Abstract
The transcriptional co-factor Friend of GATA1 (FOG-1) has been shown to interact with subunits of the nucleosome remodelling and histone deacetylase (NuRD) complex through a specific motif located at its N-terminus. To test the importance of FOG-1/NuRD interaction for haematopoiesis in vivo, we generated mice with a mutation that specifically disrupts FOG-1/NuRD interaction (FOG-1R3K5A). Homozygous FOG-1R3K5A mice were found to have splenomegaly, extramedullary erythropoiesis, granulocytosis and thrombocytopaenia secondary to a block in megakaryocyte maturation. FOG-1R3K5A/R3K5A megakaryocytes and erythroid progenitors expressed increased levels of GATA2, showing that FOG-1/NuRD interaction is required for the earlier described ‘GATA Switch'. In addition, ablation of FOG-1/NuRD interaction led to inappropriate expression of mast cell and eosinophil-specific genes in the megakaryocyte and erythroid lineages. Chromatin immunoprecipitation experiments revealed that the NuRD complex was not properly recruited to a mast cell gene promoter in FOG-1R3K5A/R3K5A megakaryocytes, suggesting that FOG-1/NuRD interaction is required for the direct suppression of mast cell gene expression. Taken together, these results underscore the importance of the FOG-1/NuRD interaction for the re-enforcement of lineage commitment during erythropoiesis and megakaryopoiesis in vivo.
Keywords: chromatin remodelling, haematopoiesis, lineage commitment, mast cell, transcriptional repression
Introduction
Chromatin remodelling is increasingly being recognized as an important event during the differentiation of haematopoietic stem cells (HSCs) into the various cell lineages of the haematopoietic system. It has been postulated that a dynamic chromatin structure is necessary for HSCs to maintain their pluripotency (Ng et al, 2007). This dynamic chromatin structure allows genes that will be necessary for the differentiation of the HSC to be maintained in a ‘primed' or ‘poised' state, in which gene expression is low or absent, but can be rapidly activated on differentiation (Laslo et al, 2006). A number of different protein complexes have been described to remodel the chromatin structure to allow for the activation or repression of gene expression (Neely and Workman, 2002). One such complex, the Nucleosome Remodelling and histone Deacetylase (NuRD) complex, is highly expressed in the HSC (Yoshida et al, 2008). The NuRD complex is a large, multi-subunit complex that possesses both chromatin remodelling ATPase activity and histone deacetylase activity in the same complex, and it is generally believed to mediate transcriptional repression (Denslow and Wade, 2007; Molli et al, 2008). Targeted disruption of Mi2β, the ATPase subunit of the NuRD complex, in the HSC resulted in an overproduction of proerythroblasts, but severe anaemia because of a failure of these proerythroblasts to properly mature (Yoshida et al, 2008). Gene expression profiling revealed that most of the genes dependent on Mi2β for repression in the HSC were expressed in the differentiated lineages derived from HSC, suggesting that the NuRD complex is necessary for maintaining the repressed state of these ‘primed' target genes. However, these results must be interpreted with care, given that the NuRD complex is known to interact with many transcriptional regulators during development.
The NuRD complex has been shown to associate with several transcription factors known to be important for haematopoiesis (Cismasiu et al, 2005; Sridharan and Smale, 2007). One such factor is Friend of GATA1 (FOG-1, also known as Zfpm1), a multi-zinc-finger protein critical for development of erythrocytes and megakaryocytes (Tsang et al, 1997, 1998; Hong et al, 2005). Although FOG-1 does not bind DNA directly, it physically interacts with members of the GATA family of DNA-binding transcriptional activators (Tsang et al, 1997), factors known to be important in haematopoiesis. Germline deletion of GATA1 in mice leads to embryonic lethality caused by an arrest of erythroid development at the proerythroblast stage, providing direct evidence that GATA1 is essential for erythropoiesis (Weiss et al, 1994; Pevny et al, 1991, 1995; Fujiwara et al, 1996). Further, megakaryocyte-specific ablation of GATA1 results in defective megakaryocyte maturation (Shivdasani et al, 1997). Similarly, GATA2 is also required for haematopoiesis. GATA2-deficient embryos die of anaemia, and show defects in the self-renewal and proliferation of HSC (Tsai and Orkin, 1997; Ling et al, 2004). GATA2 has an important function in proliferation of haematopoietic progenitors, including erythroid precursors (Ling et al, 2004). On differentiation of these cells into mature erythrocytes, expression of GATA1 is up-regulated, whereas that of GATA2 is repressed. The down-regulation of GATA2 transcription is mediated by the displacement of GATA2 from its upstream enhancer by increasing levels of GATA1 (Grass et al, 2003). This ‘GATA switch', which requires GATA–FOG interaction, also mediates changes in expression of the α-globin and c-Kit genes during erythropoiesis (Anguita et al, 2004; Jing et al, 2008).
FOG-1 is also required for haematopoiesis, as FOG-1-deficient mice display erythropoietic defects that largely mimic those of GATA1-null mice, suggesting that GATA1 functions in concert with FOG-1 during the red cell development (Tsang et al, 1998). Moreover, both mice and human beings with mutations in GATA1 that block binding to FOG-1 also display defects in erythropoiesis, confirming the importance of the GATA-1/FOG-1 interaction (Crispino et al, 1999; Nichols et al, 2000). As shown by transient transfection assays, FOG-1 can activate or repress GATA1-mediated gene transcription in a promoter and cell context-dependent manner (Tsang et al, 1997; Fox et al, 1999; Lu et al, 1999; Svensson et al, 2000; Robert et al, 2002). To modulate GATA-mediated transcription, FOG proteins are believed to recruit other transcriptional co-factors to the GATA/FOG complex. These co-factors include C-terminal binding protein (CtBP) and the NuRD chromatin remodelling complex. CtBP-1 and -2 interact specifically with FOG-1 through a motif located in the C-terminal half of FOG-1 (Fox et al, 1999). However, although transient transfection assays suggested that the FOG-1/CtBP interaction is important for mediating transcriptional repression, mice harbouring a mutant FOG-1 with a disrupted CtBP-binding motif showed normal haematopoietic development, indicating that the FOG-1/CtBP interaction is not required in vivo for FOG-1 function (Katz et al, 2002). Recently, FOG-1 has also been shown to interact with the NuRD chromatin remodelling complex through a specific, 12 amino-acid motif located in the N-terminus of FOG-1 (Hong et al, 2005; Rodriguez et al, 2005; Roche et al, 2008). Although transient transfection assays suggest that the FOG-1/NuRD interaction is required for repression of specific GATA1-dependent target genes, the extent to which the FOG/NuRD interaction is required for haematopoiesis in vivo is unclear.
In this report, we describe the generation of mice with a mutation in the FOG-1 gene that abolishes FOG-1's ability to interact with the NuRD complex. These mice display defects in both erythropoiesis and megakaryopoiesis. Furthermore, gene expression analysis revealed that FOG-1/NuRD interaction is required for the ‘GATA Switch' during terminal differentiation of megakaryocyte and erythroid precursors. In addition, the FOG-1/NuRD interaction is required to repress mis-expression of mast cell-specific genes in erythroid and megakaryocyte lineages. Taken together, these results suggest that the FOG-1/NuRD interaction is necessary for re-enforcement of lineage commitment during erythroid and megakaryocyte development.
Results
Generation of FOG-1R3K5A/R3K5A mice
FOG proteins physically interact with subunits of the NuRD chromatin remodelling complex through a conserved, 12-amino-acid motif, termed the FOG repression motif, located at the N-terminus of both FOG-1 and FOG-2 (Lin et al, 2004; Hong et al, 2005; Roche et al, 2008). This motif interacts with the MTA as well as RbAP subunits of the NuRD complex. Single amino-acid mutations within this motif greatly attenuate all of these interactions and, most importantly, the ability of FOG-1 to repress GATA-mediated gene transcription (Hong et al, 2005; Roche et al, 2008). As a first step to probe the importance of FOG-1/NuRD interaction in vivo, we generated an FOG-1 cDNA encoding two point mutations in the FOG repression motif (asparagine 3 to alanine and lysine 5 to alanine, hereafter referred to as FOG-1R3K5A) to disrupt the interaction between FOG-1 and subunits of the NuRD complex (Figure 1A). To test the effectiveness in disruption of the FOG-1/NuRD interaction, we used an in vitro binding assay with purified glutathione-S-transferase (GST)-FOG-1 fusion protein and in vitro translated, 35S-labelled MTA1. As we have earlier shown, MTA1 and the N-terminal 12 amino acids of FOG-1 strongly interact in an MTA1 concentration-dependent manner. However, mutation of R3 and K5 to alanine abolished the ability of FOG-1 to bind MTA1 at all concentrations of MTA1 tested (Figure 1B). To determine the effect of this double mutation on the ability of FOG-1 to repress GATA-1-mediated transcription, we transiently transfected NIH 3T3 fibroblasts with expression vectors for GATA1, wild-type or mutant FOG-1 (see Supplementary Figure 1), and a reporter construct containing −69/+4 bp region of the mast cell-specific FcɛR1β promoter driving expression of human growth hormone. This promoter is strongly activated by GATA1 and repressed by co-expression of FOG-1 (Maeda et al, 2006). As shown in Figure 1C, GATA1 transactivated the FcɛR1β promoter ∼150-fold, whereas the addition of wild-type FOG-1 resulted in a 10-fold repression of GATA1-mediated transactivation. In contrast, expression of FOG-1R3K5A failed to significantly inhibit GATA-1 transactivation of this promoter, showing the importance of FOG-1/NuRD interactions for transcriptional repression.
Figure 1.
Mutations in the N-terminus of FOG-1 abrogate FOG-mediated repression of GATA1 activity. In (A), a schematic of the FOG-1 protein, with its nine zinc-finger domains indicated by loops. The sequence of the first 12 amino acids of FOG-1 is indicated, with the sequence of the R3K5A mutation shown below. In (B), quantitation of in vitro binding assays using purified GST (circles), GST-FOG-1 (triangles) or GST-FOG-1R3K5A (diamonds) fusion proteins with increasing amounts in vitro translated MTA1. In (C), NIH 3T3 fibroblasts were transfected with a reporter construct containing the mast cell-specific FcɛR1β promoter driving expression of human growth hormone. Fibroblasts were also transfected with expression vectors encoding GATA1, FOG-1 and FOG-1R3K5A as indicated. Forty-eight hours after transfection, cell media was assayed for human growth hormone expression as described in Materials and methods. The results are reported as the mean±s.e.m. (n=6). A * indicates statistical significance (P=0.006).
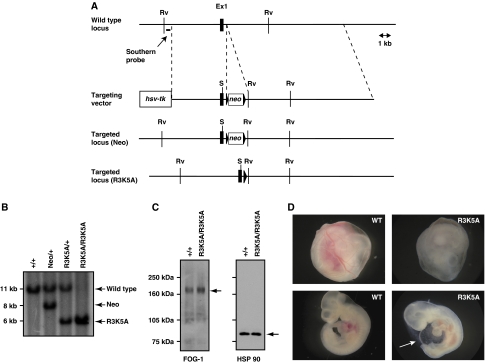
To test our hypothesis that the interaction between FOG-1 and the NuRD complex is important in vivo, we used homologous recombination in murine embryonic stem (ES) cells to generate an ES cell line carrying an R3K5A double mutation in exon 1 of the FOG-1 gene (Figure 2A). Correctly targeted ES cell clones were injected into murine blastocysts to generate chimeric mice, and offspring from these mice were screened for germline transmission of the mutant allele. Two independent ES cell clones achieved germline transmission to establish two independent mouse lines. These mice were then crossed with Prm-Cre transgenic mice to remove the floxed neomycin cassette from intron 1 of the FOG-1 gene in each mouse line. Southern analysis using probes to the 5′ and 3′ end of the targeted region confirmed that the FOG-1 locus was intact and the neomycin cassette excised (Figure 2B and data not shown). Further, genomic sequencing of exon 1 confirmed the presence of the R3K5A mutations within the genome of each line. As a final check on the integrity of the targeted FOG-1 locus, we performed western analysis on whole bone marrow cell lysates using an anti-FOG-1 antibody (Figure 2C). The expression level of FOG-1 in the homozygous mutant mice was comparable with that of their wild-type littermates, strongly suggesting that the insertion of the R3K5A double mutation did not alter the regulation of FOG-1 gene expression.
Figure 2.
Generation of mice with an FOG-1 mutation that disrupts FOG-1/NuRD interaction. In (A), a schematic of the targeting strategy used to generate the FOG-1R3K5A mutation. In (B), southern analysis of genomic DNA from mice containing the FOG-1 alleles shown in (A). Arrows indicate the position of the expected fragment for each allele. In (C), western analysis of whole bone marrow cell lysates from wild-type and FOG-1R3K5A/R3K5A mice using an antibody against FOG-1 (left panel), or HSP 90 (right panel) as a loading control. In (D), photographs of wild-type (left panels) or FOG-1R3K5A/R3K5A (right panels) embryos at E10.5 with their yolk sac intact (top panels), or dissected free (bottom panels). Note the pale yolk sack and pericardial oedema (arrow) of the FOG-1R3K5A/R3K5A embryo.
Abnormal erythropoiesis in FOG-1R3K5A/R3K5A mice
Earlier work has shown that FOG-1-null mice have defects in both erythropoiesis and megakaryopoiesis, and homozygous mutant embryos die by embryonic day (E) 12.5 (Tsang et al, 1998). On the basis of our model that the FOG-1/NuRD interaction is essential for FOG-1's function in haematopoiesis, we hypothesized that the FOG-1R3K5A/R3K5A mice may have defects in haematopoiesis similar to those of the FOG-1-null embryos. We genotyped the offspring of heterozygous crosses and found that homozygous mutant mice were underrepresented at weaning, suggesting a partial embryonic lethality (Table 1). To determine the timing of this lethality, viable embryos from timed mating of heterozygous mice were genotyped between E10.5 and E12.5. The majority of homozygous mutant embryos were indistinguishable from their wild-type littermates. However, a few mutant embryos showed a pale yolk sac with intact blood vessels, reminiscent of the FOG-1-null embryonic phenotype, suggesting that the lethality was due to defective erythropoiesis (Figure 2D). Given the low numbers of affected embryos, the remainder of this report is focused on the characterization of the phenotype of the adult mutant mice.
Table 1.
FOG-1R3K5A/R3K5A mice have a partial embryonic lethality
| +/+ | Genotype | R3K5A/R3K5A | |
|---|---|---|---|
| +/R3K5A | |||
| E10.5 | 12 (31%) | 20 (51%) | 7 (18%) |
| E12.5 | 11 (31%) | 17 (49%) | 7 (20%) |
| Weaning | 61 (32 %) | 92 (48%) | 38 (20%) |
| Embryos from intercrosses of FOG-1+/R3K5A mice were genotyped by PCR. The ratio of wild-type, heterozygous and homozygous offspring indicates a partial lethality in FOG-1R3K5A/R3K5A mice as early as E10.5. | |||
To examine erythropoiesis in adult FOG-1R3K5A/R3K5A mice, we first analysed complete blood counts of peripheral blood collected from wild-type and homozygous mutant mice at 6–8 weeks of age (Table 2). Although red cell numbers, haemoglobin concentrations and haematocrits were not significantly different between wild-type and mutant mice, red cell width distribution, an index of the variation of red cell volume within the red cell population, was increased. Gross dissection of mutant mice revealed striking splenomegaly, with spleen weight to body weight ratios increased in mutant mice by 200% (Figure 3A and B). Histologic sections of mutant spleens showed that there was a prominent expansion of the red pulp in the FOG-1R3K5A/R3K5A mice, with a preponderance of erythrocytes within the red pulp (Figure 3C). To quantify the expansion of erythroid progenitors in the FOG-1R3K5A/R3K5A mice, we performed in vitro erythroid colony-forming assays from bone marrow and spleen (Figure 3D). Although there was no statistically significant difference in the number of erythroid colony-forming units (CFU-E) in the bone marrow (46±12 versus 27±6, respectively, P=0.23), the number of CFU-E's was increased >20-fold in spleens of FOG-1R3K5A/R3K5A mice (2.1±1.8 versus 43±14, P=0.028). This observation is consistent with the histology of the spleen and argues that there is substantial extramedullary haematopoiesis in the FOG-1R3K5A/R3K5A mice.
Table 2.
Complete peripheral blood count of wild-type and FOG-1R3K5A/R3K5A mice
| +/+ | R3K5A/R3K5A | P-value | |
|---|---|---|---|
| WBC (k/μl) | 6.57±0.31 | 10.69±1.75 | 0.043 |
| NE (k/μl) | 0.84±0.12 | 2.65±0.70 | 0.029 |
| LY (k/μl) | 5.32±0.25 | 6.79±0.97 | 0.17 |
| MO (k/μl) | 0.358±0.029 | 1.153±0.507 | 0.15 |
| EO (k/μl) | 0.035±0.018 | 0.077±0.036 | 0.33 |
| BA (k/μl) | 0.010±0.004 | 0.018±0.014 | 0.59 |
| RBC (M/μl) | 9.67±0.24 | 10.42±0.31 | 0.085 |
| Hb (g/dl) | 14.82±0.20 | 15.48±0.23 | 0.051 |
| HCT (%) | 48.4±1.8 | 49.1±1.4 | 0.76 |
| MCV (fL) | 50.2±2.2 | 47.4±2.2 | 0.39 |
| MCH (pg) | 15.35±0.26 | 14.90±0.33 | 0.31 |
| RDW (%) | 18.35±0.37 | 20.20±0.64 | 0.032 |
| PLT (k/μl) | 578±32 | 334±45 | 0.0012 |
| MPV (fL) | 3.70±0.14 | 6.03±0.19 | <0.0001 |
| A comparison of complete blood counts from wild-type and FOG-1R3K5A/R3K5A mice at 6–8 weeks of age demonstrates that FOG-1R3K5A/R3K5A mice have a granulocytosis, increased red cell distribution width and thrombocytopaenia (n=6). | |||
Figure 3.
Aberrant erythropoiesis in FOG-1R3K5A/R3K5A mice. In (A), gross morphology of spleens from wild-type and mutant mice reveal splenomegaly in the FOG-1R3K5A/R3K5A mice. In (B), quantitation of spleen/body weight ratio of wild-type (n=11) and FOG-1R3K5A/R3K5A (n=15) spleens. In panel (C), haematoxylin and eosin staining of splenic sections from wild-type (left panel) and FOG-1R3K5A/R3K5A (right panel) mice. Note the dramatic increase in red pulp (RP) seen in the FOG-1R3K5A/R3K5A spleens. ‘WP' indicates white pulp. In (D), erythroid colony-formation assays using cells from both bone marrow and spleen of wild-type or FOG-1R3K5A/R3K5A mice. The results are reported as the mean±s.e.m. (n=4). A * indicates statistical significance (P<0.05). In (E), representative FACS analysis of whole bone marrow or spleen from wild-type or FOG-1R3K5A/R3K5A mice using the erythroid markers CD71 and Ter119. Numbers beside each gate (red boxes) indicate the percentage of the total number of cells analysed.
As erythroid progenitors mature, they increase expression of the transferrin receptor and down-regulate the cell surface marker CD71 (Socolovsky et al, 2001). To evaluate the effect of the FOG-1 mutation on the maturation of erythroid cells, we performed flow cytometry to measure the expression of CD71 and Ter119 in cells of the bone marrow and spleen of wild-type and FOG-1 mutant animals (Figure 3E; Supplementary Table 1). In the bone marrow of FOG-1R3K5A/R3K5A mice, we observed a 1.8-fold decrease in the proportion of Ter119hi CD71lo cells, which represent the most mature orthochromatic population (19.4±2.3% versus 10.7±1.8%, P=0.027). In the spleen, we saw a dramatic four-fold increase in the proportion of Ter119hi CD71hi basophilic erythroid progenitors (5.3±1.7% versus 20.6±4.5%, P=0.02). Taken together, these observations suggest that FOG-1 mutant bone marrow erythroid progenitors are failing to give rise to sufficient numbers of mature erythroid cells, either because of a block in terminal differentiation or to decreased survival of maturing erythroblasts. Surprisingly, erythropoiesis in the spleen seems to proceed through the orthochromatic stages, as evidenced by the high percentage of Ter119hi CD71lo cells. These results show that the FOG-1/NuRD interaction is required for normal erythropoiesis, but that the animals compensate for the defect by increasing splenic production of erythrocytes.
Enhanced granulopoiesis in FOG-1R3K5A/R3K5A mice
In addition to the defects in erythropoiesis, we also observed alterations in granulocyte development in FOG-1R3K5A/R3K5A mice. First, peripheral neutrophil counts were elevated 3.1-fold in mutant mice (Table 2). Consistent with this observation, histologic examination of mutant bone marrow showed increased numbers of granulocytes (Figure 4A, insert). Flow cytometry on whole bone marrow cells using the granulocyte markers Mac-1 and Gr-1 also showed an increase in the fraction of double positive cells in FOG-1R3K5A/R3K5A mice when compared with wild-type littermates (63±8% versus 54±6%, P=0.044; Figure 4B). To determine whether the increase in granulocytes was due to an increase in the number of granulocyte progenitors, we performed in vitro myeloid colony-formation assays in the presence of IL-3, G-CSF and GM-CSF (Figure 4C). In both bone marrow and spleen, there was no statistically significant difference observed in the number of GM colonies formed between cells from wild-type and FOG-1R3K5A/R3K5A mice. However, the number of myeloid colonies observed in IL-3 alone was significantly higher in both the bone marrow (336±8 versus 445±32, P=0.03) and in particular the spleen (15.3±1.8 versus 318±48, P=0.003; Figure 4D). This result shows that IL-3 responsive progenitors are expanded in both the bone marrow and the spleen of the mutant animals.
Figure 4.
Increased granulopoiesis in FOG-1R3K5A/R3K5A mice. In (A), haematoxylin and eosin staining of sections through the sternum of wild-type (left panel) and FOG-1R3K5A/R3K5A (right panel) mice. Note the paucity of megakaryocytes (arrows) and increased number of granulocytes in the FOG-1R3K5A/R3K5A bone marrow (see inset). In (B), representative FACS analysis of whole bone marrow from wild-type (left panel) or FOG-1R3K5A/R3K5A mice (right panel) using the granulocytic markers GR-1 and Mac-1. In (C) and (D), granulocyte-macrophage colony-forming assays using bone marrow or spleen from wild-type or FOG-1R3K5A/R3K5A mice in the presence (C) or absence (D) of GM-CSF. The results are reported as the mean±s.e.m. (n=4) and * indicates a statistically significant difference in means (P<0.03).
FOG-1/NuRD interaction is required for megakaryopoiesis
Given the requirement for FOG-1 in formation of megakaryocytes, we next sought to characterize megakaryopoiesis in FOG-1R3K5A/R3K5A mice. As shown in Table 2, peripheral platelet counts were significantly reduced in mutant mice (578±32 versus 334±45, P=0.001) and mean platelet volume was strikingly increased (3.70±0.14 versus 6.03±0.19 fL, P<0.001). Peripheral blood smears confirmed these findings (Figure 5A). Also consistent with these observations, histologic examination of the bone marrow revealed a paucity of megakaryocytes in mutant mice (Figure 4A). To quantitate these observations, we determined the fraction of CD41-positive cells in bone marrow and spleen of wild-type or FOG-1R3K5A/R3K5A mice by flow cytometry (Figure 5B). We observed a significant decrease in the fraction of CD41-positive cells in the bone marrow of mutant mice (2.0±0.6% versus 6.1±0.7%, P=0.004; Supplementary Table 2). Furthermore, FOG-1R3K5A/R3K5A megakaryocytes show a reduced polyploidization (34.5% versus 17.5%; Figure 5C), indicating abnormal megakaryocytic maturation. To determine whether the number of megakaryocyte progenitors was also abnormal in FOG-1R3K5A/R3K5A mice, we performed in vitro colony-forming assays using cells from bone marrow or spleen. As shown in Figure 5D, we observed similar numbers of megakaryocyte colonies (CFU-MK) in wild-type and FOG-1R3K5A/R3K5A bone marrow. In contrast, we observed a 5.4-fold increase in the number of CFU-MKs present in the mutant spleen (6.5±1.8 versus 35±7, P=0.007), consistent with the increased erythroid and IL-3-dependent colonies also detected in this site of extramedullary haematopoiesis. However, although the number of colonies present in the FOG-1R3K5A/R3K5A mice was similar to or greater than those of wild-type mice, the colonies derived from mutant bone marrow cells were much smaller and contained significantly fewer acetylcholinesterase (Ach)-positive cells when compared with those derived from wild-type bone marrow (10.4±1.0 versus 30.7±2.7, P<0.0001; Figure 5E and F). Taken together, these observations show that the FOG-1/NuRD interaction is required for proliferation and maturation of megakaryocytes in vivo.
Figure 5.
Defective megakaryopoiesis in FOG-1R3K5A/R3K5A mice. In (A), May–Grünwald–Giemsa staining of peripheral blood smears from wild-type (left panel) and FOG-1R3K5A/R3K5A (right panel) mice. FOG-1R3K5A/R3K5A mice have much fewer, but larger platelets in the peripheral blood than their wild-type littermates (arrows). In (B), representative FACS analysis of whole bone marrow (top panels) or spleen (bottom panels) from wild-type (left panels) or FOG-1R3K5A/R3K5A (right panels) mice using the megakaryocyte marker CD41. Numbers within each gate (red boxes) indicate the percentage of the total number of cells analysed. In (C), polyploidy analysis of megakaryocytes from wild-type (left) or FOG-1R3K5A/R3K5A (right) mice. In (D), megakaryocyte colony-forming assays using cells from bone marrow or spleen from wild-type or FOG-1R3K5A/R3K5A mice. The results are reported as the mean±s.e.m. (n=4) and * indicates a statistically significant difference in means (P<0.007). In (E), photographs of megakaryocyte colonies derived from wild-type (left panel) and FOG-1R3K5A/R3K5A (right panel) megakaryocyte colony-forming assays stained for acetylcholinesterase (Ach) expression (brown). Note the decreased numbers of Ach+ cells per colony seen in colonies derived from FOG-1R3K5A/R3K5A mice as quantitated in (F).
Failure of the GATA switch in FOG-1R3K5A/R3K5A mice
To gain further insight into the mechanisms underlining the observed platelet defects, we compared gene expression in wild-type and FOG-1R3K5A/R3K5A megakaryocytes. Lin-depleted bone marrow cells were cultured in vitro and total RNA was prepared from BSA-gradient purified megakaryocytes. Microarray analysis revealed the down-regulation of a number of megakaryocyte-specific genes and the up-regulation of several mast cell-specific genes (data not shown). Quantitative RT–PCR was used to confirm a selection of mis-regulated genes identified on microarray analysis (Figure 6A). The megakaryocyte-specific genes glycoprotein IIb (GPIIb) and platelet factor 4 (PF4) were down-regulated 2.3- and 3.1-fold in FOG-1R3K5A/R3K5A megakaryocytes, respectively, consistent with the megakaryocytic defects observed in the mutant mice. The expressions of c-Myc, PU.1 and Runx1 were unchanged. Interestingly, the mast cell-specific genes Cpa3, Mcpt4, FcɛR1α and FcɛR1β were up-regulated 3.5- to 9.5-fold. Further, the eosinophilic transcriptional regulator CEBP-β was elevated 2.4-fold. Similar, but less dramatic, gene expression changes were seen examining gene expression in CD71hiTer119hi erythroid cells isolated from FOG-1R3K5A/R3K5A mice (Figure 6B). Together, these results show that FOG-1R3K5A/R3K5A megakaryocytes and erythrocytes failed to fully commit to their respective lineages because of an inability to fully repress gene expression of related lineages.
Figure 6.
Up-regulation of GATA2 and mast cell-specific genes in FOG-1R3K5A/R3K5A megakaryocytes. In (A), results of quantitative RT–PCR on RNA prepared from primary megakaryocyte cultures derived from wild-type and FOG-1R3K5A/R3K5A bone marrow using primers specific for the indicated genes. The results are reported as the mean±s.e.m. of the ratio of expression levels between FOG-1R3K5A/R3K5A and wild-type megakaryocytes. A * indicates a statistically significant difference from 1.00. In (B), results of quantitative RT–PCR on total RNA from CD71hiTer119hi erythroblasts using primers specific for the indicated genes. In (C), Lin bone marrow derived cells from FOG-1R3K5A/R3K5A mice were transduced with a retrovirus expressing an shRNA directed against GATA2 or with a control retrovirus. Three days after transduction, RNA was prepared and subject to quantitative RT–PCR with primers specific for the indicated genes. The results are reported as the mean±s.e.m. of the ratio of gene expression levels between FOG-1R3K5A/R3K5A cells transduced with the anti-GATA2 shRNA encoding retrovirus (white bars) or cells transduced with the control virus (black bars) relative to wild-type cells transduced with the control virus. A * indicates a statistically significant difference in relative expression between anti-GATA2 shRNA retrovirus transduced cells and control virus transduced cells.
As the GATA family members GATA1 and GATA2 are known to be critical in these lineage decisions, we also examined the expression level of each of these factors. GATA2 is highly expressed in the common myeloid progenitor and in more restricted progenitors, such as proerythroblasts, but then is down-regulated on terminal differentiation of these cells into megakaryocytes and mature erythroid cells. This down-regulation of GATA2 is known as the ‘GATA switch' (Grass et al, 2003; Jing et al, 2008). In BSA-gradient purified FOG-1R3K5A/R3K5A megakaryocytes, this GATA switch did not occur, as GATA2 levels were up-regulated six-fold, whereas GATA1 levels were decreased two-fold. Similarly, GATA2 was up-regulated 1.8-fold and GATA1 decreased 50-fold in purified FOG-1 mutant CD71hiTer119hi erythroblasts (Figure 6B). These results suggest that in the absence FOG-1/NuRD interaction, the GATA switch fails to function efficiently, leading to the failure to fully commit to the megakaryocyte or erythroid lineage.
Attenuation of GATA2 levels in FOG-1R3K5A/R3K5A megakaryocytes does not restore lineage-specific gene expression
To explore the importance of GATA1/FOG-1-mediated down-regulation of GATA-2 in megakaryocyte development, we used a commercially available retrovirus encoding an shRNA directed against murine GATA2 to decrease GATA2 mRNA levels in transduced cells. This shRNA virus or a control virus lacking this shRNA was used to infect enriched haematopoietic progenitor cells from wild-type and FOG-1R3K5A/R3K5A bone marrow. These cells were then cultured in the presence of puromycin to select for transduced cells, and then in the presence of thrombopoietin to allow for the differentiation of megakaryocytes. The resulting megakaryocytes were harvested, total RNA was prepared and quantitative RT–PCR was performed to assess relative levels of gene expression (Figure 6C). Under these conditions, we found that GATA2 mRNA levels were elevated 1.9-fold in FOG-1R3K5A/R3K5A megakaryocytes transduced with the control retrovirus when compared with wild-type megakaryocytes. In FOG-1R3K5A/R3K5A megakaryocytes transduced with a retrovirus encoding an anti-GATA2 shRNA, GATA2 mRNA levels were reduced to levels seen in wild-type megakaryocytes (Column 2, Figure 6C). Despite normalization of GATA2 levels in transduced FOG-1R3K5A/R3K5A megakaryocytes, mast cell-specific gene expression remained elevated and megakaryocyte-specific gene expression remained reduced. These results suggest that loss of lineage-restricted gene expression cannot be solely explained by the mis-regulation of GATA2 in megakaryocytes. Instead, FOG-1 must have a more direct effect on the activation of megakaryocyte-specific gene expression and the repression of mast cell gene expression in developing megakaryocytes.
Failed recruitment of the NuRD chromatin remodelling subunit, MTA2, to the FcɛR1β locus
To show that the failure to repress mast cell gene expression in FOG-1R3K5A/R3K5A megakaryocytes was due to the failure to recruit subunits of NuRD complex, we initially attempted to perform chromatin immunoprecipitation experiments using cells isolated from whole bone marrow of wild-type and FOG-1R3K5A/R3K5A mice. However, these experiments were technically challenging and did not yield reproducible results, perhaps in part because of a heterogeneous cell population within the bone marrow and the difficulty of obtaining enough material to effectively perform these experiments. Therefore, we took advantage of an earlier described FOG-1-deficient haematopoietic cell line (Cantor et al, 2002). This cell line has been earlier shown to be capable of differentiating into megakaryocytes when transduced with a retrovirus programming the expression of FOG-1. We transduced this cell line with a retrovirus programming expression of a bi-cistronic mRNA encoding eGFP and wild-type FOG-1 or FOG-1 containing the R3A and K5A mutations that abrogate NuRD interaction. After infection, transduced cells were selected for GFP expression, expanded and differentiated into megakaryocytes by the addition of thrombopoietin into the culture medium. Western analysis of cell lysates showed that FOG-1 protein levels were comparable in each sample (see Supplementary Figure 2). Five days after transfection, cells were harvested and chromatin immunoprecipitation was performed on the FcɛR1β promoter using an antibody to the MTA2 subunit of the NuRD complex (Figure 7). In cells infected with wild-type FOG-1, we found significant chromatin immunoprecipitation of the promoter region of the FcɛR1β gene with an anti-MTA2 antibody. As a control, no significant immunoprecipitation of the GAPDH promoter was observed (Figure 7A). These results show that NuRD subunit MTA2 is recruited to this promoter and are consistent with earlier in vitro promoter analysis implicating GATA1 and FOG-1 in the transcriptional regulation of this gene (Maeda et al, 2006). In cells infected with a retrovirus encoding FOG-1R3K5A, however, an antibody to MTA2 could no longer immunoprecipitate this promoter region (Figure 7B), consistent with our in vitro results showing that the R3A and K5A mutations of FOG-1 abrogate the FOG-1/MTA interaction (see Figure 1). Together, these results suggest that the recruitment of the NuRD subunit MTA2 by FOG-1 is required to directly repress this mast cell-specific gene within developing megakaryocytes.
Figure 7.
Recruitment of MTA2 to the FcɛR1β gene promoter is disrupted by the FOG-1R3K5A mutation. An FOG-1-deficient haematopoietic cell line was transduced with a retrovirus encoding wild-type FOG-1 (A) or FOG-1R3K5A (B). Stably transduced cells were selected and differentiated along the megakaryocyte lineage using thrombopoietin and then subject to chromatin immunoprecipitation using an antibody against the NuRD subunit MTA2 (black bars) or control IgG (white bars) followed by quantitative RT–PCR with primers specific for the FcɛR1β promoter or the GAPDH promoter. The results are reported the mean±s.e.m. of immunoprecipitation relative to total input chromatin.
Discussion
Chromatin remodelling is being increasingly appreciated as an important process during development. In this study, we have investigated the in vivo importance of FOG-1/NuRD interaction during haematopoiesis. This work is the first report of a specific disruption of the interaction of NuRD complex with one of its binding partners and provides further evidence for the importance of the NuRD complex during haematopoiesis. We have shown that targeted mutation of the N-terminal repression domain of FOG-1, which disrupts FOG-1's ability to recruit the NuRD complex (Hong et al, 2005), results in several haematopoietic defects including an increased white cell count, abnormal erythroid development and thrombocytopaenia. Our results show that FOG-1/NuRD interaction is not required for the specification of the erythroid or megakaryocyte lineage, as these cells are found in FOG-1R3K5A/R3K5A mice. However, FOG-1/NuRD interaction does seem to be required to suppress extraneous mast and eosinophil-specific gene expression in the megakaryocyte and erythroid lineages, thus ‘re-enforcing' the lineage decision in these cells by refinement and repression of related lineage-specific genes. Taken together, these results point to a crucial function for FOG-1/NuRD interaction in several steps of haematopoiesis.
Granulocyte differentiation and the FOG-1/NuRD complex
The increased number of granulocytes seen in the bone marrow and peripheral circulation of the FOG-1R3K5A/R3K5A mice suggests an earlier unappreciated function for FOG-1 in the regulation of granulocyte development. It has been earlier reported that FOG-1 is expressed in the HSC at low levels and is up-regulated in myelo-erythroid progenitors (Querfurth et al, 2000). The increase in IL3-dependent progenitors in the FOG-1 mutant mice may be due to the inability of GATA2 to be down-regulated in the HSC. Consistent with this notion, forced over-expression of GATA2 in FDCP-mix cells, a multipotent HSC line, results in the differentiation of these cells down the granulocytic and monocytic pathways in a GM-CSF-independent manner (Heyworth et al, 1999). In addition, recent work by Rodrigues et al (2008) showed that the granulocyte-macrophage progenitor population was reduced in GATA2+/− mice, further supporting the notion that GATA2 is a critical regulator of granulocyte development. Thus, it is possible that FOG-1, in conjunction with the NuRD complex, blocks the differentiation of the HSC down the granulocyte lineage through attenuation of GATA2 levels.
FOG-1/NuRD interactions are required for the ‘GATA switch'
Earlier work has established that on terminal maturation of erythroblasts, GATA1 expression is up-regulated, whereas that of GATA2 is down-regulated and that this ‘GATA switch' requires FOG-1 (Pal et al, 2004). Our results further refine this model by showing that FOG-1/NuRD interaction is required for the down-regulation of GATA2. Given that the NuRD complex is known to remodel chromatin into a more condensed state, it is possible that GATA2 gene expression is repressed by an alteration in the local chromatin structure through FOG-1-mediated recruitment of the NuRD complex. Indeed, a recent report has shown that GATA2 and GATA1 form distinct looped chromatin structures on the c-Kit loci during the GATA switch (Jing et al, 2008).
Erythropoiesis and megakaryopoiesis require the FOG-1/NuRD complex
FOG-1's function in erythrocyte and megakaryocyte development was first shown by the generation of a mouse with a complete disruption in the FOG-1 gene (Tsang et al, 1998). Subsequent in vitro work with a FOG-1-deficient HSC line suggested that the N-terminus of FOG-1, containing the NuRD interaction motif, was dispensable for erythroid maturation (Cantor et al, 2002). In our FOG-1R3K5A/R3K5A mice, however, we observed a fraction of these mice dying in utero between E10.5 and E12.5, likely from anaemia. Further, adult FOG-1R3K5A/R3K5A mice have a perturbation in erythrocyte development as shown by FACS analysis as well as splenomegaly because of extramedullary erythropoiesis, highlighting the importance of the N-terminus of FOG-1 and its interaction with the NuRD complex for proper erythrocyte development (Figure 3).
In addition to erythropoiesis, FOG-1 is also critical for megakaryocyte development (Tsang et al, 1998). Rescue experiments in an FOG-1-deficient haematopoietic cell line suggested that the N-terminus of FOG-1 was critical for the function of FOG-1 during megakaryocyte development (Tsang et al, 1998; Cantor et al, 2002). Consistent with these earlier observations, we also have observed defects in megakaryopoiesis in our FOG-1R3K5A/R3K5A mice. However, in contrast to the absence of megakaryocyte progenitors seen in the FOG-1-null mice, we have detected normal numbers of progenitors, but a block in the maturation of these progenitors. Taken together, our observations suggest an NuRD-independent function of FOG-1 during megakaryocyte specification and a later, NuRD-dependent function in the maturation of megakaryocytes.
FOG-1/NuRD complex in the refinement of lineage-specific gene expression during erythropoiesis and megakaryopoiesis
As shown in Figure 6, we observed inappropriate expression of mast cell and eosinophil-specific genes within FOG-1R3K5A/R3K5A megakaryocytes and erythroid progenitors, suggesting that the FOG-1/NuRD interaction is required to repress extraneous gene expression within these lineages. This is consistent with the observation that down-regulation of FOG-1 is required for mast cell development. Sugiyama et al (2008) showed that expression of FOG-1 in mature mast cells led to the repression of mast cell-specific genes such as Fcɛ receptor β-chain. However, recently published work using retrovirally transduced in vitro cell lines suggested that the N-terminus of FOG-1 was not required for mast cell gene repression, implying that FOG-1/NuRD interaction is not required for repression of mast cell-specific gene expression (Cantor et al, 2008). Our results suggest that FOG-1/NuRD interaction is required for this repression through a direct recruitment of the NuRD complex by FOG-1 to the mast cell-specific FcɛR1β promoter (Figure 7). The difference between our results and those of Cantor et al may be due to the 150-fold over-expression of the N-terminally deleted FOG-1 protein in the in vitro system used by Cantor et al. Our results are consistent with a model in which in the megakaryocyte–erythroid progenitor, FOG-1, recruits the NuRD complex to silence those mast and eosinophilic genes that were in a transcriptionally ‘poised' or ‘primed' state in its progenitor, the common myeloid progenitor, to re-enforce lineage commitment.
Materials and methods
Generation of FOG-1R3K5A/R3K5A mice
A targeting vector harbouring an R3K5A double mutation in the first exon of the FOG-1 gene was generated by recombineering (Liu et al, 2003) using a BAC clone (RP22-43G10, CHORI) containing exon 1 of the FOG-1 locus identified by screening the RPCI-22 Mouse BAC Library (Invitrogen). The targeting vector was then linearized and electroporated into 129 S6/SvEv ES cells, and ES cell clones were screened for homologous recombination using Southern analysis. Correctly targeted ES cell clones were injected into C57BL/6 blastocysts and the resulting chimeric mice were further bred to C57BL/6 mice to achieve germline transmission. The germline transmitters were bred with Prm-Cre mice (Jackson Laboratory) to excise neomycin cassette from the FOG-1 locus and generate the FOG-1R3K5A allele.
Histological analysis and blood counts
Sternums and spleens from 6–8-week-old mice were isolated and fixed in 10% neutral formalin. Serial sections were stained with haematoxylin and eosin, and peripheral blood smears were stained with May–Grünwald–Giemsa (Sigma-Aldrich). A complete blood count was performed using an HEMAVET HV950FS multispecies haematology system (Drew Scientific). The results from six homozygous mutant mice and six wild-type littermates were compared statistically using the unpaired Student's t-test.
Flow cytometric analysis and cell sorting
Bone marrow and spleen cells were harvested from 8–12-week-old wild-type and FOG-1R3K5A/R3K5A mice (n=4 per genotype). For the erythroid population, cells were stained with conjugated anti-CD71 and Ter119 antibodies. For megakaryocyte and granulocyte/monocyte populations, cells were first treated with Red Blood Cell Lysis buffer (0.15M NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA, pH 7.4), washed and then stained with anti-CD41, or anti-Gr-1 and Mac-1 antibodies conjugated to APC, PE or FITC (BD Biosciences). Finally, surface marker expression was analysed using an FACScan flow cytometer (BD Biosciences) and FlowJo software.
Colony-forming assays
Primary mouse bone marrow and spleen cells were harvested from FOG-1R3K5A/R3K5A mice (n=4) and their wild-type littermates (n=4) and enriched for progenitors with the EasySep negative-selection mouse haematopoietic progenitor-enrichment kit (StemCell Technologies). To evaluate erythroid colony formation, we plated 25 000 cells in MethoCult 3234 medium (StemCell Technologies), supplemented with IL-3, SCF and 10 U/ml EPO. Erythroid and mixed colonies were enumerated after 6 days. To evaluate myeloid colony formation, 25 000 cells were plated in MethoCult 3234, supplemented with IL-3, IL-6, M-CSF and GM-CSF, and mixed colonies were counted after 6 days. To evaluate megakaryocyte colony formation, 25 000 cells were plated in MegCult medium (StemCell Technologies), supplemented with IL-3, IL-6, IL-11 and thrombopoietin. After 6 days, colonies were stained for Ach activity and quantitated (StemCell Technologies).
In vitro megakaryocytes culture and quantitative RT–PCR analysis
Primary mouse bone marrow cells were enriched for haematopoietic progenitors (StemCell Technologies) and cultured in expansion media (DMEM/RPMI 1:1, 2 mM L-glutamine, 7.5% NaHCO3) for 2 days, followed by differentiation in the presence of 10 ng/ml thrombopoietin for 3 days. The resulting megakaryocytes were analysed by FACS or purified using a BSA gradient. Total RNA was prepared using Trizol (Invitrogen) and subject to real-time PCR analysis as described (Flagg et al, 2007) using primers: Runx1 (5′-GCACTCTGGTCACCGTCAT and 5′-ATGGTAGGTGGCAACTTGTG), Mcpt4 (5′-GTAATTCCTCTGCCTCGTCCT and 5′-CCCAAGGGTTATTAGAAGAGCTC), Cpa3 (5′-ACACAGGATCGAATGTGGAG and 5′-TAATGCAGGACTTCATGAGC), C/EBP-β (5′-ACTTCTACTACGAGCCCGACTG and 5′-AAGAGGTCGGAGAGGAAGTCGT), FcɛR1α (5′-GAGCCCCGTCTCCATTAGAGA and 5′-CTGCCTAAGATAGCCCTTGCA), and FcɛR1β (5′-GGCTGCTTTGTGGCTTCTTT and 5′-AAGGCCAGGATGGTGAGAAA). Primers for GATA1, c-Myc, GPIIb, PF4 (Muntean and Crispino, 2005), GATA2 (Hong et al, 2005) and PU.1 (Shepherd and Hassell, 2001) have been earlier described.
Transient transfection and promoter analysis
pXM-GATA1 and pcDNA3-FOG-1 have been described earlier (Wang et al, 2002). A 73 bp fragment of the FcɛR1β promoter (−69/+4) was amplified from mouse genomic DNA using the PCR and the primers 5′-AAAGTCGACAAGAGAAAGGAGTCACTGATATC and 5′-AAAGGATCCATTAATTGGGCTATCCAGGAATG and subsequently cloned into the Sal I and BamHI site of the human growth hormone reporter p0GH. Transient transfection into NIH 3T3 fibroblasts and promoter analysis was performed as described earlier (Svensson et al, 1999).
In vitro binding assay
In vitro binding assays were performed as described earlier (Roche et al, 2008) using purified GST, GST-FOG-1 or GST-FOG-1R3K5A fusion proteins and different concentrations of in vitro translated, 35S-labelled MTA1 protein. The resultant complexes were purified, resolved by SDS–PAGE and quantified using a Molecular Dynamics Storm 860 phosphoimager and ImageQuant software.
Western analysis
Whole bone marrow cells from five mutant mice and five wild-type mice were pooled and red blood cells removed through incubation with Red Blood Cell Lysis buffer. Subsequently, 4 × 106 cells from wild-type and mutant bone marrow were lysed, and proteins separated using 7% SDS–PAGE followed by transfer onto a nitrocellulose membrane. Western blotting was performed using an anti-FOG-1 antibody and anti-HSP-90 antibody (M20 and H114, respectively, Santa Cruz).
Retroviral-mediated GATA2 knockdown in cultured megakaryocytes
Retroviral supernatants were produced using the plasmid pSM2c encoding an shRNA against murine GATA2 (Open Biosystems Inc., Clone ID V2MM_75808). Haematopoietic progenitor cells enriched from wild-type and FOG-1R3K5A/R3K5A bone marrow were cultured in expansion media (DMEM/RPMI 1:1, 2 mM L-glutamine, 7.5% NaHCO3) for 24 h before transduction with this retrovirus or a control retrovirus using the spinoculation method described earlier (Huang et al, 2007). After a second spinoculation, the cells were transferred to differentiation media (RPMI, 10% Fetal Calf Serum, 2 mM L-glutamine, SCF supernatant, 10 ng/ml thrombopoietin) and selected using puromycin for 3 days before harvesting. RNA preparation and quantitative RT–PCR were carried out as described above.
Chromatin immunoprecipitation of reconstituted FOG-1−/− cell lines
The full-length wild-type and R3K5A mutant form of FOG-1 were amplified by PCR and inserted into the Bgl II and EcoRI sites upstream the internal ribosome entry site (IRES) of the murine stem cell virus-internal ribosome entry site-green fluorescence protein (MSCV-IRES-GFP) retrovirus. MSCV-FOG1-GFP and MSCV-FOG-1R3K5A-GFP retroviruses were produced by co-transfection of 293FT cells with pIK and cell media harvested 72 h after transfection as described earlier (Tu et al, 2008). An FOG-1-deficient haematopoietic cell line (Cantor et al, 2002) was transduced with these retroviruses and GFP-positive cells were sorted and cultured in the presence of 10 ng/ml thrombopoietin for 5 days (Cantor et al, 2002). Chromatin immunoprecipitation was performed using a commercially available kit (Millipore). Briefly, 9 × 107 cells were subjected to 1% formaldehyde cross-linking for 10 min at room temperature. The cells were collected and their cytoplasmic membranes lysed. The resulting nuclei were collected, lysed and subjected to sonication. After pre-clearance for 2 h, supernatants were incubated with an anti-MTA2 (C20, Santa Cruz) antibody or an IgG control antibody and protein G sepharose beads (GE Healthcare) at 4°C overnight. The beads were washed and eluted with elution buffer (100 mM NaHCO3, 1% SDS). The protein/DNA cross-links were reversed by incubation at 67°C overnight, and subsequently DNA was purified. Quantitative PCR was performed using SYBR Green with primers flanking the GATA-binding sites in the FcɛR1β promoter (5′-ACTGATATCAATCAGCCTGGAGAC and 5′-GGCAGTTAAGATGGGTTGGCTC) (Maeda et al, 2003) or with primers flanking the GAPDH locus as described earlier (Hong et al, 2005). Quantitative PCR was performed in triplicate for each of two independent experiments.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplemental Figure Legends
Supplementary Tables
Review Process File
Acknowledgments
We thank Dr Chun Tu for her guidance in retroviral production and transduction and Dr Alan Cantor for providing us with the FOG-1-deficient cell line. This work was funded by grants from the National Institutes of Health (R01-HL071063 to ECS and P50GM081892 to JDC) and a post-doctoral fellowship from the American Heart Association (to ZG).
Footnotes
The authors declare that they have no conflict of interest.
References
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR (2004) Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH (2008) Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med 205: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH (2002) Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol 22: 4268–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D (2005) BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24: 6753–6764 [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH (1999) Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell 3: 219–228 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Flagg AE, Earley JU, Svensson EC (2007) FOG-2 attenuates endothelial-to-mesenchymal transformation in the endocardial cushions of the developing heart. Dev Biol 304: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J 18: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA 93: 12355–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH (2003) GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci USA 100: 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyworth C, Gale K, Dexter M, May G, Enver T (1999) A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev 13: 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J 24: 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Richmond TD, Muntean AG, Barber DL, Weiss MJ, Crispino JD (2007) STAT1 promotes megakaryopoiesis downstream of GATA-1 in mice. J Clin Invest 117: 3890–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA (2008) Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SG, Cantor AB, Orkin SH (2002) Interaction between FOG-1 and the corepressor C-terminal binding protein is dispensable for normal erythropoiesis in vivo. Mol Cell Biol 22: 3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H (2006) Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755–766 [DOI] [PubMed] [Google Scholar]
- Lin AC, Roche AE, Wilk J, Svensson EC (2004) The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem 279: 55017–55023 [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E (2004) GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med 200: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG (2003) A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN (1999) FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol 19: 4495–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Nishiyama C, Tokura T, Akizawa Y, Nishiyama M, Ogawa H, Okumura K, Ra C (2003) Regulation of cell type-specific mouse Fc epsilon RI beta-chain gene expression by GATA-1 via four GATA motifs in the promoter. J Immunol 170: 334–340 [DOI] [PubMed] [Google Scholar]
- Maeda K, Nishiyama C, Tokura T, Nakano H, Kanada S, Nishiyama M, Okumura K, Ogawa H (2006) FOG-1 represses GATA-1-dependent FcepsilonRI beta-chain transcription: transcriptional mechanism of mast-cell-specific gene expression in mice. Blood 108: 262–269 [DOI] [PubMed] [Google Scholar]
- Molli PR, Singh RR, Lee SW, Kumar R (2008) MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene 27: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Crispino JD (2005) Differential requirements for the activation domain and FOG-interaction surface of GATA-1 in megakaryocyte gene expression and development. Blood 106: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Workman JL (2002) The complexity of chromatin remodeling and its links to cancer. Biochim Biophys Acta 1603: 19–29 [DOI] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Georgopoulos K (2007) Ikaros and chromatin regulation in early hematopoiesis. Curr Opin Immunol 19: 116–122 [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet 24: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH (2004) Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci USA 101: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Lin CS, D′Agati V, Simon MC, Orkin SH, Costantini F (1995) Development of hematopoietic cells lacking transcription factor GATA-1. Development (Cambridge, England) 121: 163–172 [DOI] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D′Agati V, Orkin SH, Costantini F (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349: 257–260 [DOI] [PubMed] [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C (2000) Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev 14: 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert NM, Tremblay JJ, Viger RS (2002) Friend of GATA (FOG)-1 and FOG-2 differentially repress the GATA-dependent activity of multiple gonadal promoters. Endocrinology 143: 3963–3973 [DOI] [PubMed] [Google Scholar]
- Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC (2008) The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol 44: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NP, Boyd AS, Fugazza C, May GE, Guo Y, Tipping AJ, Scadden DT, Vyas P, Enver T (2008) GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood 112: 4862–4873 [DOI] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J (2005) GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24: 2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd T, Hassell JA (2001) Role of Ets transcription factors in mammary gland development and oncogenesis. J Mammary Gland Biol Neoplasia 6: 129–140 [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH (1997) A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J 16: 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF (2001) Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood 98: 3261–3273 [DOI] [PubMed] [Google Scholar]
- Sridharan R, Smale ST (2007) Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem 282: 30227–30238 [DOI] [PubMed] [Google Scholar]
- Sugiyama D, Tanaka M, Kitajima K, Zheng J, Yen H, Murotani T, Yamatodani A, Nakano T (2008) Differential context-dependent effects of friend of GATA-1 (FOG-1) on mast-cell development and differentiation. Blood 111: 1924–1932 [DOI] [PubMed] [Google Scholar]
- Svensson EC, Huggins GS, Dardik FB, Polk CE, Leiden JM (2000) A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. J Biol Chem 275: 20762–20769 [DOI] [PubMed] [Google Scholar]
- Svensson EC, Tufts RL, Polk CE, Leiden JM (1999) Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci USA 96: 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Orkin SH (1997) Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89: 3636–3643 [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev 12: 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90: 109–119 [DOI] [PubMed] [Google Scholar]
- Tu C, Ortega-Cava CF, Chen G, Fernandes ND, Cavallo-Medved D, Sloane BF, Band V, Band H (2008) Lysosomal cathepsin B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in v-Src fibroblasts. Cancer Res 68: 9147–9156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Crispino JD, Letting DL, Nakazawa M, Poncz M, Blobel GA (2002) Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J 21: 5225–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH (1994) Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev 8: 1184–1197 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K (2008) The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplemental Figure Legends
Supplementary Tables
Review Process File