesis affecting stem cell homeostasis, and erythroid and myeloid compartments (Yoshida et al, 2008). However, interpretation of these findings in the context of erythroid gene regulation is difficult as Mi-2β-depleted cells can express increased amounts of Mi-2α (Williams et al, 2004). Other unknowns include the intactness of the NuRD complex in the absence of a key subunit (Kaji et al, 2006), and the effects, direct or indirect, that result from interactions of Mi-2β with diverse nuclear factors (Denslow and Wade, 2007; Manavathi and Kumar, 2007; Bowen et al, 2004).
The generation of the FOG-1 mutation in mice not only established an essential function of NuRD in the transcriptional circuitry surrounding GATA-1, but yielded an unexpected requirement of NuRD for activation of transcription. The mutant animals will be useful for future studies examining the GATA-1/FOG-1/NuRD axis during various aspects in development involving FOG-1. For example, recent reports implicated GATA-1, FOG-1 and NuRD in the developmental silencing of foetal haemoglobin expression in human beings (Chen et al, 2008; Harju-Baker et al, 2008). Breeding the human β-globin gene cluster (Gaensler et al, 1993) into the FOG-1 mutant mice will allow for testing this model in vivo.
Materials and methods
Plasmids
GST–FOG-1(45), GAL4–FOG-1(45) and GAL4–VP16 were generated by PCR and subcloning into pGEX-6P1 (Amersham Biosciences) and pcDNA3-Gal4 (gift from M Poncz), respectively. pcDNA3-GAL4 contains amino-acids 1-147 of GAL4. Related constructs with point mutations were generated with Site-Directed Mutagenesis (Stratagene). pGEM7Zf(+)-rMTA1 and pcDNA3CF-mMTA2 were gifts from W-M Yang (Yao and Yang, 2003). CMV-RbAp48 was a gift from N Barlev. pcDNA3-RbAp46 was generated by RT–PCR from murine RNA and inserted between the BamH1 and EcoR1 sites of pcDNA3 (Invitrogen). Reporter vector G5-TK-luciferase was gift from W-M Yang (Yao and Yang, 2003).
G1E cell culture, transfections, antibodies and ChIP
G1E cells were maintained as described (Weiss et al, 1997). Where indicated, cells were treated with 1 μM oestradiol (21 h). siRNAs were transiently transfected into 3T3 cells with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). siRNAs directed against MTA-1 (M-047751-00; L-047751-01), MTA-2 (M-046370-00; L-046370-00) and MTA-3 (M-058830-00; L-058830-01) were from Dharmacon, whereas control siRNAs were purchased from Ambion (AM4613). After 24 h, reporter plasmids were transiently transfected into 3T3 cells with Lipofectamine reagent according to the manufacturer's instructions (Invitrogen). Antibodies against MTA-1 (sc-9446), MTA-2 (sc-9447), FOG-1 (sc-9361) and GATA-1 (sc-265) RbAp46 (sc-8272) were from Santa Cruz, CA. Mi-2β antibodies A301-081A and A301-082A were from Bethyl Laboratories, TX. ChIP assays were performed as described earlier (Letting et al, 2003) using real-time PCR with SYBR Green on an ABI Prism 7900 system (PE Applied Biosystems). ChIP primer sequences are listed below. The results were normalized to unprecipitated chromatin (input).
Haematological analysis
Periorbital blood samples were collected in EDTA-containing capillary tubes (catalogue no. 07 6011; RAM Scientific Inc.). A measure of 300 μl blood was analysed on an Advia 2120 haematology analyzer using the mouse archetype software (Siemens Health Diagnostics Inc., Deerfield, IL). In addition, blood smears were stained with Wright Giemsa Stain.
Isolation and culture and isolation of primary MKs
Foetal livers were obtained from E13.5 embryos and cultured in serum-free medium containing 1% Nutridoma-SP (Roche Applied Science), 2 mM L-glutamine, 100 U/ml penicillin and streptomycin, 50 μM β-mercaptoethanol and 50 ng/ml thrombopoietin (R&D Systems) for 5 days (Tong et al, 2007). Megakaryocytes of various maturation stages were harvested in CATCH buffer (Ca2+- and Mg2+-free phosphate-buffered saline (PBS) containing 3.5% bovine serum albumin (BSA), 1 mM adenosine, 2 mM theophylline and 0.38% sodium citrate) by centrifugation at 70 g for 10 min. Cells were stained with May-Grünwald-Giemsa (Sigma Aldrich) and acetyl-choline esterase.
Bone marrow-derived megakaryocytes were isolated from the femurs and tibias and cultured either in the serum-free medium as above or in DMEM medium containing 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin and streptomycin, and 50 ng/ml of Tpo for 5 days. Cells were collected by centrifugation at 37 g (400 rpm) for 10 min, resuspended in 5 ml PBS and spun at 9 g (200 r.p.m.) for 30 min at room temperature over a BSA step gradient containing 1.5% BSA and 3% BSA in PBS (Pang et al, 2006).
Flow cytometry
Erythroid cells were analysed using PE-conjugated anti-mouse TER-119, FITC-conjugated anti-mouse CD71 antibodies from BD Biosciences, MD, and APC-conjugated anti-mouse Kit, 7-AAD from eBioscience, CA. Total spleen, bone marrow and foetal liver-derived cells were incubated with anti-CD16/32 antibody (BD Biosciences) and 1% FBS to block non-specific binding to the Fc receptor and stained with the specific antibodies. To detect apoptotic cells, foetal liver cell suspensions were stained with APC-conjugated anti-mouse TER-119, PE-conjugated anti-mouse CD71 antibodies from BD Biosciences and 7-AAD from eBioscience, CA, and subsequently labelled using AnnexinV-FITC Apoptosis Detection Kit (Abcam, UK). For cell cycle analysis, foetal liver cells were labelled with APC-conjugated anti-mouse TER-119 and FITC-conjugated anti-mouse CD71 antibodies (BD Biosciences), permeabilized using BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences) and resuspended in PBS 1% serum containing 50 μg/ml propidium iodide (BD Biosciences). Data were acquired on a FACSCalibur flow cytometer (BD Biosciences) and analysed using FlowJo v8.7.1 (TreeStar, OR).
Colony assays
Bone marrow-derived cells were plated at a density of 1 × 105 cells/ml in methylcellulose medium (M3234; Stem Cell Technologies, BC, Canada) supplemented with 3 U/ml rhEpo (Amgen, CA), 10 ng/ml rmIL6 (Peprotech Inc., NJ), 10 ng/ml rmIL-3 (Invitrogen, CA) and 50 ng/ml rmSCF (Invitrogen). The BFU-E and myeloid colonies were scored by light microscopy 10–14 days after plating. For CFU-E assays, cells were cultured at a density of 2.5 × 104 cells/ml in methylcellulose medium containing 2 U/ml rhEpo. Colonies were scored after 3 days of culture. For the CFU-Mk assay, BM cells were cultured at a density of 1 × 105 cells/ml in MegaCult-C medium in the presence of 20 ng/ml rmIL6, 10 ng/ml rmIL-3 and 50 ng/ml rhTPO, as per manufacturers' instructions (Stem Cell Technologies). CFU-Mk colonies were fixed on day 8 and stained for acetyl-cholinesterase activity and counterstained with Harris' haematoxylin (Sigma, MO) before scoring of colonies.
Electron microscopy
Platelets were isolated by differential centrifugation of whole blood (Eslin et al, 2004); 1 ml of mouse blood was drawn from the inferior vena cava by using a 23G needle/1 ml syringe with 200 μl of acid-citrate-dextrose for anti-coagulation. The total volume was adjusted to 3 ml using modified Tyrode's calcium-free buffer (Eslin et al, 2004). Platelet-rich plasma (PRP) was obtained by centrifugation at 900 rpm (200 g) for 4.5 min at room temperature. Prostagladin E1 (PGE1) was added to the supernatant PRP fraction to a final concentration of 1 μmol/l. The platelets were pelleted at 2000 r.p.m. (800 g) for 10 min at room temperature and gently resuspended in 100 μl of 1% paraformaldehyde (EM grade) fixing solution. The volume was adjusted to 1 ml for the electron microscopy study. The final concentration of platelets ranged between 3 × and 5 × 10E8/ml for wild-type mice. EM was performed at the University of Pennsylvania Biomedical Imaging Facilty.
Supplementary Material
Supplementary Figures S1-S6
Supplementary Figure S7
Supplementary Figures S8-S11
Supplementary Table S1
Supplementary Table S2
Supplementary Figure Legends
Review Process File
Acknowledgments
We thank Tobias Raabe and Adele Button for expert technical assistance, and Eric Svensson for sharing results before publication. This work was supported by NIH grants DK54937 and DK58044 to GAB, HL40387 to MP and HL064632 to EEM.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bowen NJ, Fujita N, Kajita M, Wade PA (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH (2008) Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med 205: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH (2002) Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol 22: 4268–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH (2002) GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci USA 99: 9237–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Deng Z, Kim AY, Blobel GA, Lieberman PM (2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol Cell Biol 21: 476–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Luo HY, Basran RK, Hsu TH, Mang DW, Nuntakarn L, Rosenfield CG, Patrinos GP, Hardison RC, Steinberg MH, Chui DH (2008) A T-to-G transversion at nucleotide -567 upstream of HBG2 in a GATA-1 binding motif is associated with elevated hemoglobin F. Mol Cell Biol 28: 4386–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL (2001) Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2: 769–779 [DOI] [PubMed] [Google Scholar]
- Crispino JD (2005) GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol 16: 137–147 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH (1999) Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell 3: 219–228 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Eslin DE, Zhang C, Samuels KJ, Rauova L, Zhai L, Niewiarowski S, Cines DB, Poncz M, Kowalska MA (2004) Transgenic mice studies demonstrate a role for platelet factor 4 in thrombosis: dissociation between anticoagulant and antithrombotic effect of heparin. Blood 104: 3173–3180 [DOI] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S (2005) GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol 25: 1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J 18: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freson K, Devriendt K, Matthijs G, Van Hoof A, De Vos R, Thys C, Minner K, Hoylaerts MF, Vermylen J, Van Geet C (2001) Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood 98: 85–92 [DOI] [PubMed] [Google Scholar]
- Gaensler KM, Kitamura M, Kan YW (1993) Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human beta-globin locus in transgenic mice. Proc Natl Acad Sci USA 90: 11381–11385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines P, Geiger JN, Knudsen G, Seshasayee D, Wojchowski DM (2000) GATA-1- and FOG-dependent activation of megakaryocytic alpha IIB gene expression. J Biol Chem 275: 34114–34121 [DOI] [PubMed] [Google Scholar]
- Garriga-Canut M, Orkin SH (2004) Transforming acidic coiled-coil protein 3 (TACC3) controls friend of GATA-1 (FOG-1) subcellular localization and regulates the association between GATA-1 and FOG-1 during hematopoiesis. J Biol Chem 279: 23597–23605 [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH (2003) GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA 100: 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Jing H, Kim SI, Martowicz ML, Pal S, Blobel GA, Bresnick EH (2006) Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol 26: 7056–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, Workman JL (2007) Activation domains drive nucleosome eviction by SWI/SNF. EMBO J 26: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harju-Baker S, Costa FC, Fedosyuk H, Neades R, Peterson KR (2008) Silencing of Agamma-globin gene expression during adult definitive erythropoiesis mediated by GATA-1-FOG-1-Mi2 complex binding at the -566 GATA site. Mol Cell Biol 28: 3101–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J 24: 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak CE, Mahajan MC, Luscombe NM, Gerstein M, Weissman SM, Snyder M (2002) GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc Natl Acad Sci USA 99: 2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH (2005) Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci USA 102: 17065–17070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA (2008) Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Boyer ME, Kang JA, Wickrema A, Cantor AB, Bresnick EH (2007) Friend of GATA-1-independent transcriptional repression: a novel mode of GATA-1 function. Blood 109: 5230–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH (2002) Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc Natl Acad Sci USA 99: 11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B (2006) The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 8: 285–292 [DOI] [PubMed] [Google Scholar]
- Katz SG, Cantor AB, Orkin SH (2002) Interaction between FOG-1 and the corepressor C-terminal binding protein is dispensable for normal erythropoiesis in vivo. Mol Cell Biol 22: 3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SG, Williams A, Yang J, Fujiwara Y, Tsang AP, Epstein JA, Orkin SH (2003) Endothelial lineage-mediated loss of the GATA cofactor friend of GATA 1 impairs cardiac development. Proc Natl Acad Sci USA 100: 14030–14035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer SM, Ohlemiller KK, Yang J, McDill BW, Kohlhase J, Rauchman M (2003) Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum Mol Genet 12: 2221–2227 [DOI] [PubMed] [Google Scholar]
- Lauberth SM, Bilyeu AC, Firulli BA, Kroll KL, Rauchman M (2007) A phosphomimetic mutation in the Sall1 repression motif disrupts recruitment of the nucleosome remodeling and deacetylase complex and repression of Gbx2. J Biol Chem 282: 34858–34868 [DOI] [PubMed] [Google Scholar]
- Lauberth SM, Rauchman M (2006) A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem 281: 23922–23931 [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA (2004) Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci USA 101: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letting DL, Rakowski C, Weiss MJ, Blobel GA (2003) Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol Cell Biol 23: 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lin Q, Wang W, Wade P, Wong J (2002) Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev 16: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Roche AE, Wilk J, Svensson EC (2004) The N termini of friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem 279: 55017–55023 [DOI] [PubMed] [Google Scholar]
- Manavathi B, Kumar R (2007) Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem 282: 1529–1533 [DOI] [PubMed] [Google Scholar]
- Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH (2005) Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem 280: 1724–1732 [DOI] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R (2005) Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol 25: 6747–6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K (2007) Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity 27: 723–734 [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet 24: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH (2004) Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA 101: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang L, Xue HH, Szalai G, Wang X, Wang Y, Watson DK, Leonard WJ, Blobel GA, Poncz M (2006) Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood 108: 2198–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C (2000) Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev 14: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC (2008) The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol 44: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ (2003) GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol 23: 5031–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS (2007) The chromatin remodeling factor Mi-2alpha acts as a novel co-activator for human c-Myb. J Biol Chem 282: 13994–14005 [DOI] [PubMed] [Google Scholar]
- Smith CL (2008) A shifting paradigm: histone deacetylases and transcriptional activation. Bioessays 30: 15–24 [DOI] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF (2001) Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood 98: 3261–3273 [DOI] [PubMed] [Google Scholar]
- Tong W, Ibarra YM, Lodish HF (2007) Signals emanating from the membrane proximal region of the thrombopoietin receptor (mpl) support hematopoietic stem cell self-renewal. Exp Hematol 35: 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA (2009) SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119 [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 12: 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstappen G, van Grunsven LA, Michiels C, Van de Putte T, Souopgui J, Van Damme J, Bellefroid E, Vandekerckhove J, Huylebroeck D (2008) Atypical Mowat-Wilson patient confirms the importance of the novel association between ZFHX1B/SIP1 and NuRD corepressor complex. Hum Mol Genet 17: 1175–1183 [DOI] [PubMed] [Google Scholar]
- Wang X, Crispino JD, Letting DL, Nakazawa M, Poncz M, Blobel GA (2002) Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J 21: 5225–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH (1994) Novel insights into erythroid development revealed through in vitro differentiation of GATA-1- embryonic stem cells. Genes Dev 8: 1184–1197 [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH (1997) Erythroid-cell-specific properities of transcription factor GATA-1 revealed by phenotypic rescue of a gene targeted cell line. Mol Cell Biol 17: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104: 3136–3147 [DOI] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K (2004) The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity 20: 719–733 [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM (2003) The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. J Biol Chem 278: 42560–42568 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K (2008) The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Canas B, Pappin D, Kouzarides T (2002) Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem 277: 11621–11624 [DOI] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF (2003) Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood 102: 3938–3946 [DOI] [PubMed] [Google Scholar]
- Zweier C, Horn D, Kraus C, Rauch A (2006) Atypical ZFHX1B mutation associated with a mild Mowat-Wilson syndrome phenotype. Am J Med Genet A 140: 869–872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S6
Supplementary Figure S7
Supplementary Figures S8-S11
Supplementary Table S1
Supplementary Table S2
Supplementary Figure Legends
Review Process File